Microbial Community and Enzyme Activity of Forest Plantation, Natural Forests, and Agricultural Land in Chilean Coastal Cordillera Soils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Soil Sampling
2.2. Soil Enzyme Activities
2.3. DNA Extraction
2.4. Microbial Community Analyses
2.5. Statistical Analysis
3. Results
3.1. Soil Physicochemical Properties
3.2. Enzyme Activities
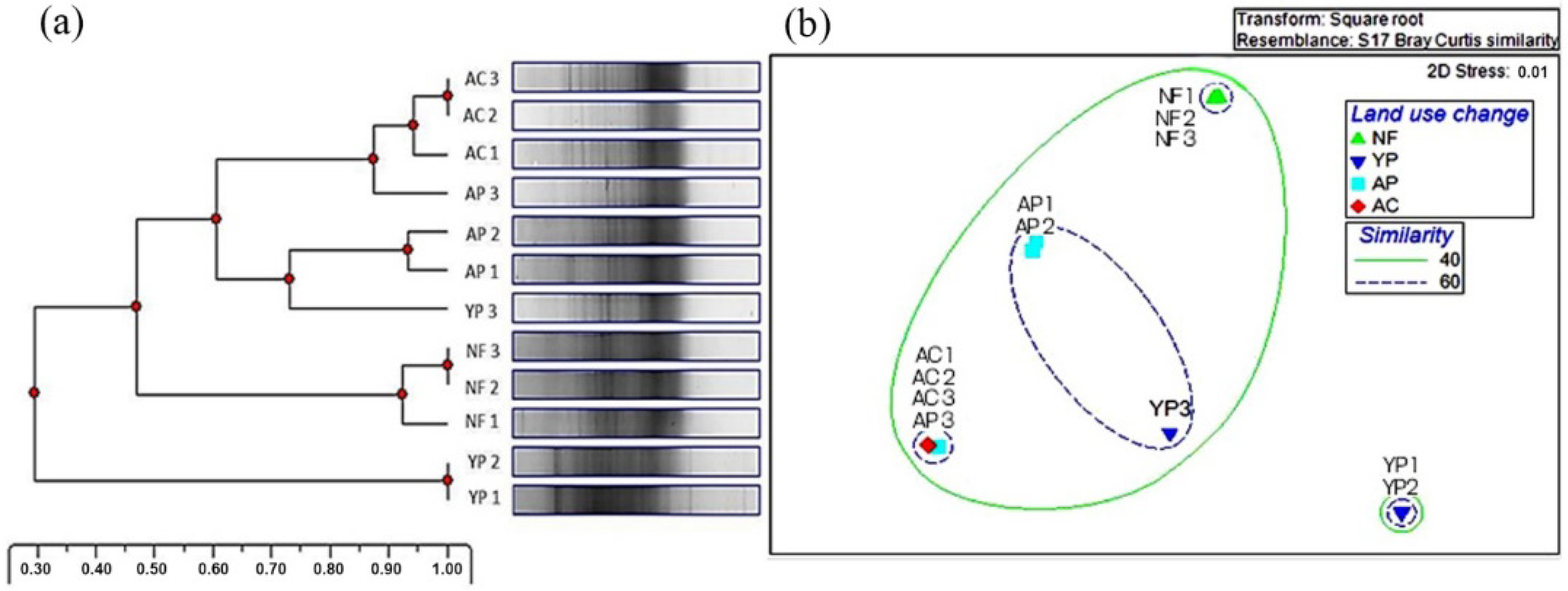
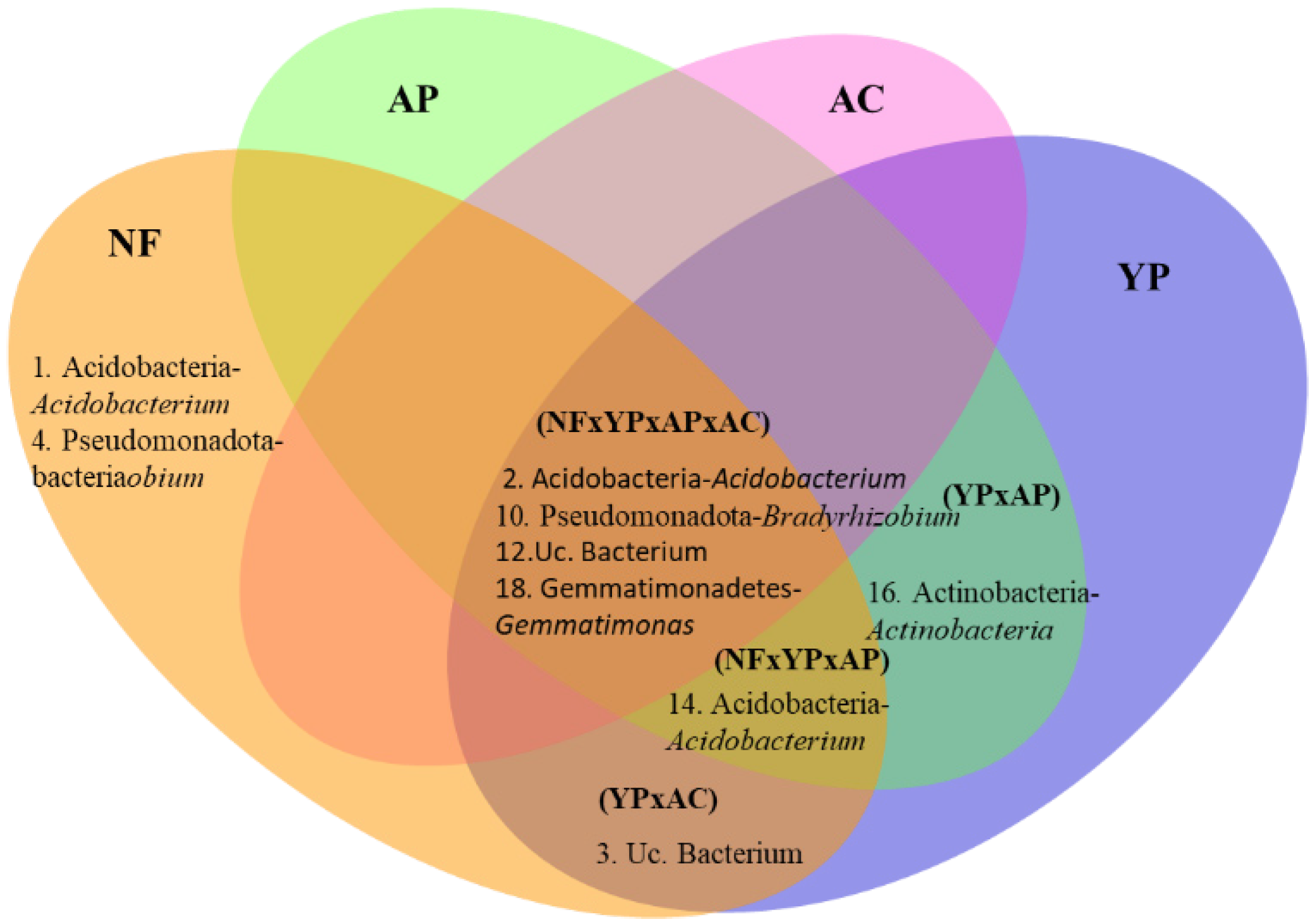
3.3. Bacterial and Fungal Community Structure, Biodiversity Index, and Species Composition
3.4. Drivers of Richness, Shannon–Wiener Index of Bacterial, Fungal, and Enzyme Activities
4. Discussion
4.1. Soil Microbial Diversity and Enzyme Activities Relationships with Physical and Chemical Soil Characteristics
4.2. Models That Explain Variation in Biodiversity of Bacterial and Fungi and Enzymatic Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gibbs, H.K.; Salmon, J.M. Mapping the World’s Degraded Lands. Appl. Geogr. 2015, 57, 12–21. [Google Scholar] [CrossRef]
- Oyarzún, C.E.; Hervé-Fernandez, P.; Frêne, C. Effects of Land Use Changes and Management Practices on Water Yield in Native Forests and Exotic Plantations in Southern Chile; NOVA Publishers: New York, NY, USA, 2012. [Google Scholar]
- Mori, A.S.; Kitagawa, R. Retention Forestry as a Major Paradigm for Safeguarding Forest Biodiversity in Productive Landscapes: A Global Meta-Analysis. Biol. Conserv. 2014, 175, 65–73. [Google Scholar] [CrossRef]
- Van der Plas, F.; Manning, P.; Allan, E.; Scherer-Lorenzen, M.; Verheyen, K.; Wirth, C.; Zavala, M.A.; Hector, A.; Ampoorter, E.; Baeten, L.; et al. Jack-of-All-Trades Effects Drive Biodiversity–Ecosystem Multifunctionality Relationships in European Forests. Nat. Commun. 2016, 7, 11109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldrian, P. Forest Microbiome: Diversity, Complexity and Dynamics. FEMS Microbiol. Rev. 2017, 41, 109–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merino, C.; Jofré, I.; Matus, F. Soil Redox Controls CO2, CH4 and N2O Efflux from White-Rot Fungi in Temperate Forest Ecosystems. J. Fungi 2021, 7, 621. [Google Scholar] [CrossRef]
- Lladó, S.; López-Mondéjar, R.; Baldrian, P. Forest Soil Bacteria: Diversity, Involvement in Ecosystem Processes, and Response to Global Change. Microbiol. Mol. Biol. Rev. 2017, 81, e00063-16. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, P.; Anderson, I.C.; Singh, B.K. Microbial Modulators of Soil Carbon Storage: Integrating Genomic and Metabolic Knowledge for Global Prediction. Trends Microbiol. 2013, 21, 641–651. [Google Scholar] [CrossRef]
- Van der Wal, A.; Geydan, T.D.; Kuyper, T.W.; de Boer, W. A Thready Affair: Linking Fungal Diversity and Community Dynamics to Terrestrial Decomposition Processes. FEMS Microbiol. Rev. 2013, 37, 477–494. [Google Scholar] [CrossRef] [Green Version]
- Van der Heijden, M.G.A.; Martin, F.M.; Selosse, M.-A.; Sanders, I.R. Mycorrhizal Ecology and Evolution: The Past, the Present, and the Future. New Phytol. 2015, 205, 1406–1423. [Google Scholar] [CrossRef]
- Reed, S.C.; Cleveland, C.C.; Townsend, A.R. Functional Ecology of Free-Living Nitrogen Fixation: A Contemporary Perspective. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 489–512. [Google Scholar] [CrossRef]
- Uroz, S.; Oger, P.; Lepleux, C.; Collignon, C.; Frey-Klett, P.; Turpault, M.-P. Bacterial Weathering and Its Contribution to Nutrient Cycling in Temperate Forest Ecosystems. Res. Microbiol. 2011, 162, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Donoso, C. Bosques Templados de Chile y Argentina: Variacion, Estructura y Dinamica; Segunda; Universitaria: Santiago, Chile, 1994. [Google Scholar]
- Neira, E.; Verscheure, H.; Revenga, C. Chile’s Frontier Forests: Conserving A Global Treasure; University Austral of Chile: Valdivia, Chile, 2022; ISBN 1-56973-517-4. [Google Scholar]
- Miranda, A.; Altamirano, A.; Cayuela, L.; Lara, A.; González, M. Native Forest Loss in the Chilean Biodiversity Hotspot: Revealing the Evidence. Reg Environ. Change 2017, 17, 285–297. [Google Scholar] [CrossRef]
- Torres, R.; Azócar, G.; Rojas, J.; Montecinos, A.; Paredes, P. Vulnerability and Resistance to Neoliberal Environmental Changes: An Assessment of Agriculture and Forestry in the Biobio Region of Chile (1974–2014). Geoforum 2015, 60, 107–122. [Google Scholar] [CrossRef]
- Echeverria, C.; Coomes, D.; Salas, J.; Rey-Benayas, J.M.; Lara, A.; Newton, A. Rapid Deforestation and Fragmentation of Chilean Temperate Forests. Biol. Conserv. 2006, 130, 481–494. [Google Scholar] [CrossRef]
- Echeverría, C.; Newton, A.C.; Lara, A.; Benayas, J.M.R.; Coomes, D.A. Impacts of Forest Fragmentation on Species Composition and Forest Structure in the Temperate Landscape of Southern Chile. Glob. Ecol. Biogeogr. 2007, 16, 426–439. [Google Scholar] [CrossRef] [Green Version]
- Huber, A.; Iroumé, A.; Mohr, C.; Frêne, C. Efecto de Plantaciones de Pinus Radiata y Eucalyptus Globulus Sobre El Recurso Agua En La Cordillera de La Costa de La Región Del Biobío, Chile. Bosque (Valdivia) 2010, 31, 219–230. [Google Scholar] [CrossRef]
- Nahuelhual, L.; Carmona, A.; Aguayo, M.; Echeverria, C. Land Use Change and Ecosystem Services Provision: A Case Study of Recreation and Ecotourism Opportunities in Southern Chile. Landsc. Ecol. 2014, 29, 329–344. [Google Scholar] [CrossRef]
- Rodríguez-Echeverry, J.; Echeverría, C.; Oyarzún, C.; Morales, L. Impact of Land-Use Change on Biodiversity and Ecosystem Services in the Chilean Temperate Forests. Landsc. Ecol. 2018, 33, 439–453. [Google Scholar] [CrossRef]
- He, R.; Yang, K.; Li, Z.; Schädler, M.; Yang, W.; Wu, F.; Tan, B.; Zhang, L.; Xu, Z. Effects of forest conversion on soil microbial communities depend on soil layer on the eastern Tibetan Plateau of China. PLoS ONE 2017, 12, e0186053. [Google Scholar] [CrossRef] [Green Version]
- Jiang-shan, Z.; Jian-fen, G.; Guang-shui, C.; Wei, Q. Soil Microbial Biomass and Its Controls. J. For. Res. 2005, 16, 327–330. [Google Scholar] [CrossRef]
- Voříšková, J.; Brabcová, V.; Cajthaml, T.; Baldrian, P. Seasonal Dynamics of Fungal Communities in a Temperate Oak Forest Soil. New Phytol. 2014, 201, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Corral-Pazos-de-Provens, E.; Rapp-Arrarás, Í.; Domingo-Santos, J.M. Estimating Textural Fractions of the USDA Using Those of the International System: A Quantile Approach. Geoderma 2022, 416, 115783. [Google Scholar] [CrossRef]
- Hirzel, J.; Retamal-Salgado, J.; Walter, I.; Matus, I. Effect of Soil Cadmium Concentration on Three Chilean Durum Wheat Cultivars in Four Environments. Arch. Agron. Soil Sci. 2018, 64, 162–172. [Google Scholar] [CrossRef]
- Sadzawka, A.; Carrasco, M.; Grez, R.; Mora, M.; Flores, H.; Neaman, A. Métodos de Análisis Recomendados Para Los Suelos de Chile; Actas INIA N° 34; Revision 2006; Instituto de Investigaciones Agropecuarias: Santiago, Chile, 2006. [Google Scholar]
- Rivas, Y.; Retamal-Salgado, J.; Knicker, H.; Matus, F.; Rivera, D. Neutral Sugar Content and Composition as a Sensitive Indicator of Fire Severity in the Andisols of an Araucaria–Nothofagus Forest in Southern Chile. Sustainability 2021, 13, 12061. [Google Scholar] [CrossRef]
- Hirzel, J.; Retamal-Salgado, J.; Walter, I.; Matus, I. Fixation Coefficient and Soil Residual Effect of Cadmium Application on Three Biannual Crop Rotations at Four Locations in Chile. J. Soil Sci. Plant Nutr. 2019, 19, 450–462. [Google Scholar] [CrossRef]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; U.S. Department of Agriculture: Washington, DC, USA, 1954. [Google Scholar]
- Hirzel, J.; Retamal-Salgado, J.; Walter, I.; Matus, I. Residual Effect of Cadmium Applications in Different Crop Rotations and Environments on Durum Wheat Cadmium Accumulation. J. Soil Water Conserv. 2019, 74, 41–50. [Google Scholar] [CrossRef]
- Rivas, Y.; Rivera, D.; Gallardo, R.; Lagos, E.; Yevenes, M.; Zambrano, F.; Mendoza, J. Water Availability, Quality, and Use in Rural Communities of the Chilean Coastal Range. J. Soil Water Conserv. 2020, 75, 75–90. [Google Scholar] [CrossRef]
- Pinto-Poblete, A.; Retamal-Salgado, J.; López, M.D.; Zapata, N.; Sierra-Almeida, A.; Schoebitz, M. Combined Effect of Microplastics and Cd Alters the Enzymatic Activity of Soil and the Productivity of Strawberry Plants. Plants 2022, 11, 536. [Google Scholar] [CrossRef]
- Khadem, A.; Raiesi, F.; Besharati, H.; Khalaj, M.A. The Effects of Biochar on Soil Nutrients Status, Microbial Activity and Carbon Sequestration Potential in Two Calcareous Soils. Biochar 2021, 3, 105–116. [Google Scholar] [CrossRef]
- Jorquera, M.A.; Hernández, M.; Martínez, O.; Marschner, P.; de la Luz Mora, M. Detection of Aluminium Tolerance Plasmids and Microbial Diversity in the Rhizosphere of Plants Grown in Acidic Volcanic Soil. Eur. J. Soil Biol. 2010, 46, 255–263. [Google Scholar] [CrossRef]
- Fromin, N.; Hamelin, J.; Tarnawski, S.; Roesti, D.; Jourdain-Miserez, K.; Forestier, N.; Teyssier-Cuvelle, S.; Gillet, F.; Aragno, M.; Rossi, P. Statistical Analysis of Denaturing Gel Electrophoresis (DGE) Fingerprinting Patterns. Environ. Microbiol. 2002, 4, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Salas, C.; Donoso, P.J.; Vargas, R.; Arriagada, C.A.; Pedraza, R.; Soto, D.P. The Forest Sector in Chile: An Overview and Current Challenges. J. For. 2016, 114, 562–571. [Google Scholar] [CrossRef] [Green Version]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial Diversity and Soil Functions. Eur. J. Soil Sci. 2003, 54, 655–670. [Google Scholar] [CrossRef]
- Gans, J.; Wolinsky, M.; Dunbar, J. Computational Improvements Reveal Great Bacterial Diversity and High Metal Toxicity in Soil. Science 2005, 309, 1387–1390. [Google Scholar] [CrossRef]
- Singh, B.K.; Quince, C.; Macdonald, C.A.; Khachane, A.; Thomas, N.; Al-Soud, W.A.; Sørensen, S.J.; He, Z.; White, D.; Sinclair, A.; et al. Loss of Microbial Diversity in Soils Is Coincident with Reductions in Some Specialized Functions. Environ. Microbiol. 2014, 16, 2408–2420. [Google Scholar] [CrossRef] [PubMed]
- García, R.A.; Pauchard, A.; Escudero, A. French Broom (Teline Monspessulana) Invasion in South-Central Chile Depends on Factors Operating at Different Spatial Scales. Biol. Invasions 2014, 16, 113–124. [Google Scholar] [CrossRef]
- Liu, J.; Ding, C.; Zhang, W.; Wei, Y.; Zhou, Y.; Zhu, W. Litter Mixing Promoted Decomposition Rate through Increasing Diversities of Phyllosphere Microbial Communities. Front. Microbiol. 2022, 13, 1009091. [Google Scholar] [CrossRef] [PubMed]
- Seaton, F.M.; George, P.B.L.; Lebron, I.; Jones, D.L.; Creer, S.; Robinson, D.A. Soil Textural Heterogeneity Impacts Bacterial but Not Fungal Diversity. Soil Biol. Biochem. 2020, 144, 107766. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil Microbiomes and Climate Change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef]
- Rawls, W.J.; Pachepsky, Y.A.; Ritchie, J.C.; Sobecki, T.M.; Bloodworth, H. Effect of Soil Organic Carbon on Soil Water Retention. Geoderma 2003, 116, 61–76. [Google Scholar] [CrossRef]
- Widyati, E.; Nuroniah, H.S.; Tata, H.L.; Mindawati, N.; Lisnawati, Y.; Darwo; Abdulah, L.; Lelana, N.E.; Mawazin; Octavia, D.; et al. Soil Degradation Due to Conversion from Natural to Plantation Forests in Indonesia. Forests 2022, 13, 1913. [Google Scholar] [CrossRef]
- Sowerby, A.; Emmett, B.; Beier, C.; Tietema, A.; Peñuelas, J.; Estiarte, M.; Van Meeteren, M.J.M.; Hughes, S.; Freeman, C. Microbial Community Changes in Heathland Soil Communities along a Geographical Gradient: Interaction with Climate Change Manipulations. Soil Biol. Biochem. 2005, 37, 1805–1813. [Google Scholar] [CrossRef]
- Pérez-Hernández, J.; Gavilán, R.G. Impacts of Land-Use Changes on Vegetation and Ecosystem Functioning: Old-Field Secondary Succession. Plants 2021, 10, 990. [Google Scholar] [CrossRef] [PubMed]
- Vinhal-Freitas, I.C.; Corrêa, G.F.; Wendling, B.; Bobuľská, L.; Ferreira, A.S. Soil Textural Class Plays a Major Role in Evaluating the Effects of Land Use on Soil Quality Indicators. Ecol. Indic. 2017, 74, 182–190. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere Microbiome Assemblage Is Affected by Plant Development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [Green Version]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three Genomes from the Phylum Acidobacteria Provide Insight into the Lifestyles of These Microorganisms in Soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef] [Green Version]
- Priyanka, J.; Koel, M. Diversity Study of Nitrate Reducing Bacteria from Soil Samples-A Metagenomics Approach. J. Comput. Sci. Syst. Biol. 2015, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.; Singh, L.; Singh, D. Unboxing the Black Box—One Step Forward to Understand the Soil Microbiome: A Systematic Review. Microb. Ecol. 2023, 85, 669–683. [Google Scholar] [CrossRef]
- Zeng, Y.; Baumbach, J.; Barbosa, E.G.V.; Azevedo, V.; Zhang, C.; Koblížek, M. Metagenomic Evidence for the Presence of Phototrophic Gemmatimonadetes Bacteria in Diverse Environments. Environ. Microbiol. Rep. 2016, 8, 139–149. [Google Scholar] [CrossRef]
- Baltz, R.H. Renaissance in Antibacterial Discovery from Actinomycetes. Curr. Opin. Pharmacol. 2008, 8, 557–563. [Google Scholar] [CrossRef]
- Wang, J.; Hu, A.; Meng, F.; Zhao, W.; Yang, Y.; Soininen, J.; Shen, J.; Zhou, J. Embracing Mountain Microbiome and Ecosystem Functions under Global Change. New Phytol. 2022, 234, 1987–2002. [Google Scholar] [CrossRef] [PubMed]
- Pescador, D.S.; Delgado-Baquerizo, M.; Fiore-Donno, A.M.; Singh, B.K.; Bonkowski, M.; Maestre, F.T. Ecological Clusters of Soil Taxa within Bipartite Networks Are Highly Sensitive to Climatic Conditions in Global Drylands. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20210387. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Jackson, R.B.; Hollinger, D.Y.; Phillips, O.L.; Nowak, R.S.; Norby, R.J.; Oren, R.; Reich, P.B.; Lüscher, A.; Mueller, K.E.; et al. Contrasting Responses of Woody and Grassland Ecosystems to Increased CO2 as Water Supply Varies. Nat. Ecol. Evol. 2022, 6, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Harmon, M.E.; Franklin, J.F.; Swanson, F.J.; Sollins, P.; Gregory, S.V.; Lattin, J.D.; Anderson, N.H.; Cline, S.P.; Aumen, N.G.; Sedell, J.R.; et al. Ecology of Coarse Woody Debris in Temperate Ecosystems. In Advances in Ecological Research; Advances in Ecological Research: Classic Papers; Academic Press: Cambridge, MA, USA, 2004; Volume 34, pp. 59–234. [Google Scholar]
- Lindenmayer, D.B.; Franklin, J.F. Conserving Forest Biodiversity: A Comprehensive Multiscaled Approach; Island Press: Washington, DC, USA, 2002; ISBN 978-1-55963-935-4. [Google Scholar]









| Ecosystem | Soil Horizon | θfc | PWP | ρb | Clay | Silt | Sand | Soil Survey (USDA) |
|---|---|---|---|---|---|---|---|---|
| % | g cm−3 | % | ||||||
| NF | A-h | 40.3 b | 29.4 d | 1.25 a | 50.11 a | 23.33 b | 26.55 d | Clay |
| −0.8 | −0.4 | −0.06 | −0.31 | −0.23 | −0.28 | |||
| B-h | 42.2 b | 31.9 c | 1.37 a | 34.81 d | 15.27 a | 49.91 a | Sandy clay loam | |
| −0.8 | −0.4 | −0.03 | −0.32 | −0.23 | −0.28 | |||
| Yp | A-h | 27.4 d,e | 19.6 f | 1.42 a | 48.38 b | 37.09 d | 14.54 g | Clay |
| −0.8 | −1.4 | −0.04 | −0.32 | −0.25 | −0.33 | |||
| B-h | 24.0 e | 17.9 f | 1.46 a | 50.94 b | 29.91 e | 19.15 f | Sandy clay loam | |
| −0.7 | −0.4 | −0.23 | −0.31 | −0.24 | −0.58 | |||
| AP | A-h | 33.3 c | 28.2 d | 1.32 a | 36.38 c | 48.73 b | 14.89 f,g | Silty clay loam |
| −1.5 | −0.4 | −0.39 | −0.31 | −0.26 | −0.33 | |||
| B-h | 43.7 b | 54.0 b | 1.23 a | 15.44 f | 42.41 c | 42.14 a | Loam | |
| −1.4 | −0.4 | −0.01 | −0.31 | −0.23 | −0.58 | |||
| AC | A-h | 30.9 c,d | 22.5 e | 1.25 a | 32.27 e | 51.52 a | 16.21 f | Silty clay loam |
| −0.7 | −0.5 | −0.01 | −0.33 | −0.23 | −0.58 | |||
| B-h | 66.6 a | 54.0 a | 1.39 a | 13.14 g | 50.65 a | 36.21 c | Silty loam | |
| −1.9 | −0.4 | −0.01 | −0.31 | −0.23 | −0.33 | |||
| Ecosystem | Soil Horizon | MOS | pH | Base Cations | P- PO4 Olsen | N-N03− | Al | K | Ca | Mg | S | Fe | Mn | Zn | Cu | B |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (%) | Cmol(+) kg−1 | mg Kg−1 | Saturation (%) | mg Kg−1 | ||||||||||||
| NF | A-h | 27.5 a | 5.6 a | 24.2 a | 7.5 a | 13.9 a | 0.23 c | 5.8 c | 64.8 a | 23.0 a | 10.9 a | 72.7 a | 280.7 a | 16.0 a | 3.5 a | 1.2 a |
| −1.6 | −0.2 | −2.5 | −7.7 | −0.78 | −4.5 | −1.9 | −8.2 | −1.5 | −5.5 | −10.8 | −24.5 | −9.3 | −1.5 | −0.2 | ||
| B-h | 3.5 b | 5.4 a | 4.0 b | 1.1 a | 1.7 b,c | 42.9 a | 7.5 b,c | 32.3 b | 22.0 a | 2.6 a | 31.4 a,b | 33.3 b | 0.1 b | 0.8 b,c | 0.4 b | |
| −1.6 | −0.2 | −2.5 | −0.5 | −0.4 | −10.6 | −2.3 | −8.5 | −3.2 | −1.2 | −19.3 | −20.1 | −0.2 | −0.4 | −0.2 | ||
| YP | A-h | 6.1 b | 5.8 a | 10.3 b | 2.1 a | 1.5 b,c | 16.8 a,b,c | 6.2 c | 47.3 a,b | 27.9 a | 1.5 b | 31.8 a | 63.1 b | 1.0 b | 0.7 b,c | 0.7 a,b |
| −1.6 | −0.3 | −6.1 | −3.6 | −1.3 | −5.6 | −2.2 | −17 | −2.3 | −2.3 | −33.7 | −24.1 | −1.5 | −0.5 | −0.2 | ||
| B-h | 1.4 b | 5.7 a | 5.9 b | 0.2 a | 0.6 c | 31.1 a,b | 5.7 c | 29.5 b | 30.7 a | 1.5 b | 2.4 b | 2.1 b | 0.0 b | 0.1 c | 0.3 b | |
| −1.6 | −0.2 | −1.6 | −0.4 | −0.3 | −15.9 | −2.8 | −17 | −2.6 | −0.3 | −1.4 | −24.3 | −0.1 | −0.2 | −0.2 | ||
| AP | A-h | 4.4 b | 5.7 a | 10.9 b | 1.7 | 1.8 b,c | 8.9 b,c | 10.1 b,c | 57.4 a,b | 21.6 a | 1.3 b | 74.8 a | 40.1 b | 0.8 b | 1.5 b | 0.6 a,b |
| −1.7 | −0.1 | −3.2 | −0.1 | −0.7 | −5.6 | −2.2 | −8.7 | −1.8 | −1 | −19.4 | −15.1 | −0.2 | −0.3 | −0.2 | ||
| B-h | 3.4 b | 5.7 a | 9.4 b | 1.2 a | 3.5 b,c | 11.9 a | 10.4 b,c | 52.6 a,b | 23.3 a | 1.3 b | 53.0 a | 27.7 b | 0.7 b | 1.0 b,c | 0.5 a,b | |
| −2.4 | −0.4 | −2 | −0.8 | −2.8 | −6.9 | −0.6 | −4.1 | −1.2 | −1.6 | −28.8 | −24.4 | −0.8 | −0.6 | −0.2 | ||
| AC | A-h | 3.9 b | 5.9 a | 14.5 a,b | 5.0 b | 2.6 b | 7.2 b,c | 52.6 a | 55.1 a,b | 31.5 a | 1.5 b | 96.2 a | 34.0 b | 1.4 b | 1.3 b,c | 0.4 b |
| −1.2 | −0.3 | −5.8 | −1.4 | −1.4 | −5.5 | −6.2 | −12.4 | −9.7 | −0.4 | −19.2 | −24.2 | −0.1 | −0.4 | −0.2 | ||
| B-h | 5.4 b | 5.7 a | 9.63 b | 1.6 b,c | 0.8 b | 10.6 b,c | 36.4 a | 28.6 b | 29.8 a | 1.4 b | 77.2 a | 24.1 b | 1.0 b | 1.2 b,c | 0.3 b | |
| −2.5 | −0.2 | −2.2 | −0.4 | −1.4 | −2.3 | −2.7 | −10.7 | −13.2 | −0.5 | −17.4 | −24.2 | −0.2 | −0.5 | −0.2 | ||
| Ecosystem | Soil Horizon | Richness | Shannon–Wiener Index | ||
|---|---|---|---|---|---|
| Bacteria | Fungi | Bacteria | Fungi | ||
| NF | A-h | 13.00 a (0.00) | 9.33 a,b (0.87) | 2.56 a (0.00) | 2.20 a,b (0.09) |
| B-h | 11.33 a (2.60) | 12.67 a (0.33) | 2.36 a,b (0.24) | 2.47 a (0.05) | |
| YP | A-h | 13.00 a (0.01) | 9.33 a,b (0.88) | 1.86 b (0.05) | 2.18 a,b (0.10) |
| B-h | 10.67 a (2.19) | 10.67 a,b (1.20) | 2.32 a,b (0.18) | 2.19 a,b (0.13) | |
| AP | A-h | 8.33 a (0.88) | 7.00 b (1.15) | 2.11 a,b (0.10) | 1.91 a,b (0.17) |
| B-h | 11.67 a (1.76) | 7.67 a,b (0.67) | 2.45 a,b (0.16) | 1.94 a,b (0.11) | |
| AC | A-h | 8.00 a (0.01) | 7.33 b (0.88) | 2.08 a,b (0.00) | 1.87 b (0.11) |
| B-h | 11.00 a (1.53) | 11.67 a,b (1.86) | 2.36 a,b (0.15) | 2.25 a,b (0.15) | |
| Model | Coefficient | t-Value | D.f. | p-Value | R2 | AIC |
|---|---|---|---|---|---|---|
| Null Model | 23 | |||||
| (a) Bacteria Richness (RSE:2.95) | 5 | 0.007 * | 0.55 | 115.07 | ||
| Intercept | 9.29 | 6.89 | 1.90 × 10−7 *** | |||
| N-NO3 (mg Kg−1) | 1.21 | 2.20 | 0.047 * | |||
| P Olsen (mg Kg−1) | 0.89 | 2.36 | 0.029 * | |||
| Ca (mg Kg−1) | −0.58 | −3.54 | 0.002 ** | |||
| Available S (mg Kg−1) | 1.05 | 1.73 | 0.101 | |||
| Zn (mg Kg−1) | −2.07 | −2.85 | 0.010 * | |||
| (b) Shannon Bacteria (RSE:2.29) | 5 | 0.007 | 0.56 | 3.115 | ||
| Intercept | 2.21 | 16.89 | 1.75 × 10−12 *** | |||
| N-NO3 (mg Kg−1) | 0.10 | 1.97 | 0.064 | |||
| P Olsen (mg Kg−1) | 0.08 | 2.24 | 0.038 * | |||
| Ca (mg Kg−1) | −0.05 | −3.20 | 0.005 ** | |||
| Available S (mg Kg−1) | 0.09 | 1.47 | 0.158 | |||
| Zn (mg Kg−1) | −0.18 | −2.65 | 0.016 * | |||
| (c) Fungi Richness (RSE:1.84) | 3 | 0.0013 | 0.53 | 113.4 | ||
| Intercept | 11.64 | 7.12 | 6.72 × 10−7 *** | |||
| N-NO3 (mg Kg−1) | −0.25 | −2.56 | 0.0185 * | |||
| Silt (%) | −0.13 | −4.16 | 4.86 × 10−4 *** | |||
| Field capacity (θfc) % | 0.09 | 3.12 | 5.42 × 10−3 ** | |||
| (d) Shannon Fungi (RSE:0.19) | 2 | 0.001** | 0.65 | −5.5 | ||
| Intercept | 2.52 | 15.17 | 1.07 × 10−11 *** | |||
| N-NO3 (mg Kg−1) | −0.08 | −3.29 | 0.004 ** | |||
| P Olsen (mg Kg−1) | 0.06 | 2.67 | 0.015 * | |||
| Silt (%) | −0.13 | −2.49 | 0.022 * | |||
| Sand (%) | −0.11 | 2.18 | 0.042 * | |||
| Field capacity (θfc) % | 0.01 | 3.20 | 0.005 ** | |||
| (e) Cellulase activity (RSE:8.40) | 5 | 1.70 × 10−5 *** | 0.82 | 180.6 | ||
| Intercept | 40.79 | 5.38 | 4.91 × 10−5 *** | |||
| N-N03 (mg Kg−1) | −6.16 | −3.09 | 0.006 ** | |||
| P Olsen (mg Kg−1) | −4.49 | −3.49 | 0.002 ** | |||
| Mn (mg Kg−1) | 0.16 | −2.37 | 0.023 * | |||
| Zn (mg Kg−1) | 13.43 | 6.51 | 5.30 × 10−6 *** | |||
| Sand (%) | −0.39 | 2.32 | 0.033 * | |||
| Field capacity (θfc) % | −0.45 | −2.95 | 0.008 ** | |||
| (f) Urease activity (RSE:54.5) | 7 | 1.09 × 10−6 *** | 0.89 | 276.2 | ||
| Intercept | 175.82 | 2.41 | 0.025 * | |||
| MOS (mg Kg−1) | −28.19 | −3.07 | 0.007 ** | |||
| P Olsen (mg Kg−1) | 42.64 | 4.68 | 0.000 *** | |||
| Mn (mg Kg−1) | 2.610 | 3.67 | 0.002 ** | |||
| Zn (mg Kg−1 | −36.56 | −3.17 | 0.006 ** | |||
| Cu (mg Kg−1) | 72.80 | 2.37 | 2 | 0.030 * | ||
| Sand (%) | −6.73 | −6.20 | 0.000 *** | |||
| Richness_B | 11.38 | 2.54 | 0.021 * | |||
| (g) FDA hydrolysis (RSE:72.64) | 2 | 0.000 *** | 0.62 | 276.2 | ||
| Intercept | 263.09 | 4.87 | 0.000 *** | |||
| Silt (%) | 3.334 | 2.784 | 0.011 * | |||
| Available S (mg Kg−1) | 15.493 | 3.146 | 0.004 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivas, Y.; Aponte, H.; Rivera-Salazar, D.; Matus, F.; Martínez, O.; Encina, C.; Retamal-Salgado, J. Microbial Community and Enzyme Activity of Forest Plantation, Natural Forests, and Agricultural Land in Chilean Coastal Cordillera Soils. Forests 2023, 14, 938. https://doi.org/10.3390/f14050938
Rivas Y, Aponte H, Rivera-Salazar D, Matus F, Martínez O, Encina C, Retamal-Salgado J. Microbial Community and Enzyme Activity of Forest Plantation, Natural Forests, and Agricultural Land in Chilean Coastal Cordillera Soils. Forests. 2023; 14(5):938. https://doi.org/10.3390/f14050938
Chicago/Turabian StyleRivas, Yessica, Humberto Aponte, Diego Rivera-Salazar, Francisco Matus, Oscar Martínez, Carolina Encina, and Jorge Retamal-Salgado. 2023. "Microbial Community and Enzyme Activity of Forest Plantation, Natural Forests, and Agricultural Land in Chilean Coastal Cordillera Soils" Forests 14, no. 5: 938. https://doi.org/10.3390/f14050938
APA StyleRivas, Y., Aponte, H., Rivera-Salazar, D., Matus, F., Martínez, O., Encina, C., & Retamal-Salgado, J. (2023). Microbial Community and Enzyme Activity of Forest Plantation, Natural Forests, and Agricultural Land in Chilean Coastal Cordillera Soils. Forests, 14(5), 938. https://doi.org/10.3390/f14050938








