Validation by SSRs of Morphometric Markers for Genetic Variability in Araucaria araucana (Molina) K. Koch
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Sampling and Spatial Distribution
2.2. Morphological Characterization
2.3. Genetic Characterization
2.3.1. DNA Extraction and Quantification
2.3.2. SSR Markers, PCR Amplification and Sizing
2.4. Data Analysis
3. Results
3.1. Morphometry
3.2. Genetic Diversity
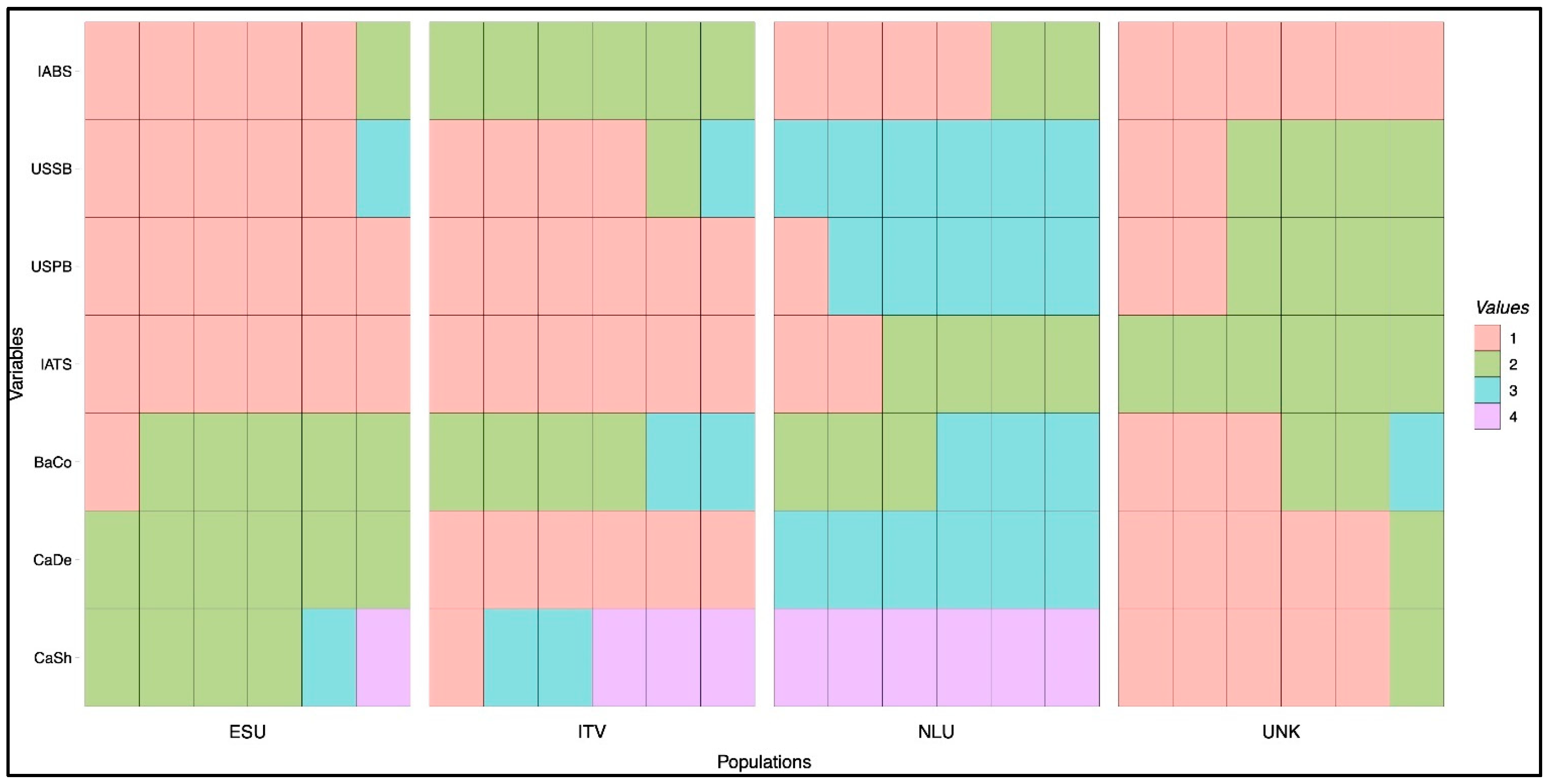
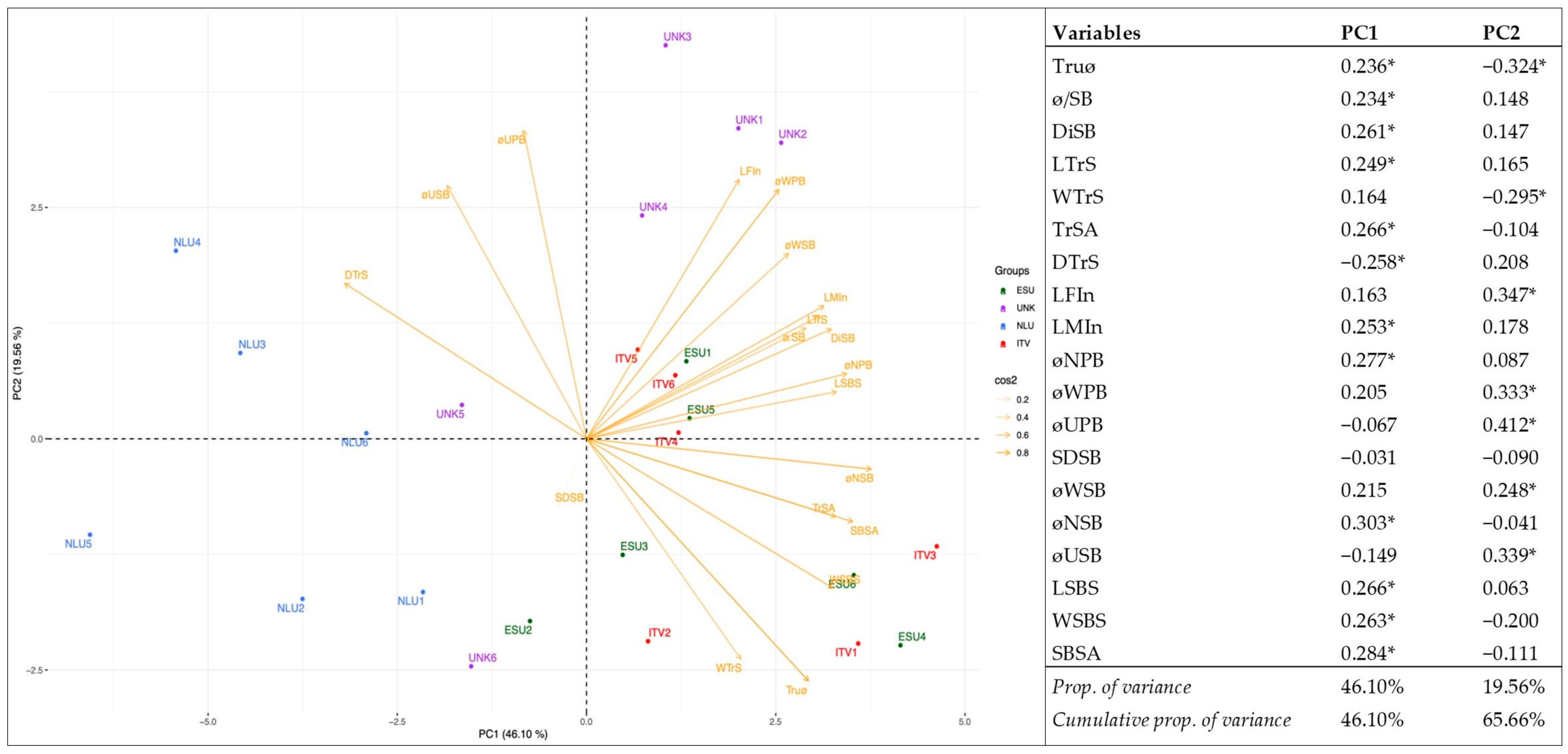
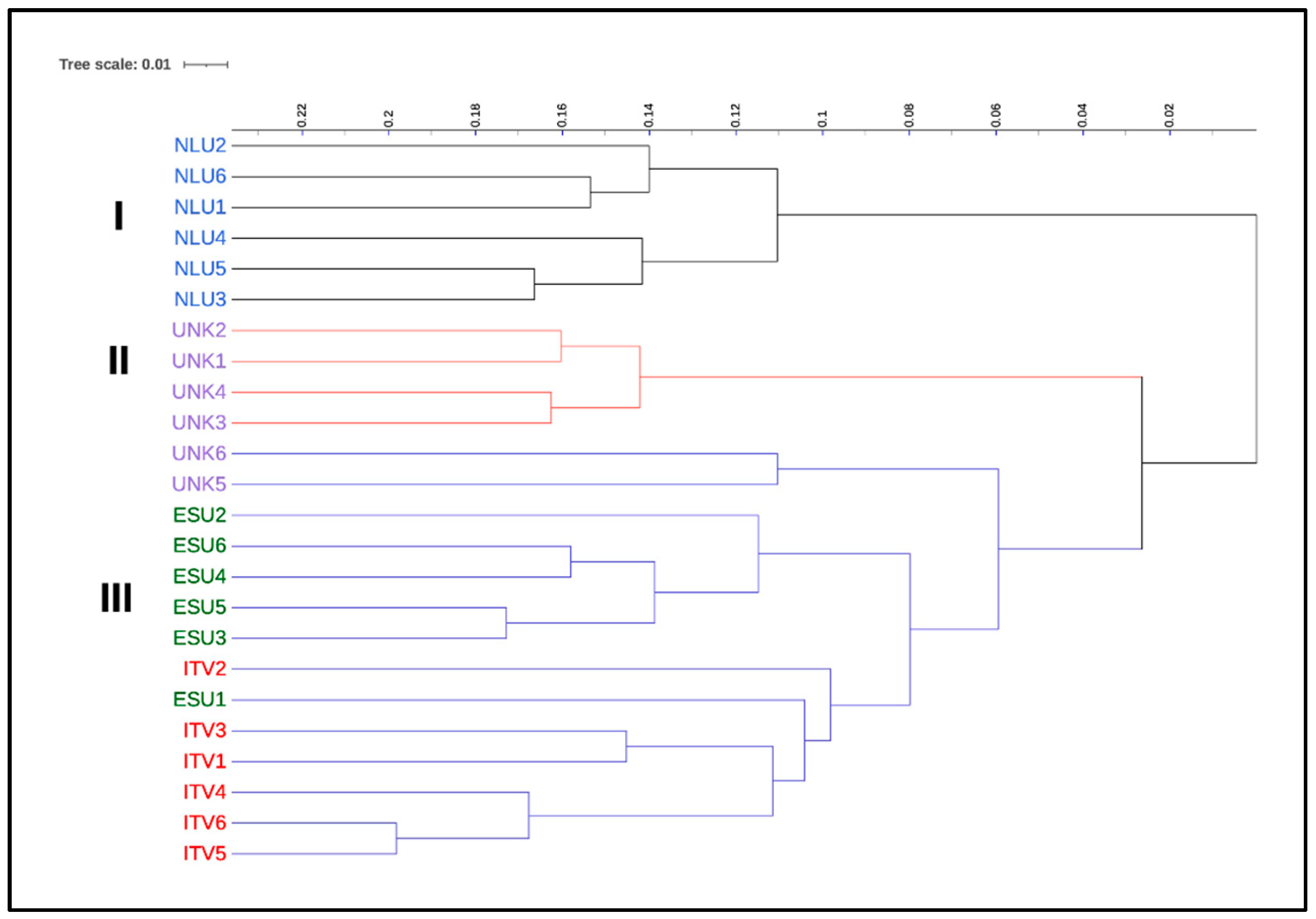
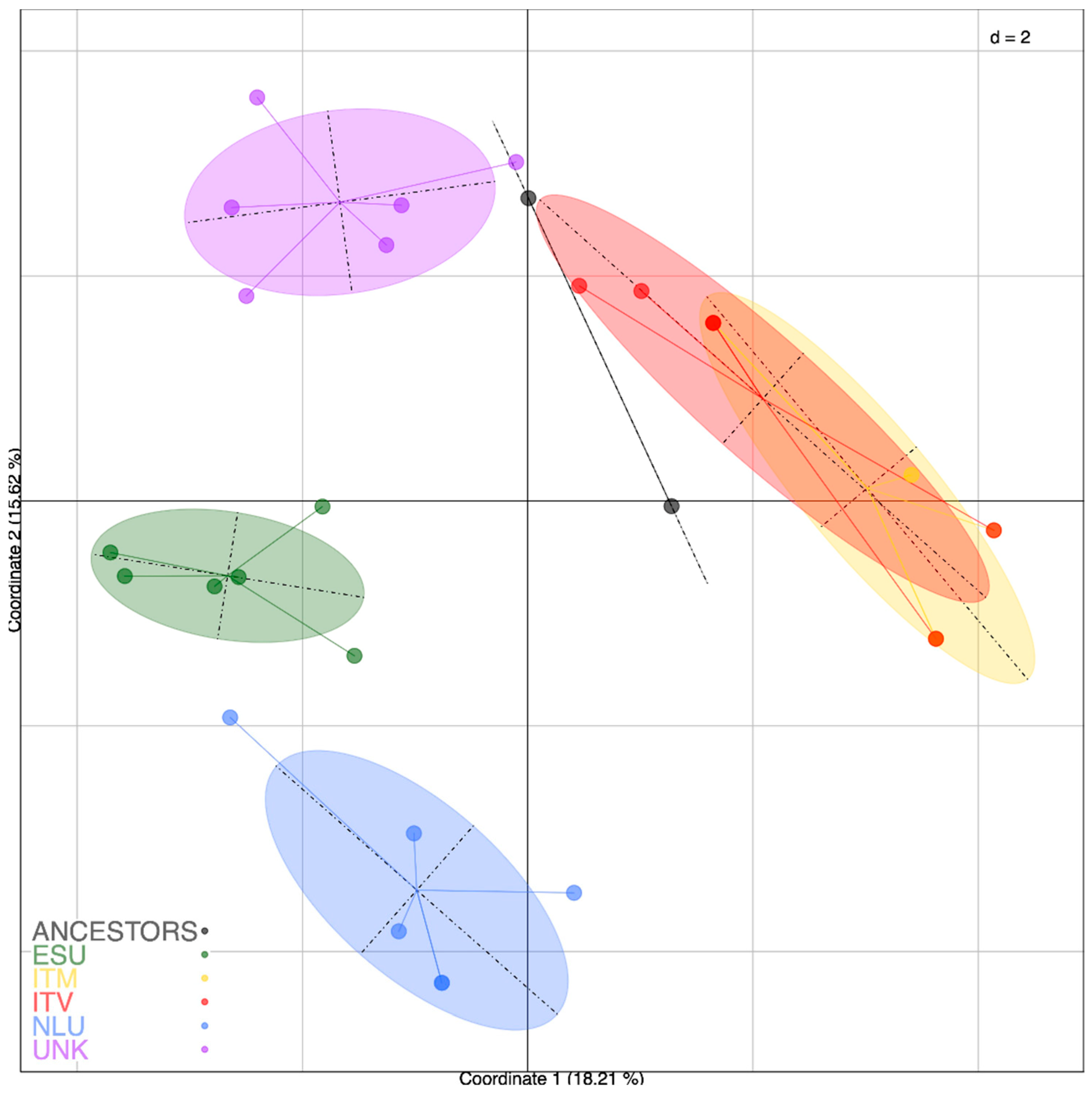
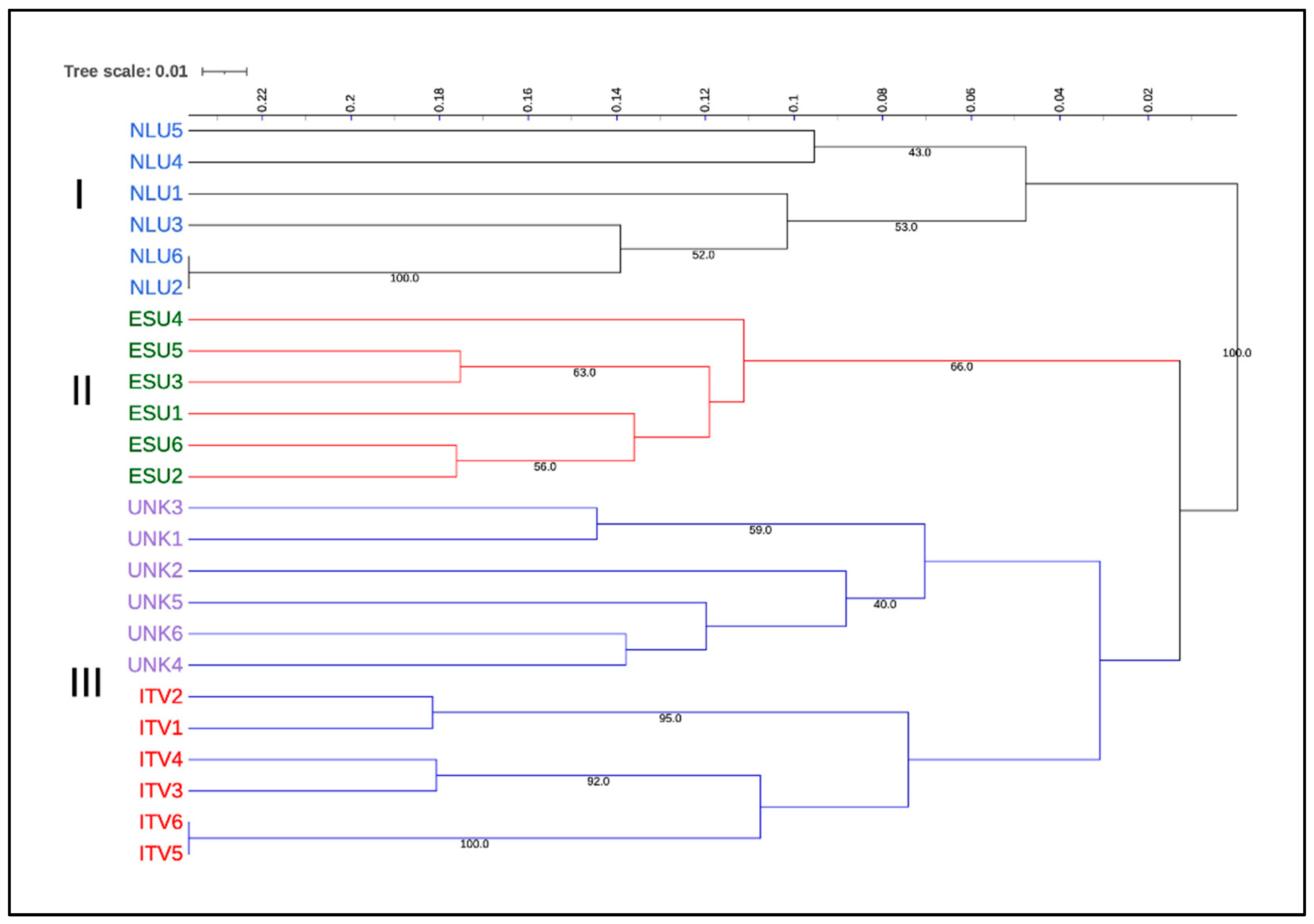
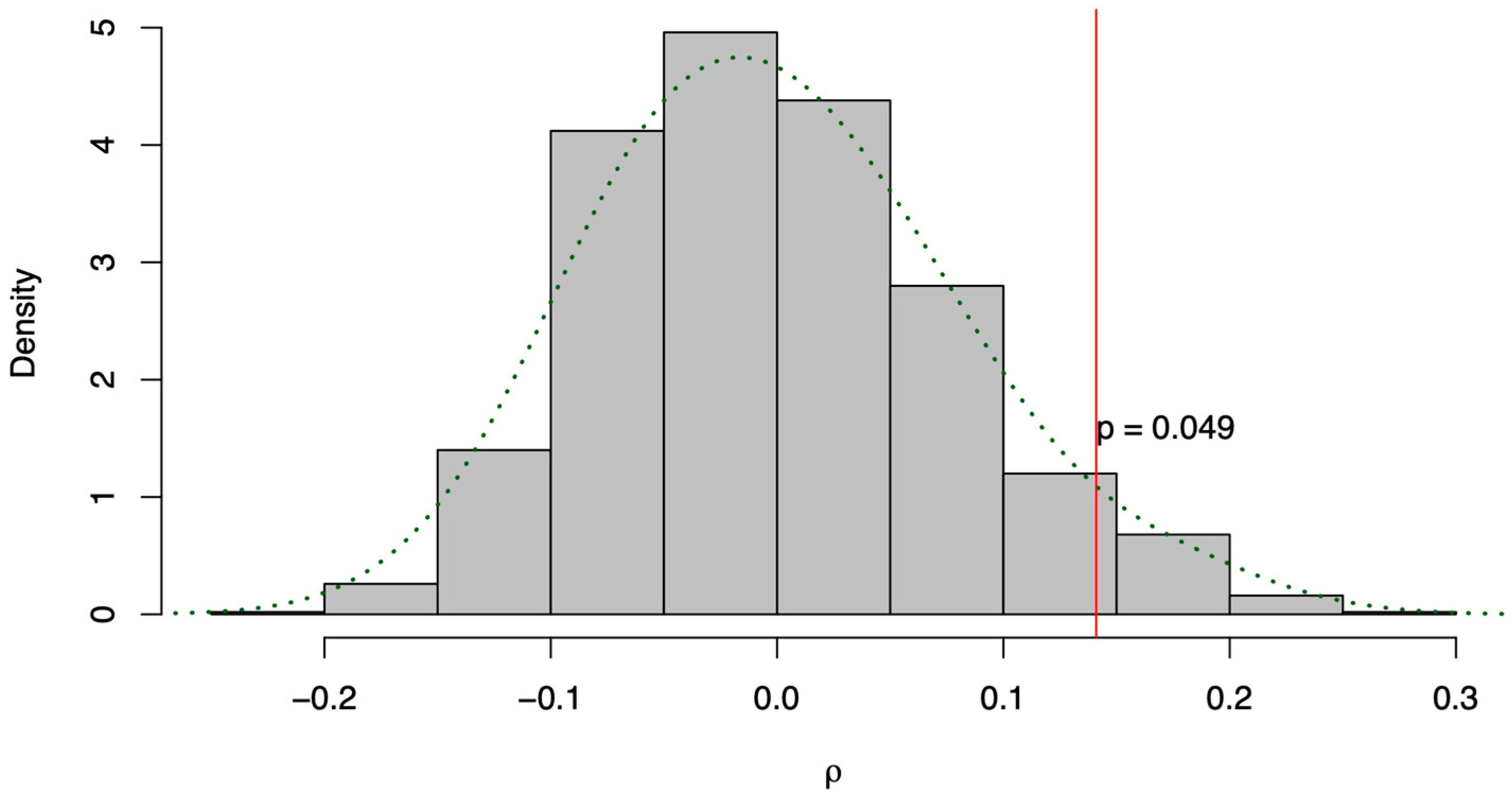
3.3. Relationship and Concordance among Morphological and Molecular Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rau, M.F. Land Use Change and Natural Araucaria Forest Degradation, Northeastern Misiones-Argentina. Ph.D. Thesis, Faculty of Forestry, University of Freiburg—Albert-Ludwigs-University, Breisgau, Germany, 2005. [Google Scholar]
- Guerra, M.P.; Silveira, V.; Reis, M.D.; Schneider, L. Exploração, Manejo e Conservação Da Araucária (Araucaria angustifolia). In Sustentável Mata Atlântica—A Exploração de Seus Recursos Florestais; Editora Senac: São Paulo, Brazil, 2002; pp. 85–101. [Google Scholar]
- González, M.; Cortés, M.; Izquierdo, G.; Gallo, L.; Echeverría, C.; Bekkesy, S.; Montaldo, P. Araucaria araucana (Molina) K. Koch. Las Especies Arbóreas de los Bosques Templados de Chile y Argentina: Autoecología; Marisa Cuneo Ediciones: Valdivia, Chile, 2006; pp. 36–53. [Google Scholar]
- Zamorano, C.; Cortés, M.; Echeverria, C.; Hechenleitner, P.; Lara, A. Experiencias de Restauración Con Especies Forestales Amenazadas En Chile. In Restauración de Bosques en America Latina; FIRE/Mundi-Prensa: Mexico City, Mexico, 2008; pp. 18–36. [Google Scholar]
- Ribeiro, M.C.; Metzger, J.P.; Martensen, A.C.; Ponzoni, F.J.; Hirota, M.M. The Brazilian Atlantic Forest: How Much Is Left, and How Is the Remaining Forest Distributed? Implications for Conservation. Biol. Conserv. 2009, 142, 1141–1153. [Google Scholar] [CrossRef]
- Lara, A.; Solari, M.; Rutherford, P.; Thiers, O.; Trecaman, R.; Molina, R.; Prieto, R.; Montory, C. Cobertura de La Vegetación Original de La Ecoregión de Los Bosques Valdivianos En Chile Hacia 1550. In Informe Técnico; Proyecto FB49 WWF-UACH: Valdivia, Chile, 1999. [Google Scholar]
- Dos Reis, M.S.; Ladio, A.; Peroni, N. Landscapes with Araucaria in South America: Evidence for a Cultural Dimension. Ecol. Soc. 2014, 19, 43. [Google Scholar] [CrossRef]
- Aagesen, D.L. The Natural and Social Geography of Araucaria araucana. Master’s Thesis, University of Minnesota, Minneapolis, MN, USA, 1993. [Google Scholar]
- Sanguinetti, J.; Kitzberger, T. Patterns and Mechanisms of Masting in the Large-seeded Southern Hemisphere Conifer Araucaria araucana. Austral Ecol. 2008, 33, 78–87. [Google Scholar] [CrossRef]
- Gedye, D. The Introduction of Araucaria araucana into the British Isles. In Yearbook 2017; International Dendrology Society: Las Cruces, NM, USA, 2017. [Google Scholar]
- Gedye, D. Araucaria The Monkey Puzzle; Orakaria Press: Huntingdon, UK, 2019; 216p. [Google Scholar]
- Antonetti, M.; Nin, S.; Burchi, G. First Insight into Araucaria araucana (Molina) K. Koch under Its Southernmost European Growing Condition: A Proposed Descriptor List for Morphological Characterization. Adv. Hortic. Sci. 2019, 33, 283–294. [Google Scholar]
- Delmastro, R.; Donoso, C. Review of Distribution, Variation and Utilization of Gene Resources of Araucaria araucana (Mol.) Koch in Chile. In Proceedings of the IUFRO Symposio em Melboramiento Genetico e Productividade de Especias Florestais de Rapido Crescimento, Sociedade Brasiliera de Silvicoltura, Águas de São Pedro, Brazil, 25–30 August 1980; pp. 133–135. [Google Scholar]
- Rafii, Z.A.; Dodd, R.S. Genetic Diversity among Coastal and Andean Natural Populations of Araucaria araucana (Molina) K. Koch. Biochem. Syst. Ecol. 1998, 26, 441–451. [Google Scholar] [CrossRef]
- Bekessy, S.; Allnutt, T.; Premoli, A.; Lara, A.; Ennos, R.; Burgman, M.; Cortes, M.; Newton, A. Genetic Variation in the Monkey Puzzle Tree (Araucaria araucana (Molina) K. Koch), Detected Using RAPD. Heredity 2002, 88, 243–249. [Google Scholar] [CrossRef]
- Gallo, L.; Izquierdo, F.; Sanguinetti, L.; Pinna, A.; Siffredi, G.; Ayesa, J.; Lopez, C.; Pelliza, A.; Strizler, N.; Peñalba, M.G. Araucaria araucana Forest Genetic Resources in Argentina. In Challenges in Managing Forest Genetic Resource for Livelihoods: Examples from Argentina and Brazil; International Plant Genetic Resources Institute: Rome, Italy, 2004; pp. 105–131. [Google Scholar]
- Ruiz, E.; González, F.; Torres-Díaz, C.; Fuentes, G.; Mardones, M.; Stuessy, T.; Samuel, R.; Becerra, J.; Silva, M. Genetic Diversity and Differentiation within and among Chilean Populations of Araucaria araucana (Araucariaceae) Based on Allozyme Variability. Taxon 2007, 56, 1221–1228. [Google Scholar] [CrossRef]
- Martín, M.; Mattioni, C.; Lusini, I.; Molina, J.; Cherubini, M.; Drake, F.; Herrera, M.; Villani, F.; Martín, L. New Insights into the Genetic Structure of Araucaria araucana Forests Based on Molecular and Historic Evidences. Tree Genet. Genomes 2014, 10, 839–851. [Google Scholar] [CrossRef]
- Marchelli, P.; Baier, C.; Mengel, C.; Ziegenhagen, B.; Gallo, L. Biogeographic History of the Threatened Species Araucaria araucana (Molina) K. Koch and Implications for Conservation: A Case Study with Organelle DNA Markers. Conserv. Genet. 2010, 11, 951–963. [Google Scholar] [CrossRef]
- Marchelli, P.; Sanguinetti, J.; Izquierdo, F.; Ziegenhagen, B.; Martín, A.; Mattioni, C.; Gallo, L.A.; Amico, I.; Bozzi, J.; Gazo, M.C. Araucaria araucana and Salix humboldtiana: Two Species Highly Appreciated by the Society with Domestication Potential. In Low Intensity Breeding of Native Forest Trees in Argentina; Springer: Berlin/Heidelberg, Germany, 2021; pp. 175–214. [Google Scholar]
- Fuentes, G.; González, F.; Saavedra, J.; López-Sepúlveda, P.; Victoriano, P.F.; Stuessy, T.F.; Ruiz-Ponce, E. Assessing Signals of Selection and Historical Demography to Develop Conservation Strategies in the Chilean Emblematic Araucaria araucana. Sci. Rep. 2021, 11, 20504. [Google Scholar] [CrossRef]
- Scott, L.J.; Shepherd, M.; Henry, R.J. Characterization of Highly Conserved Microsatellite Loci in Araucaria cunninghamii and Related Species. Plant Syst. Evol. 2003, 236, 115–123. [Google Scholar] [CrossRef]
- Salgueiro, F.; Caron, H.; De Souza, M.; Kremer, A.; Margis, R. Characterization of Nuclear Microsatellite Loci in South American Araucariaceae Species. Mol. Ecol. Notes 2005, 5, 256–258. [Google Scholar] [CrossRef]
- Schmidt, A.; Ciampi, A.; Guerra, M.; Nodari, R. Isolation and Characterization of Microsatellite Markers for Araucaria angustifolia (Araucariaceae). Mol. Ecol. Notes 2007, 7, 340–342. [Google Scholar] [CrossRef]
- Martín, M.A.; Mattioni, C.; Lusini, I.; Drake, F.; Cherubini, M.; Herrera, M.A.; Villani, F.; Martín, L.M. Microsatellite Development for the Relictual Conifer Araucaria araucana (Araucariaceae) Using Next-generation Sequencing. Am. J. Bot. 2012, 99, e213–e215. [Google Scholar] [CrossRef]
- Wei, T.; Simko, V. R Package ‘Corrplot’: Visualization of a Correlation Matrix. (Version 0.90). Statistician 56. 2021. Available online: https://github.com/tayun/corrplot (accessed on 22 May 2022).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Package ‘Factoextra.’ Extract and visualize the results of multivariate data analyses. RCRAN 2017, 76, 1–74. [Google Scholar]
- Šlenker, M.; Koutecký, P.; Marhold, K. MorphoTools2: An R Package for Multivariate Morphometric Analysis. Bioinformatics 2022, 38, 2954–2955. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An Integrated Analysis Environment for Genetic Marker Analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Chessel, D.; Dufour, A.B.; Thioulouse, J. The Ade4 Package-I-One-Table Methods. R News 2004, 4, 5–10. [Google Scholar]
- Siberchicot, A.; Julien-Laferrière, A.; Dufour, A.-B.; Thioulouse, J.; Dray, S. Adegraphics: An S4 Lattice-Based Package for the Representation of Multivariate Data. R J. 2017, 9, 198–212. [Google Scholar] [CrossRef]
- Bruvo, R.; Michiels, N.K.; D’SOUZA, T.G.; Schulenburg, H. A Simple Method for the Calculation of Microsatellite Genotype Distances Irrespective of Ploidy Level. Mol. Ecol. 2004, 13, 2101–2106. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package ‘Vegan.’ Community Ecology Package, Version 2; R Core Team: Vienna, Austria, 2013; Volume 2, pp. 1–295. [Google Scholar]
- Hijmans, R.J. Introduction to the” Geosphere” Package (Version 1.5-14). 2021. Available online: https://CRAN.R-project.org/package=geosphere (accessed on 18 March 2022).
- Balduzzi, M.; Binder, B.M.; Bucksch, A.; Chang, C.; Hong, L.; Iyer-Pascuzzi, A.S.; Pradal, C.; Sparks, E.E. Reshaping Plant Biology: Qualitative and Quantitative Descriptors for Plant Morphology. Front. Plant Sci. 2017, 8, 117. [Google Scholar] [CrossRef]
- Vidaković, M. Conifers: Morphology and Variation; Grafičko Zavod Hrvatske: Zagreb, Croatia, 1991; ISBN 86-399-0279-8. [Google Scholar]
- Moreno, A.C.; Marchelli, P.; Vendramin, G.G.; Gallo, L.A. Cross Transferability of SSRs to Five Species of Araucariaceae: A Useful Tool for Population Genetc Studis in “Araucaria araucana”. For. Syst. 2011, 20, 303–314. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a Genetic Linkage Map in Man Using Restriction Fragment Length Polymorphisms. Am. J. Hum. Genet. 1980, 32, 314. [Google Scholar]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Benowicz, A.; Stoehr, M.; Hamann, A.; Yanchuk, A.D. Estimation of the F2 Generation Segregation Variance and Relationships among Growth, Frost Damage, and Bud Break in Coastal Douglas-Fir (Pseudotsuga menziesii (Mirb.) Franco) Wide-Crosses. Ann. For. Sci. 2020, 77, 28. [Google Scholar] [CrossRef]
- Al-Ashkar, I.; Alderfasi, A.; Ben Romdhane, W.; Seleiman, M.F.; El-Said, R.A.; Al-Doss, A. Morphological and Genetic Diversity within Salt Tolerance Detection in Eighteen Wheat Genotypes. Plants 2020, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates Inc.: Sunderland, MA, USA, 1998; 980p. [Google Scholar]
| Seed Source | Seed Origin | Population Code | Cultivation Area | Coordinates | Tree Age | Tree Samples |
|---|---|---|---|---|---|---|
| Spain—fair in Valencia | Unknown | ESU | Vivai Bartolini, Pistoia | 43°53′ N; 10°55′ E; 60 m a.s.l. | 20–25 | ESU1, ESU2, ESU3, ESU4, ESU5, ESU6 |
| Unknown | Unknown | UNK | Vivai Bartolini, Pistoia | 43°53′ N; 10°55′ E; 60 m a.s.l. | 20–25 | UNK1, UNK2, UNK3, UNK4, UNK5, UNK6 |
| The Netherlands—Dutch fair stand | Unknown | NLU | Vivai Bartolini, Pistoia | 43°53′ N; 10°55′ E; 60 m a.s.l. | 20–25 | NLU1, NLU2, NLU3, NLU4, NLU5, NLU6 |
| Italy—Villa Lodolo (S. Marcello Pistoiese, Pistoia) | Ancestor trees | ITV | Azienda Capecchi, Pistoia | 43°88′ N; 10°97′ E; 60 m a.s.l. | 20–25 | ITV1, ITV2, ITV3, ITV4, ITV 5, ITV6 (F1) |
| Italy—Azienda Macchia Tommaso (La Grazie, Pistoia) | F1 of the ancestor trees | ITM | Azienda Macchia Tommaso, Pistoia | 44°0′ N; 10°52′ E; 550 m a.s.l. | 6 | ITM1, ITM2, ITM3, ITM4, ITM5, ITM6 (F2) |
| Argentina (1920) | Indigenous trees of the Argentine Andes | ARG | Villa Lodolo, S. Marcello Pistoiese, Pistoia | 44°03′ N; 10°47′ E; 623 m a.s.l. | ~100 | ARG1 = female ancestor, ARG2 = male ancestor |
| SSR Name | GenBank Accession | Forward Primer (5′-3′) | Reverse primer (5′-3′) | Repeat Motif | Size (bp) | References |
|---|---|---|---|---|---|---|
| Ara2027 | JN896693 | AGGAAGGCATTTTGGCTTGG | TGGTCATCTTAATGGTACTTTGATTG | (AC)22 | 128–156 | [25] |
| Ara5179 | JN896694 | GCTTATAGACTCGACTTGCCAC | CGGATCCACCATTTGTAACTTTG | (CA)15 | 144 | [25] |
| Ara5182 | JN896695 | TGATGTGAGCCAAAATCAAAATC | AGGAGAGAGTCATGAAGCCG | (TG)15 | 172–206 | [25] |
| Ara5595 | JN896696 | AGTCCAAAATAGACATAGGCATCC | TGGGAAAATCAAACCCTCGC | (CA)12 | 123–125 | [25] |
| Ara11382 | JN896697 | GGAAAGTAGCAAGGCCTCAAC | TGCCTAAAACATCCCTTGGAC | (AC)14 | 200–208 | [25] |
| Ara11384 | JN896698 | TGATTGATGTGATTGGCTACAAATTC | TGTTTGCATGCTTGGAGTGG | (CA)16 | 120–130 | [25] |
| Ara20681 | JN896699 | AACTAAAAACCTTAAATGCTCATCG | CAATCCTCAAATTAGCCCATGC | (GT)12 | 187–191 | [25] |
| Ag20 | AJ749964 | ACTAGGAATGGATGTTGGTG | AAGGTATGGCATCATGTCTC | (GA)12 | 180–200 | [23] |
| Ag45 | AJ749966 | CCATCCTCCATCATTCATCC | TCCCTCCCTATGTCCCAAAG | (GT)4AT(GT)7 | 170–182 | [23] |
| Ag56 | AJ749967 | CCACACTCAAAACAATAGCAGTTC | TGAAGTTGGCCAATCAGATAC | (TC)11 | 165–171 | [23] |
| Ag62 | AJ749968 | ATATGGTGGGGTGCCTACAG | TCCAATCGTTCCTCCAACAG | (TC)13 | 126–130 | [23] |
| CRCAc2 | AF522867 | ATGCATGACTAGGATGAACA | ATAGTTCTGCTTATCACATCT | (GA)23 | 190–196 | [22] |
| Aang01 | AY865575 | TGACGGGTTCACTCCTACCT | TAGGAACCCCCATTCATTTG | (CT)22 | 224–234 | [24] |
| Aang03 | AY865577 | CGCCTACCTCAATCACTGGT | TGGGACAATGTGCTTATCCA | (CT)13 | 254–260 | [24] |
| Aang07 | AY865579 | ACCTCACAGGGACACCTCAC | TTTTCATGCATGTTGCTTGC | (GA)24 | 195 | [24] |
| Aang12 | AY865581 | AAGGGTTCACAATGCTGAGG | TGGATTTTATTATGATGGTTGTTCC | (GA)23 | 192 | [24] |
| Aang14 | AY865583 | GAGCACGTGCAGATGTTGAT | CCATCCTCTCCATGACCACT | (GA)27 | 160 | [24] |
| Aang18 | AY865586 | ACACGTTTAATCAGACGAAGAAG | ATGCCACCTTTTTCAGCAAC | (TC)9 | 207 | [24] |
| Aang21 | AY865587 | GGAGACACCTCACCCCCTA | TGATGAGGGAGGATTACAAGC | (CT)12 | 188 | [24] |
| Aang22 | AY865588 | TCAACTTGCAAGGTCACCTCTA | ATGGGAGCCCCTTCTAGTGT | (GA)10 | 220 | [24] |
| Aang24 | AY865590 | CTCTCCTTCCCCTTGCTCTT | AGGTGGATCACCCACTGAAG | (CT)19 | 198–204 | [24] |
| Aang27 | AY865591 | CATGGTGGCTATTGCTCCTT | AGAAGCCATCAAAGGAGTGG | (CT)12 | 193 | [24] |
| Aang28 | AY865592 | TCCATTGCATTAGTTTGGGATA | TTTCCAATCATACATTCACCACA | (CT)11 | 136–142 | [24] |
| Aang35 | AY865594 | GGTGAAGCTTCGTTTCAAGG | CCACTTGTCTTCACCAACCA | (GA)10 | 169–173 | [24] |
| Aang37 | AY865596 | GGGGAGTTTCCATGAGATGA | TCCACTCACCACTCTGAGGA | (GA)18 | 266–268 | [24] |
| Aang45 | AY865601 | AGGCTCACATCAGGCTCACT | TGGTTTTGGTGGTCAAATCA | (CT)15 | 199–216 | [24] |
| Aang46 | AY865602 | TCCACCTACCTCAATCACTGG | TGGGACAATGTGCTTATCCA | (CT)12 | 259–267 | [24] |
| Aang47 | AY865603 | GATATGAAAAGAAGGGTTCTATGCT | TTTCTTCCATTCCTCCAAGC | (GA)15 | 180–188 | [24] |
| Morfological Parameter | Statistics | TS | ESU | UNK | NLU | ITV | |
|---|---|---|---|---|---|---|---|
| n | 24 | 6 | 6 | 6 | 6 | ||
| TRUNK | Trunk diameter (mm) Truø | Average * | 115.0 | 143.3 a | 97.5 b | 88.3 b | 140.0 a |
| SD | 32.4 | 13.6 | 16.6 | 29.9 | 23.3 | ||
| CV% | 18.8 | 9.5 | 17.1 | 33.9 | 17.1 | ||
| Density of trunk scales (mm) DTrS | Average * | 28.5 | 20.8 b | 33.2 a | 38.5 a | 19.2 b | |
| SD | 8.5 | 4.3 | 10.2 | 9.7 | 5.0 | ||
| CV% | 27.4 | 20.9 | 30.8 | 25.2 | 26.7 | ||
| Maximum width of trunk scale (mm) WTrS | Average * | 15.6 | 16.4 ab | 14.9 bc | 14.6 c | 17.5 a | |
| SD | 2.7 | 1.9 | 1.0 | 2.4 | 4.3 | ||
| CV% | 27.3 | 11.7 | 6.7 | 17.0 | 25.9 | ||
| Maximum lenght of trunk scale (mm) LTrS | Average * | 39.6 | 41.5 a | 39.5 a | 33.9 b | 43.6 a | |
| SD | 6.8 | 3.4 | 9.8 | 3.6 | 5.2 | ||
| CV% | 17.2 | 8.4 | 25.0 | 10.8 | 12.1 | ||
| Trunk scale area (mm) TrSA | Average * | 624.4 | 680.1 a | 588.4 b | 505.1 c | 749.1 a | |
| SD | 159.6 | 90.1 | 162.3 | 130.1 | 177.9 | ||
| CV% | 25.7 | 13.2 | 28.0 | 25.7 | 24.4 | ||
| Distance among scaffold branches (mm) DiSB | Average * | 335.8 | 338.3 a | 377.1 a | 258.4 b | 365.0 a | |
| SD | 74.2 | 65.5 | 78.4 | 46.3 | 32.7 | ||
| CV% | 18.0 | 19.4 | 21.4 | 18.0 | 9.0 | ||
| Diameter/number of secondary branches (mm) ø/SB | Average * | 13.1 | 13.1 b | 14.8 ab | 8.1 c | 16.4 a | |
| SD | 3.8 | 2.1 | 3.1 | 0.7 | 2.6 | ||
| CV% | 18.7 | 16.7 | 21.3 | 9.0 | 16.0 | ||
| PRIMARY BRANCH | Lenght of first internode on primary branch (cm) LFIn | Average * | 34.5 | 31.5 c | 45.8 a | 25.0 d | 38.5 b |
| SD | 13.0 | 5.3 | 19.6 | 3.8 | 5.8 | ||
| CV% | 29.2 | 16.9 | 42.9 | 15.4 | 15.3 | ||
| Lenght of median internode on primary branch (cm) LMIn | Average * | 20.9 | 21.5 b | 22.7 ab | 15 c | 24.4 a | |
| SD | 4.7 | 2.7 | 5.0 | 2.5 | 2.0 | ||
| CV% | 22.8 | 12.8 | 22.4 | 17.0 | 8.5 | ||
| Wide section of primary branch including scales (mm) øWPB | Average * | 9.4 | 9.0 b | 11.0 a | 7.9 c | 9.6 b | |
| SD | 1.5 | 1.0 | 1.5 | 1.0 | 0.8 | ||
| CV% | 16.3 | 11.7 | 13.7 | 12.5 | 8.8 | ||
| Narrow section of primary branch including scales (mm) øNPB | Average * | 7.0 | 7.2 b | 7.9 a | 5.3 c | 7.8 a | |
| SD | 1.3 | 1.4 | 0.8 | 0.6 | 0.5 | ||
| CV% | 19.3 | 19.8 | 11.1 | 12.3 | 6.8 | ||
| Apparent diameter uniformity on primary branch (mm) øUPB | Average * | 2.3 | 1.8 b | 3.0 a | 2.6 a | 1.8 b | |
| SD | 1.0 | 0.8 | 1.2 | 0.6 | 0.6 | ||
| CV% | 43.8 | 44.5 | 43.3 | 31.7 | 35.3 | ||
| SECONDARY BRANCH | Wide section of secondary branch including scales (mm) øWSB | Average * | 7.3 | 7.8 a | 7.5 a | 6.6 b | 7.3 a |
| SD | 0.8 | 1.0 | 0.7 | 0.7 | 0.3 | ||
| CV% | 11.8 | 14.1 | 10.6 | 11.7 | 5.3 | ||
| Narrow section of secondary branch including scales (mm) øNSB | Average * | 5.0 | 5.7 a | 5.0 b | 3.8 c | 5.6 a | |
| SD | 0.9 | 1.0 | 0.4 | 0.5 | 0.5 | ||
| CV% | 19.6 | 18.0 | 8.3 | 14.2 | 10.1 | ||
| Apparent diameter uniformity on secondary branch (mm) øUSB | Average * | 2.3 | 2.2 b | 2.5 ab | 2.8 a | 1.7 c | |
| SD | 0.7 | 0.7 | 0.7 | 0.7 | 0.3 | ||
| CV% | 32.9 | 32.7 | 30.7 | 26.9 | 21.0 | ||
| Scale density on secondary branch SDSB | Average * | 16.6 | 16.0 b | 13.6 c | 17.0 b | 19.9 a | |
| SD | 2.9 | 1.3 | 1.3 | 1.2 | 3.0 | ||
| CV% | 17.5 | 8.7 | 10.3 | 7.1 | 15.5 | ||
| Maximum width of secondary branch scale WSBS | Average * | 12.7 | 14.7 a | 12.6 b | 9.7 c | 13.6 c | |
| SD | 2.5 | 2.3 | 1.4 | 1.7 | 1.5 | ||
| CV% | 19.9 | 15.5 | 11.3 | 17.9 | 11.2 | ||
| Maximum lenght of secondary branch scale LSBS | Average * | 34.3 | 38.6 a | 35.2 b | 28.6 c | 34.6 b | |
| SD | 4.5 | 4.1 | 1.2 | 1.7 | 3.2 | ||
| CV% | 13.2 | 10.8 | 3.6 | 6.1 | 9.3 | ||
| Secondary branch scale area SBSA | Average * | 444.0 | 570.3 a | 451.7 b | 284.1 c | 469.8 b | |
| SD | 128.6 | 110.3 | 53.2 | 55.6 | 84.8 | ||
| CV% | 29.0 | 19.3 | 11.7 | 19.9 | 18.1 | ||
| Locus | Na | Ne | Fa | Ho | He | F | PIC |
|---|---|---|---|---|---|---|---|
| aang01 | 7.000 | 4.188 | 0.375 | 0.188 | 0.761 | 0.754 | 0.727 |
| aang03 | 2.000 | 1.600 | 0.750 | 0.000 | 0.375 | 1.000 | 0.305 |
| aang24 | 2.000 | 1.822 | 0.656 | 0.000 | 0.451 | 1.000 | 0.349 |
| aang28 | 2.000 | 1.679 | 0.719 | 0.063 | 0.404 | 0.845 | 0.323 |
| aang35 | 3.000 | 2.092 | 0.547 | 0.719 | 0.522 | −0.377 | 0.415 |
| aang45 | 2.000 | 1.822 | 0.656 | 0.000 | 0.451 | 1.000 | 0.349 |
| aang46 | 5.000 | 1.923 | 0.703 | 0.375 | 0.480 | 0.219 | 0.454 |
| aang47 | 2.000 | 1.600 | 0.750 | 0.000 | 0.375 | 1.000 | 0.305 |
| ag20 | 4.000 | 3.969 | 0.281 | 0.000 | 0.748 | 1.000 | 0.701 |
| ag45 | 3.000 | 2.136 | 0.609 | 0.313 | 0.532 | 0.412 | 0.458 |
| ag56 | 3.000 | 1.697 | 0.734 | 0.469 | 0.411 | −0.141 | 0.357 |
| ag62 | 2.000 | 1.640 | 0.734 | 0.219 | 0.390 | 0.439 | 0.314 |
| ara11382 | 2.000 | 1.822 | 0.656 | 0.063 | 0.451 | 0.861 | 0.349 |
| ara11384 | 3.000 | 2.653 | 0.500 | 0.500 | 0.623 | 0.197 | 0.552 |
| ara2027 | 7.000 | 4.171 | 0.328 | 0.031 | 0.760 | 0.959 | 0.724 |
| ara20681 | 3.000 | 2.169 | 0.563 | 0.188 | 0.539 | 0.652 | 0.447 |
| ara5182 | 2.000 | 1.679 | 0.719 | 0.313 | 0.404 | 0.227 | 0.323 |
| ara5595 | 2.000 | 1.358 | 0.844 | 0.000 | 0.264 | 1.000 | 0.229 |
| CRCAc2 | 2.000 | 1.882 | 0.625 | 0.125 | 0.469 | 0.733 | 0.359 |
| Mean | 3.053 | 2.205 | 0.618 | 0.188 | 0.495 | 0.620 | 0.423 |
| Source | df | SS | MS | Est. Var. | % | FST | Nm |
|---|---|---|---|---|---|---|---|
| Among Pops | 5 | 115.974 | 23.195 | 1.905 | 37% | ||
| Within Pops | 58 | 185.167 | 3.193 | 3.193 | 63% | ||
| Total | 63 | 301.141 | 5.098 | 100% | 0.374 *** | 0.418 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nin, S.; Antonetti, M.; Burchi, G.; Gori, M.; Bini, L. Validation by SSRs of Morphometric Markers for Genetic Variability in Araucaria araucana (Molina) K. Koch. Forests 2023, 14, 466. https://doi.org/10.3390/f14030466
Nin S, Antonetti M, Burchi G, Gori M, Bini L. Validation by SSRs of Morphometric Markers for Genetic Variability in Araucaria araucana (Molina) K. Koch. Forests. 2023; 14(3):466. https://doi.org/10.3390/f14030466
Chicago/Turabian StyleNin, Stefania, Maurizio Antonetti, Gianluca Burchi, Massimo Gori, and Lorenzo Bini. 2023. "Validation by SSRs of Morphometric Markers for Genetic Variability in Araucaria araucana (Molina) K. Koch" Forests 14, no. 3: 466. https://doi.org/10.3390/f14030466
APA StyleNin, S., Antonetti, M., Burchi, G., Gori, M., & Bini, L. (2023). Validation by SSRs of Morphometric Markers for Genetic Variability in Araucaria araucana (Molina) K. Koch. Forests, 14(3), 466. https://doi.org/10.3390/f14030466







