Estimating Stem Diameter Distributions with Airborne Laser Scanning Metrics and Derived Canopy Surface Texture Metrics
Abstract
1. Introduction
2. Materials and Methods
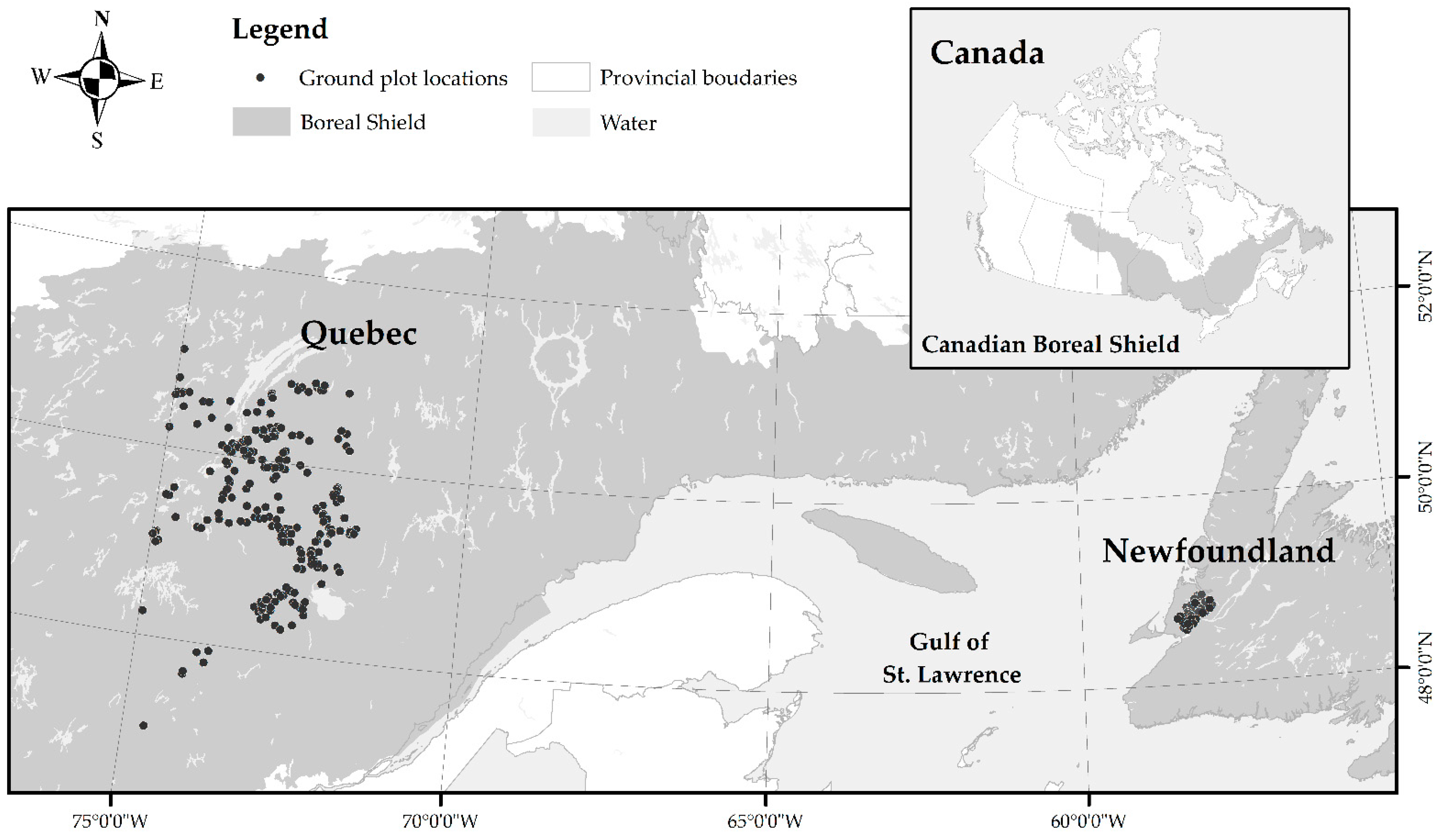
2.1. Study Area
2.2. Ground Plots
2.3. ALS Data and Metrics
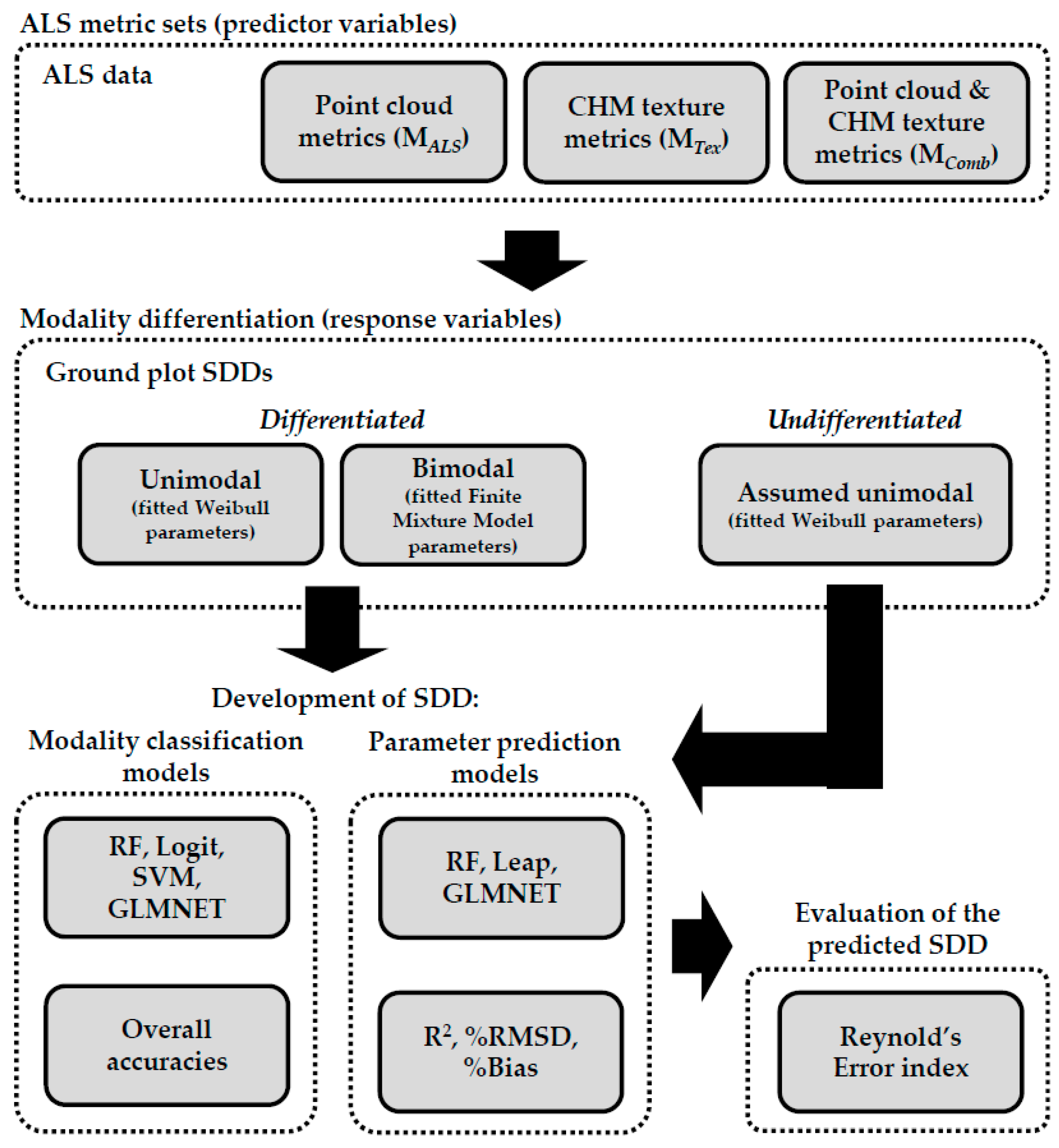
2.4. Overview of the Methods
2.5. Development of SDD Modality Classification Models
2.6. Development of SDD Prediction Models
2.7. Evaluation of the Predicted SDD
3. Results
3.1. SDD Modality Classification Models
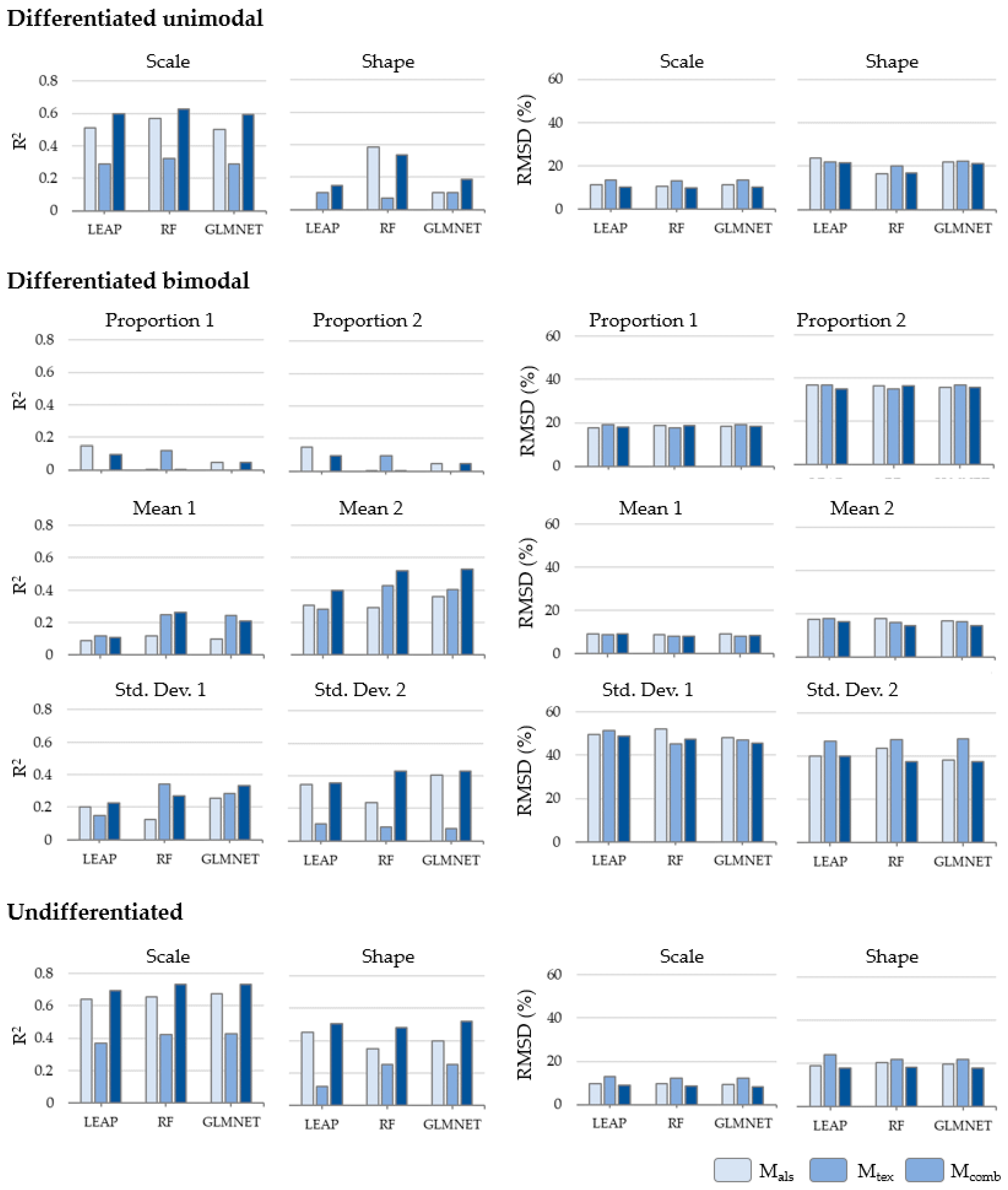
3.2. SDD Prediction Models
3.3. Goodness-of-Fit of the Predicted SDD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hyyppä, J.; Hyyppä, H.; Leckie, D.; Gougeon, F.; Yu, X.; Maltamo, M. Review of Methods of Small-Footprint Airborne Laser Scanning for Extracting Forest Inventory Data in Boreal Forests. Int. J. Remote Sens. 2008, 29, 1339–1366. [Google Scholar] [CrossRef]
- Næsset, E. Area-Based Inventory in Norway—From Innovation to an Operational Reality. In Forestry Applications of Airborne Laser Scanning; Springer: Dordrecht, The Netherlands, 2014; Volume 27, pp. 215–240. [Google Scholar]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef]
- Badreldin, N.; Sanchez-Azofeifa, A. Estimating Forest Biomass Dynamics by Integrating Multi-Temporal Landsat Satellite Images with Ground and Airborne LiDAR Data in the Coal Valley Mine, Alberta, Canada. Remote Sens. 2015, 7, 2832–2849. [Google Scholar] [CrossRef]
- Luther, J.E.; Fournier, R.A.; van Lier, O.R.; Bujold, M. Extending ALS-Based Mapping of Forest Attributes with Medium Resolution Satellite and Environmental Data. Remote Sens. 2019, 11, 1092. [Google Scholar] [CrossRef]
- Mielcarek, M.; Kamińska, A.; Stereńczak, K. Digital Aerial Photogrammetry (DAP) and Airborne Laser Scanning (ALS) as Sources of Information about Tree Height: Comparisons of the Accuracy of Remote Sensing Methods for Tree Height Estimation. Remote Sens. 2020, 12, 1808. [Google Scholar] [CrossRef]
- Cao, L.; Zhang, Z.; Yun, T.; Wang, G.; Ruan, H.; She, G. Estimating Tree Volume Distributions in Subtropical Forests Using Airborne LiDAR Data. Remote Sens. 2019, 11, 97. [Google Scholar] [CrossRef]
- Spriggs, R.A.; Coomes, D.A.; Jones, T.A.; Caspersen, J.P.; Vanderwel, M.C. An Alternative Approach to Using LiDAR Remote Sensing Data to Predict Stem Diameter Distributions across a Temperate Forest Landscape. Remote Sens. 2017, 9, 944. [Google Scholar] [CrossRef]
- Tompalski, P.; Coops, N.C.; White, J.C.; Wulder, M.A. Enriching ALS-Derived Area-Based Estimates of Volume through Tree-Level Downscaling. Forests 2015, 6, 2608–2630. [Google Scholar] [CrossRef]
- Zhang, L.; Gove, J.H.; Liu, C.; Leak, W.B. A Finite Mixture of Two Weibull Distributions for Modeling the Diameter Distributions of Rotated-Sigmoid, Uneven-Aged Stands. Can. J. For. Res. 2001, 31, 1654–1659. [Google Scholar] [CrossRef]
- Cao, Q.V. Predicting Parameters of a Weibull Function for Modeling Diameter Distribution. For. Sci. 2004, 50, 682–685. [Google Scholar] [CrossRef]
- Siipilehto, J.; Mehtätalo, L. Parameter Recovery vs. Parameter Prediction for the Weibull Distribution Validated for Scots Pine Stands in Finland. Silva Fenn. 2013, 47, 22. [Google Scholar] [CrossRef]
- Mcgarrigle, E.; Kershaw Jr, J.A.; Lavigne, M.B.; Weiskittel, A.R.; Ducey, M. Predicting the Number of Trees in Small Diameter Classes Using Predictions from a Two-Parameter Weibull Distribution. Forestry 2011, 84, 431–439. [Google Scholar] [CrossRef]
- Poudel, K.P.; Cao, Q.V. Evaluation of Methods to Predict Weibull Parameters for Characterizing Diameter Distributions. For. Sci. 2013, 59, 243–252. [Google Scholar] [CrossRef]
- Palahí, M.; Pukkala, T.; Trasobares, A. Modelling the Diameter Distribution of Pinus Sylvestris, Pinus Nigra and Pinus Halepensis Forest Stands in Catalonia Using the Truncated Weibull Function. For. Int. J. For. Res. 2006, 79, 553–562. [Google Scholar] [CrossRef]
- Duan, A.G.; Zhang, J.G.; Zhang, X.Q.; He, C.Y. Stand Diameter Distribution Modelling and Prediction Based on Richards Function. PLoS ONE 2013, 8, e62605. [Google Scholar] [CrossRef]
- Guo, H.; Lei, X.; You, L.; Zeng, W.; Lang, P.; Lei, Y. Climate-Sensitive Diameter Distribution Models of Larch Plantations in North and Northeast China. For. Ecol. Manag. 2022, 506, 119947. [Google Scholar] [CrossRef]
- West, G.B.; Enquist, B.J.; Brown, J.H. A General Quantitative Theory of Forest Structure and Dynamics. Proc. Natl. Acad. Sci. USA 2009, 106, 7040–7045. [Google Scholar] [CrossRef]
- Martin Bollandsås, O.; Næsset, E. Estimating Percentile-Based Diameter Distributions in Uneven-Sized Norway Spruce Stands Using Airborne Laser Scanner Data. Scand. J. For. Res. 2007, 22, 33–47. [Google Scholar] [CrossRef]
- Rana, P.; Vauhkonen, J.; Junttila, V.; Hou, Z.; Gautam, B.; Cawkwell, F.; Tokola, T. Large Tree Diameter Distribution Modelling Using Sparse Airborne Laser Scanning Data in a Subtropical Forest in Nepal. ISPRS J. Photogramm. Remote Sens. 2017, 134, 86–95. [Google Scholar] [CrossRef]
- Xu, Q.; Hou, Z.; Maltamo, M.; Tokola, T. Calibration of Area Based Diameter Distribution with Individual Tree Based Diameter Estimates Using Airborne Laser Scanning. ISPRS J. Photogramm. Remote Sens. 2014, 93, 65–75. [Google Scholar] [CrossRef]
- Fries, C.; Johansson, O.; Pettersson, B.; Simonsson, P. Silvicultural Models to Maintain and Restore Natural Stand Structures in Swedish Boreal Forests. For. Ecol. Manag. 1997, 94, 89–103. [Google Scholar] [CrossRef]
- Packalén, P.; Maltamo, M. Estimation of Species-Specific Diameter Distributions Using Airborne Laser Scanning and Aerial Photographs. Can. J. For. Res. 2008, 38, 1750–1760. [Google Scholar] [CrossRef]
- Bailey, R.L.; Dell, T.R. Quantifying Diameter Distributions with the Weibull Function. For. Sci. 1973, 19, 97–104. [Google Scholar] [CrossRef]
- Hafley, W.L.; Schreuder, H.T. Statistical Distributions for Fitting Diameter and Height Data in Even-Aged Stands. Can. J. For. Res. 1977, 7, 481–487. [Google Scholar] [CrossRef]
- Gorgoso-Varela, J.J.; Ponce, R.A.; Rodríguez-Puerta, F. Modeling Diameter Distributions with Six Probability Density Functions in Pinus Halepensis Mill. Plantations Using Low-Density Airborne Laser Scanning Data in Aragón (Northeast Spain). Remote Sens. 2021, 13, 2307. [Google Scholar] [CrossRef]
- Nepomuceno Cosenza, D.; Soares, P.; Guerra-Hernández, J.; Pereira, L.; González-Ferreiro, E.; Castedo-Dorado, F.; Tomé, M. Comparing Johnson’s S B and Weibull Functions to Model the Diameter Distribution of Forest Plantations through ALS Data. Remote Sens. 2019, 11, 2792. [Google Scholar] [CrossRef]
- Thomas, V.; Oliver, R.D.; Lim, K.; Woods, M. LiDAR and Weibull Modeling of Diameter and Basal Area. For. Chron. 2008, 84, 866–875. [Google Scholar] [CrossRef]
- Hao, Y.; Widagdo, F.R.A.; Liu, X.; Quan, Y.; Liu, Z.; Dong, L.; Li, F. Estimation and Calibration of Stem Diameter Distribution Using UAV Laser Scanning Data: A Case Study for Larch (Larix Olgensis) Forests in Northeast China. Remote Sens. Environ. 2022, 268, 112769. [Google Scholar] [CrossRef]
- Peuhkurinen, J.; Tokola, T.; Plevak, K.; Sirparanta, S.; Kedrov, A.; Pyankov, S. Predicting Tree Diameter Distributions from Airborne Laser Scanning, SPOT 5 Satellite, and Field Sample Data in the Perm Region, Russia. Forests 2018, 9, 639. [Google Scholar] [CrossRef]
- Maltamo, M.; Malinen, J.; Kangas, A.; Härkönen, S.; Pasanen, A.M. Most Similar Neighbour-Based Stand Variable Estimation for Use in Inventory by Compartments in Finland. Forestry 2003, 76, 449–463. [Google Scholar] [CrossRef]
- Mauro, F.; Frank, B.; Monleon, V.J.; Temesgen, H.; Ford, K.R. Prediction of Diameter Distributions and Tree-Lists in Southwestern Oregon Using LiDAR and Stand-Level Auxiliary Information. Can. J. For. Res. 2019, 49, 775–787. [Google Scholar] [CrossRef]
- Zhang, Z.; Cao, L.; Mulverhill, C.; Liu, H.; Pang, Y.; Li, Z. Prediction of Diameter Distributions with Multimodal Models Using LiDAR Data in Subtropical Planted Forests. Forests 2019, 10, 125. [Google Scholar] [CrossRef]
- Mulverhill, C.; Coops, N.C.; White, J.C.; Tompalski, P.; Marshall, P.L.; Bailey, T. Enhancing the Estimation of Stem-Size Distributions for Unimodal and Bimodal Stands in a Boreal Mixedwood Forest with Airborne Laser Scanning Data. Forests 2018, 9, 95. [Google Scholar] [CrossRef]
- Saad, R.; Wallerman, J.; Lämås, T. Estimating Stem Diameter Distributions from Airborne Laser Scanning Data and Their Effects on Long Term Forest Management Planning. Scand. J. For. Res. 2015, 30, 186–196. [Google Scholar] [CrossRef]
- Strunk, J.L.; McGaughey, R.J. Stand Validation of Lidar Forest Inventory Modeling for a Managed Southern Pine Forest. Can. J. For. Res. 2023, 53, 1–19. [Google Scholar] [CrossRef]
- Peuhkurinen, J.; Mehtätalo, L.; Maltamo, M. Comparing Individual Tree Detection and the Areabased Statistical Approach for the Retrieval of Forest Stand Characteristics Using Airborne Laser Scanning in Scots Pine Stands. Can. J. For. Res. 2011, 41, 583–598. [Google Scholar] [CrossRef]
- Arias-Rodil, M.; Diéguez-Aranda, U.; Álvarez-González, J.G.; Pérez-Cruzado, C.; Castedo-Dorado, F.; González-Ferreiro, E. Modeling Diameter Distributions in Radiata Pine Plantations in Spain with Existing Countrywide LiDAR Data. Ann. For. Sci. 2018, 75, 36. [Google Scholar] [CrossRef]
- Tomppo, E.; Katila, M. Satellite Image-Based National Forest Inventory of Finland for Publication in the Igarss’91 Digest. In Proceedings of the IGARSS’91 Remote Sensing: Global Monitoring for Earth Management, Espoo, Finland, 3–6 June 1991; Volume 3, pp. 1141–1144. [Google Scholar]
- Maltamo, M.; Kangas, A. Methods Based on K-Nearest Neighbor Regression in the Prediction of Basal Area Diameter Distribution. Can. J. For. Res. 1998, 28, 1107–1115. [Google Scholar] [CrossRef]
- Næsset, E. Predicting Forest Stand Characteristics with Airborne Scanning Laser Using a Practical Two-Stage Procedure and Field Data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Bollandsås, O.M.; Maltamo, M.; Gobakken, T.; Næsset, E. Comparing Parametric and Non-Parametric Modelling of Diameter Distributions on Independent Data Using Airborne Laser Scanning in a Boreal Conifer Forest. Forestry 2013, 86, 493–501. [Google Scholar] [CrossRef]
- Peuhkurinen, J.; Maltamo, M.; Malinen, J. Estimating Species-Specific Diameter Distributions and Saw Log Recoveries of Boreal Forests from Airborne Laser Scanning Data and Aerial Photographs: A Distribution-Based Approach. Silva Fenn. 2008, 42, 625–641. [Google Scholar] [CrossRef]
- Strunk, J.L.; Gould, P.J.; Packalen, P.; Poudel, K.P.; Andersen, H.-E.E.; Temesgen, H. An Examination of Diameter Density Prediction with K-NN and Airborne Lidar. Forests 2017, 8, 444. [Google Scholar] [CrossRef]
- Kangas, A.; Maltamo, M. Percentile Based Basal Area Diameter Distribution Models for Scots Pine, Norway Spruce and Birch Species. Silva Fenn. 2000, 34, 371–380. [Google Scholar] [CrossRef]
- Liu, C.; Beaulieu, J.; Prégent, G.; Zhang, S.Y. Applications and Comparison of Six Methods for Predicting Parameters of the Weibull Function in Unthinned Picea Glauca Plantations. Scand. J. For. Res. 2009, 24, 67–75. [Google Scholar] [CrossRef]
- Borders, B.E.; Souter, R.A.; Bailey, R.L.; Ware, K.D. Percentile-Based Distributions Characterize Forest Stand Tables. For. Sci. 1987, 33, 570–576. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C. Fitting Irregular Diameter Distributions of Forest Stands by Weibull, Modified Weibull, and Mixture Weibull Models. J. For. Res. 2006, 11, 369–372. [Google Scholar] [CrossRef]
- Tarp-Johansen, M.J. Stem Diameter Estimation from Aerial Photographs. Scand. J. For. Res. 2002, 17, 369–376. [Google Scholar] [CrossRef]
- Gobakken, T.; Næsset, E. Estimation of Diameter and Basal Area Distributions in Coniferous Forest by Means of Airborne Laser Scanner Data. Scand. J. For. Res. 2004, 19, 529–542. [Google Scholar] [CrossRef]
- Shang, C.; Treitz, P.; Caspersen, J.; Jones, T. Estimating Stem Diameter Distributions in a Management Context for a Tolerant Hardwood Forest Using ALS Height and Intensity Data. Can. J. Remote Sens. 2017, 43, 79–94. [Google Scholar] [CrossRef]
- Hou, Z.; Xu, Q.; Tokola, T. Use of ALS, Airborne CIR and ALOS AVNIR-2 Data for Estimating Tropical Forest Attributes in Lao PDR. ISPRS J. Photogramm. Remote Sens. 2011, 66, 776–786. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Its’Hak, D. Textural Features for Image Classification. IEEE Trans. Syst. Man. Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Tuceryan, M.; Jain, A.K. Texture Analysis. In Handbook of Pattern Recognition and Computer Vision; Chen, C.H., Pau, L.F., Wang, P.S.P., Eds.; World Scientific: Singapore, 1999; pp. 207–248. [Google Scholar]
- van Ewijk, K.; Treitz, P.; Woods, M.; Jones, T.; Caspersen, J. Forest Site and Type Variability in ALS-Based Forest Resource Inventory Attribute Predictions over Three Ontario Forest Sites. Forests 2019, 10, 226. [Google Scholar] [CrossRef]
- Tuominen, S.; Pekkarinen, A. Performance of Different Spectral and Textural Aerial Photograph Features in Multi-Source Forest Inventory. Remote Sens. Environ. 2005, 94, 256–268. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O. Investigating the Robustness of the New Landsat-8 Operational Land Imager Derived Texture Metrics in Estimating Plantation Forest Aboveground Biomass in Resource Constrained Areas. ISPRS J. Photogramm. Remote Sens. 2015, 108, 12–32. [Google Scholar] [CrossRef]
- Ozdemir, I.; Donoghue, D.N.M. Modelling Tree Size Diversity from Airborne Laser Scanning Using Canopy Height Models with Image Texture Measures. For. Ecol. Manag. 2013, 295, 28–37. [Google Scholar] [CrossRef]
- Niemi, M.T.; Vauhkonen, J. Extracting Canopy Surface Texture from Airborne Laser Scanning Data for the Supervised and Unsupervised Prediction of Area-Based Forest Characteristics. Remote Sens. 2016, 8, 582. [Google Scholar] [CrossRef]
- Rowe, J.S. Forest Regions of Canada. Based on W. E. D. Halliday’s “A Forest Classification for Canada” 1937; Publication No 1300; Department of the Environment, Canadian Forestry Service: Ottawa, ON, Canada, 1972. [Google Scholar]
- Torgo, L. Data Mining with R: Learning with Case Studies, 2nd ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2017; ISBN 978-1482234893. [Google Scholar]
- Ellison, A.M. Effect of Seed Dimorphism on the Density-Dependent Dynamics of Experimental Populations of Atriplex Triangularis (Chenopodiaceae). Am. J. Bot. 1987, 74, 1280–1288. [Google Scholar] [CrossRef]
- Freeman, J.B.; Dale, R. Assessing Bimodality to Detect the Presence of a Dual Cognitive Process. Behav. Res. Methods 2013, 45, 83–97. [Google Scholar] [CrossRef]
- Pfister, R.; Schwarz, K.A.; Janczyk, M.; Dale, R.; Freeman, J. Good Things Peak in Pairs: A Note on the Bimodality Coefficient. Front. Psychol. 2013, 4, 700. [Google Scholar] [CrossRef]
- Roussel, J.-R.; Auty, D. LidR: Airborne LiDAR Data Manipulation and Visualization for Forestry Applications. Available online: https://cran.r-project.org/web/packages/lidR/index.html (accessed on 15 May 2020).
- R Core Team R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 22 May 2020).
- Hall-Beyer, M. Practical Guidelines for Choosing GLCM Textures to Use in Landscape Classification Tasks over a Range of Moderate Spatial Scales. Int. J. Remote Sens. 2017, 38, 1312–1338. [Google Scholar] [CrossRef]
- Bouvier, M.; Durrieu, S.; Fournier, R.A.; Renaud, J.-P.P. Generalizing Predictive Models of Forest Inventory Attributes Using an Area-Based Approach with Airborne LiDAR Data. Remote Sens. Environ. 2015, 156, 322–334. [Google Scholar] [CrossRef]
- Peduzzi, A.; Wynne, R.H.; Fox, T.R.; Nelson, R.F.; Thomas, V.A. Estimating Leaf Area Index in Intensively Managed Pine Plantations Using Airborne Laser Scanner Data. For. Ecol. Manag. 2012, 270, 54–65. [Google Scholar] [CrossRef]
- Pope, G.; Treitz, P. Leaf Area Index (LAI) Estimation in Boreal Mixedwood Forest of Ontario, Canada Using Light Detection and Ranging (LiDAR) and Worldview-2 Imagery. Remote Sens. 2013, 5, 5040–5063. [Google Scholar] [CrossRef]
- Goetz, S.; Steinberg, D.; Dubayah, R.; Blair, B. Laser Remote Sensing of Canopy Habitat Heterogeneity as a Predictor of Bird Species Richness in an Eastern Temperate Forest, USA. Remote Sens. Environ. 2007, 108, 254–263. [Google Scholar] [CrossRef]
- van Ewijk, K.Y.; Treitz, P.M.; Scott, N.A. Characterizing Forest Succession in Central Ontario Using Lidar-Derived Indices. Photogramm. Eng. Remote Sens. 2011, 77, 261–269. [Google Scholar] [CrossRef]
- Pretzsch, H. Description and Analysis of Stand Structures. In Forest Dynamics, Growth and Yield; Springer: Berlin/Heidelberg, Germany, 2010; pp. 223–289. ISBN 9783540883067. [Google Scholar]
- Jenness, J.S. Calculating Landscape Surface Area from Digital Elevation Models. Wildl. Soc. Bull. 2004, 32, 829–839. [Google Scholar] [CrossRef]
- Woods, M.; Pitt, D.; Penner, M.; Lim, K.; Nesbitt, D.; Etheridge, D.; Treitz, P. Operational Implementation of a LiDAR Inventory in Boreal Ontario. For. Chron. 2011, 87, 512–528. [Google Scholar] [CrossRef]
- Beets, P.N.; Reutebuch, S.; Kimberley, M.O.; Oliver, G.R.; Pearce, S.H.; McGaughey, R.J. Leaf Area Index, Biomass Carbon and Growth Rate of Radiata Pine Genetic Types and Relationships with LiDAR. Forests 2011, 2, 637–659. [Google Scholar] [CrossRef]
- Solberg, S.; Brunner, A.; Hanssen, K.H.; Lange, H.; Næsset, E.; Rautiainen, M.; Stenberg, P. Mapping LAI in a Norway Spruce Forest Using Airborne Laser Scanning. Remote Sens. Environ. 2009, 113, 2317–2327. [Google Scholar] [CrossRef]
- Hopkinson, C.; Chasmer, L. Testing LiDAR Models of Fractional Cover across Multiple Forest Ecozones. Remote Sens. Environ. 2009, 113, 275–288. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and Variable Selection via the Elastic Net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. Kernlab—An S4 Package for Kernel Methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. The Elements of Statistical Learning: Data Mining Inference and Prediction, 2nd ed.; Springer Series in Statistics New York; Springer: New York, NY, USA, 2001; Volume 1, ISBN 978-0-387-21606-5. [Google Scholar]
- Delignette-Muller, M.L.; Dutang, C. Fitdistrplus: An R Package for Fitting Distributions. J. Stat. Softw. 2015, 64, 1–34. [Google Scholar] [CrossRef]
- Yu, Y. MixR: An R Package for Finite Mixture Modeling for Both Raw and Binned Data. J. Open Source Softw. 2022, 7, 4031. [Google Scholar] [CrossRef]
- Furnival, G.M.; Wilson, R.W. Regressions by Leaps and Bounds. Technometrics 1974, 16, 499–511. [Google Scholar] [CrossRef]
- Lumley, T. Leaps: Regression Subset Selection, Based on Fortran Code by Alan Miller, R Package Version 3.1. Available online: https://cran.r-project.org/web/packages/leaps/leaps.pdf (accessed on 18 May 2020).
- Land, A.H.; Doig, A.G. An Automatic Method of Solving Discrete Programming Problems. Econometrica 1960, 28, 497–520. [Google Scholar] [CrossRef]
- Oliveira, S.; Oehler, F.; San-Miguel-Ayanz, J.; Camia, A.; Pereira, J.M.C. Modeling Spatial Patterns of Fire Occurrence in Mediterranean Europe Using Multiple Regression and Random Forest. For. Ecol. Manag. 2012, 275, 117–129. [Google Scholar] [CrossRef]
- Reynolds, M.R.; Burk, T.E.; Huang, W.-C. Goodness-of-Fit Tests and Model Selection Procedures for Diameter Distribution Models. For. Sci. 1988, 34, 373–399. [Google Scholar] [CrossRef]
- Coomes, D.A.; Duncan, R.P.; Allen, R.B.; Truscott, J. Disturbances Prevent Stem Size-Density Distributions in Natural Forests from Following Scaling Relationships. Ecol. Lett. 2003, 6, 980–989. [Google Scholar] [CrossRef]
- Pippuri, I.; Kallio, E.; Maltamo, M.; Peltola, H.; Packalén, P. Exploring Horizontal Area-Based Metrics to Discriminate the Spatial Pattern of Trees and Need for First Thinning Using Airborne Laser Scanning. Forestry 2012, 85, 305–314. [Google Scholar] [CrossRef]


| Group | Metric | Units | Description |
|---|---|---|---|
| Mals | MAX | m | Maximum height |
| MEAN | m | Mean height [68] | |
| P25, P75, P90 | m | Height percentiles. E.g., P25 is the height of the 25th percentile. [69] | |
| SKEW | Skewness | ||
| VAR | Variance [68] | ||
| COVAR | % | Coefficient of variation: standard deviation/mean [70] | |
| VDR | Vertical Distribution Ration: (MAX-MEAN)/MAX [71] | ||
| VCI | Vertical Complexity Index [72] | ||
| ENT | Entropy: normalized Shannon diversity index [73] | ||
| RI | Rumple Index of roughness [74] | ||
| D2, D5, D8 | % | Proportion of all vegetation returns found in sections divided within the range of heights of all returns for each plot. [75] | |
| COVER | Ratio of the number of vegetated returns above 2 m to the total number of ground and vegetated returns [76] | ||
| LPI | Light Penetration Index, Ground returns/(Ground returns + Canopy returns). [69] | ||
| LPI1st | Light Penetration Index (first returns): Ground first returns/(Ground returns + Canopy returns) [77] | ||
| FR | First return ratio: number of first return heights below a specified height threshold/total number of first return heights [68] | ||
| RR | All return ratio: all returns < 2 m/all returns [78] | ||
| LAI | Sum of Leaf Area Density [68] | ||
| cvLAI | Coefficient of variation of Leaf Area Density [68] | ||
| Mtex | CON | Contrast (edge texture) [67] | |
| COR | Correlation (interior textures) [67] | ||
| DIS | Dissimilarity (edge textures) [67] | ||
| HOM | Homogeneity (interior textures) [67] | ||
| MEAN | Mean (interior textures) [67] | ||
| Mcomb | Combination of all metrics (Mals and Mtex) | ||
| ALS Metric Set | RF | SVM | Logit | GLMNET |
|---|---|---|---|---|
| Model development | ||||
| Mals | 74 | 72 | 71 | 74 |
| Mtex | 73 | 72 | 68 | 68 |
| Mcomb | 74 | 71 | 70 | 74 |
| Test case | ||||
| Mals | 72 | 73 | 72 | 71 |
| Mtex | 66 | 72 | 74 | 73 |
| Mcomb | 71 | 71 | 72 | 71 |
| Model Set | |||
|---|---|---|---|
| Plot Dataset | n | Differentiated | Undifferentiated |
| Model development | |||
| Differentiated as unimodal | 215 | 50.4 | 50.3 |
| Differentiated as bimodal | 92 | 65 | 74 |
| Undifferentiated modality | 307 | 54.8 | 57.4 |
| Test case | |||
| Differentiated as unimodal | 88 | 50.8 | 50.5 |
| Differentiated as bimodal | 32 | 59.1 | 67 |
| Undifferentiated modality | 120 | 53 | 54.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallagher-Duval, X.; van Lier, O.R.; Fournier, R.A. Estimating Stem Diameter Distributions with Airborne Laser Scanning Metrics and Derived Canopy Surface Texture Metrics. Forests 2023, 14, 287. https://doi.org/10.3390/f14020287
Gallagher-Duval X, van Lier OR, Fournier RA. Estimating Stem Diameter Distributions with Airborne Laser Scanning Metrics and Derived Canopy Surface Texture Metrics. Forests. 2023; 14(2):287. https://doi.org/10.3390/f14020287
Chicago/Turabian StyleGallagher-Duval, Xavier, Olivier R. van Lier, and Richard A. Fournier. 2023. "Estimating Stem Diameter Distributions with Airborne Laser Scanning Metrics and Derived Canopy Surface Texture Metrics" Forests 14, no. 2: 287. https://doi.org/10.3390/f14020287
APA StyleGallagher-Duval, X., van Lier, O. R., & Fournier, R. A. (2023). Estimating Stem Diameter Distributions with Airborne Laser Scanning Metrics and Derived Canopy Surface Texture Metrics. Forests, 14(2), 287. https://doi.org/10.3390/f14020287







