Bioactive Compounds Concentrations and Stability in Leaves of Ilex paraguariensis Genotypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Sample Preparation
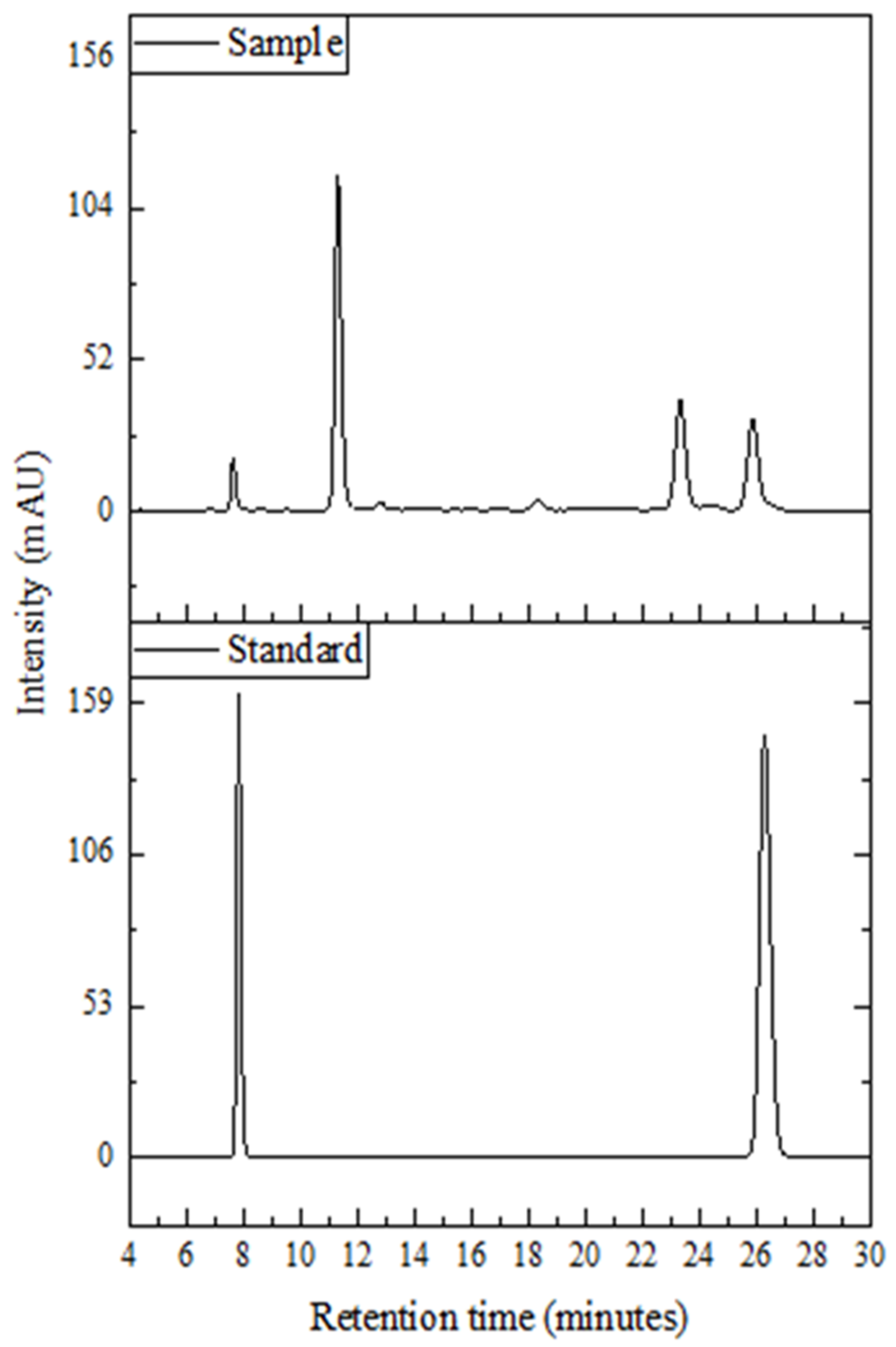
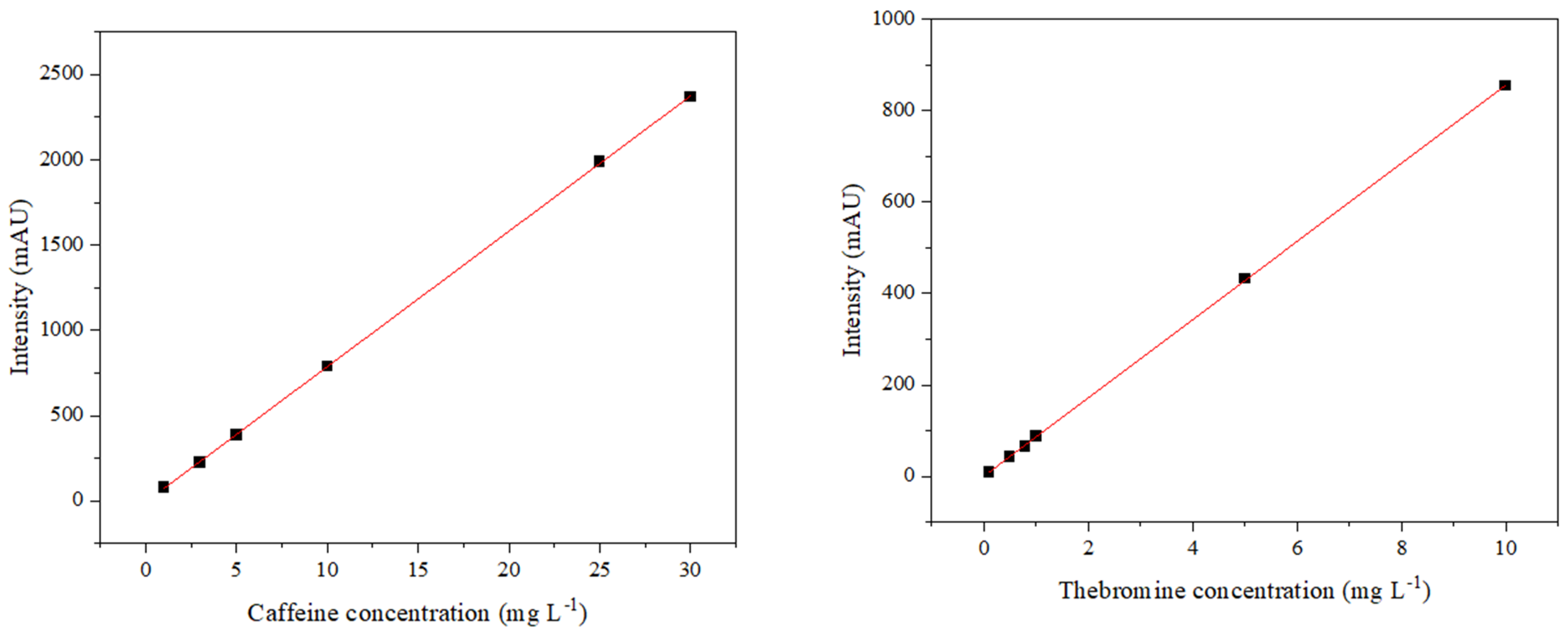
2.2. Determination of Methylxanthines
2.3. Determination of Total Phenolic Compounds
2.4. Determination of Moisture and Proteins
2.5. Statistical Analysis
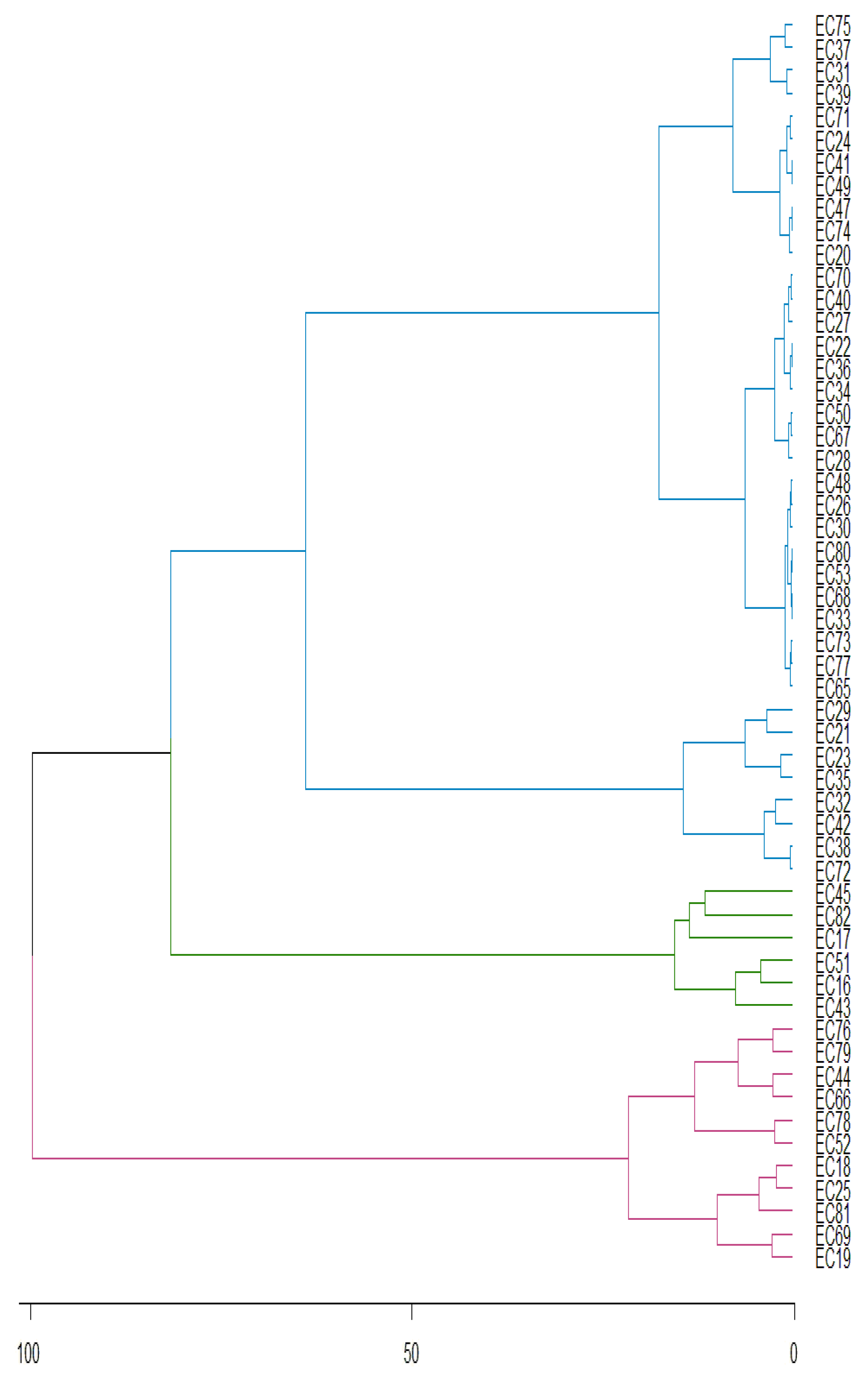
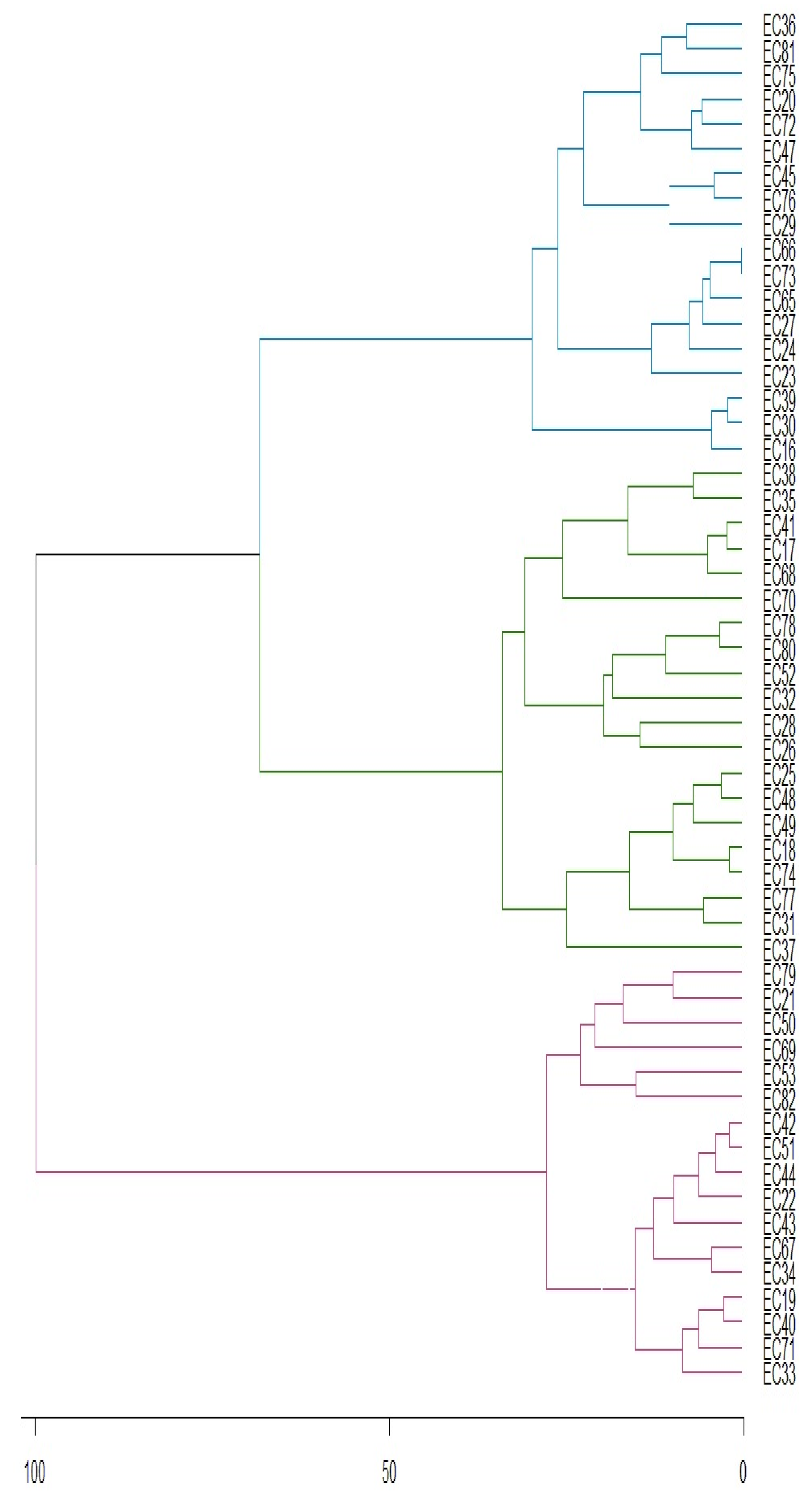
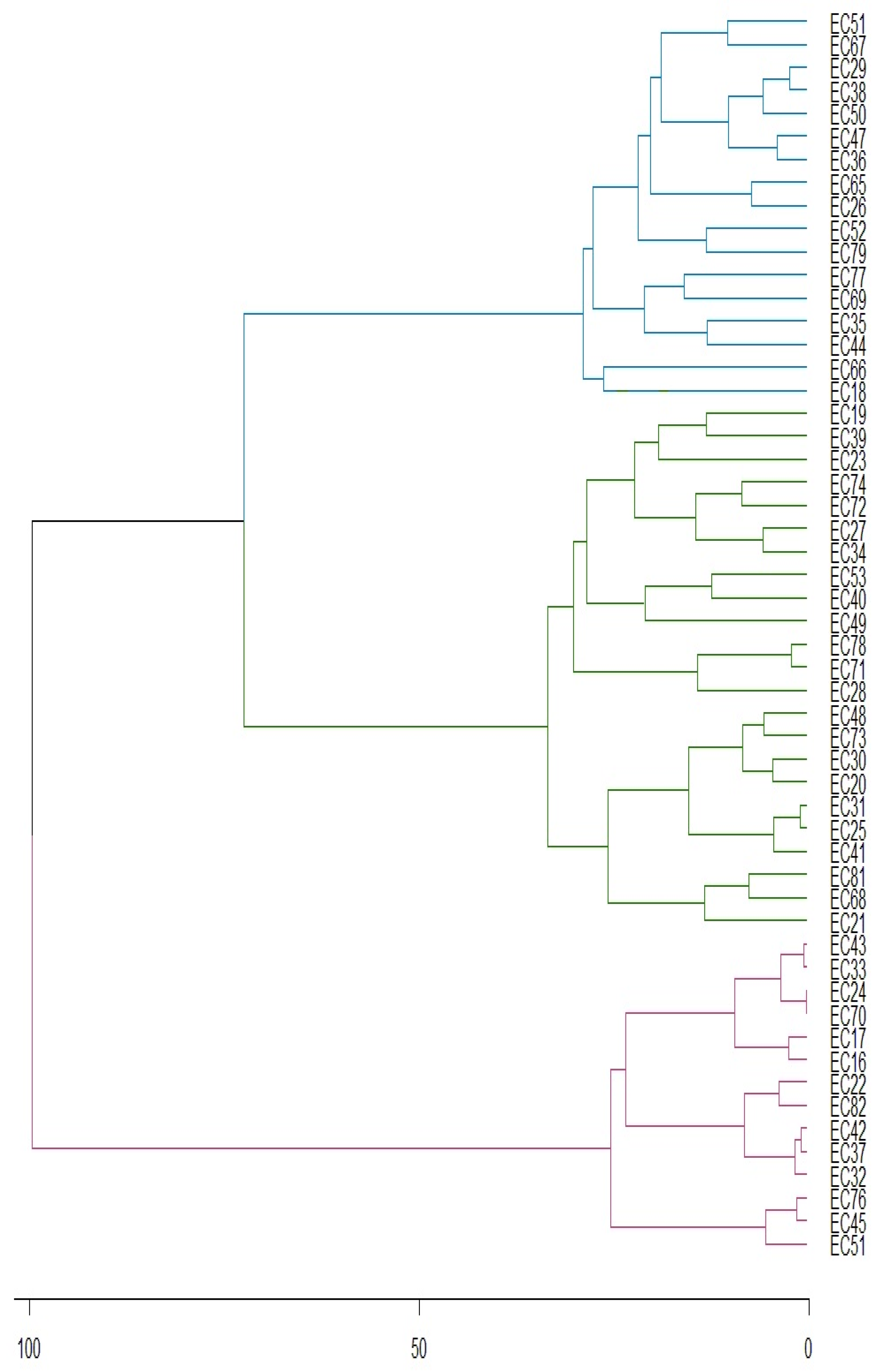
3. Results
3.1. Caffeine
3.2. Theobromine
3.3. Total Phenolic Compounds
3.4. Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heck, C.I.; de Mejia, E.G. Yerba mate tea (Ilex paraguariensis): A comprehensive review on chemistry, health implications, and technological considerations. J. Food Sci. 2007, 72, 138–151. [Google Scholar] [CrossRef]
- Gan, R.Y.; Zhang, D.; Wang, M.; Corke, H. Health benefits of bioactive compounds from the genus ilex, a source of traditional caffeinated beverages. Nutrients 2018, 10, 1682. [Google Scholar] [CrossRef]
- De Godoy, R.C.B.; Chambers, E.; Yang, G. Development of a preliminary sensory lexicon for mate tea. J. Sens. Stud. 2020, 35, e12570. [Google Scholar] [CrossRef]
- Cardozo Junior, E.L.; Morand, C. Interest of mate (Ilex paraguariensis A. St.-Hil.) as a new natural functional food to preserve human cardiovascular health—A review. J. Funct. Foods 2016, 21, 440–454. [Google Scholar] [CrossRef]
- Wendling, I.; Sturion, J.A.; Stuepp, C.A.; Reis, C.A.F.; Ramalho, M.A.P.; Resende, M.D.V.D. Early selection and classification of yerba mate progenies. Pesq. Agropecu. Bras. 2018, 53, 279–286. [Google Scholar] [CrossRef]
- Sturion, J.Á.; De Resende, M.D.V. Melhoramento Genético da Erva-Mate, 1st ed.; Pesquisa Agropecuária Brasileira (Embrapa Florestas): Colombo, Brasil, 2010; 274p. [Google Scholar]
- Anesini, C.; Turner, S.; Cogoi, L.; Filip, R. Study of the participation of caffeine and polyphenols on the overall antioxidant activity of mate (Ilex paraguariensis). Food Sci. Technol. 2012, 45, 299–304. [Google Scholar] [CrossRef]
- Debat, H.J.; Grabiele, M.; Aguilera, P.M.; Bubillo, R.E.; Otegui, M.B.; Ducasse, D.A.; Marti, D.A. Exploring the genes of yerba mate (Ilex paraguariensis A. St.-Hil.) by N.G.S and de novo transcriptome assembly. PLoS ONE 2014, 9, e109835. [Google Scholar] [CrossRef] [PubMed]
- Arbeláez, L.F.G.; Fantinelli, J.C.; Pardo, A.C.; Caldiz, C.I.; Ríos, J.L.; Schinella, G.R.; Mosca, S.M. Effect of an Ilex paraguariensis (yerba mate) extract on infarct size in isolated rat hearts: The mechanisms involved. Food Funct. 2016, 7, 816–824. [Google Scholar] [CrossRef]
- De Lima, G.G.; Ruiz, H.Z.; Matos, M.; Helm, C.V.; de Liz, M.V.; Magalhães, W.L.E. Prediction of yerba mate caffeine content using near infrared spectroscopy. Spectrosc. Lett. 2019, 52, 282–287. [Google Scholar] [CrossRef]
- Oellig, C.; Schunck, J.; Schwack, W. Determination of caffeine, theobromine and theophylline in Mate beer and Mate soft drinks by high-performance thin-layer chromatography. J. Chromatogr. 2018, A1533, 208–212. [Google Scholar] [CrossRef]
- Poswal, F.S.; Russell, G.; Mackonochie, M.; MacLennan, E.; Adukwu, E.C.; Rolfe, V. Herbal Teas and their Health Benefits: A Scoping Review. Plant Foods Hum. Nutr. 2019, 74, 266–276. [Google Scholar] [CrossRef]
- Gullón, B.; Eibes, G.; Moreira, M.T.; Herrera, R.; Labidi, J.; Gullon, P. Yerba mate waste: A sustainable resource of antioxidant compounds. Ind. Crops Prod. 2018, 113, 398–405. [Google Scholar] [CrossRef]
- Butiuk, A.P.; Martos, M.A.; Adachi, O.; Hours, R.A. Study of the chlorogenic acid content in yerba mate (Ilex paraguariensis St. Hil.): Effect of plant fraction, processing step and harvesting season. J. Appl. Res. Med. Aromat. Plants 2016, 3, 27–33. [Google Scholar] [CrossRef]
- Gil, M.; Wianowska, D. Chlorogenic acids—Their properties, occurrence and analysis. Ann. Univ. Mariae Curie-Sklodowska Sect. AA Chem. 2017, 72, 61. [Google Scholar] [CrossRef]
- Kaltbach, P.; Ballert, S.; Kabrodt, K.; Schellenberg, I. New HPTLC methods for analysis of major bioactive compounds in mate (Ilex paraguariensis) tea. J. Food Compos. Anal. 2020, 92, 103568. [Google Scholar] [CrossRef]
- Alvares, A.A.; Stape, J.L.; Sentelhas, P.C.; Gonçalves, J.L.M.; Sparovek, G. Köppen’s climate classification map for Brazil. Meteorol. Z. 2014, 22, 711–728. [Google Scholar] [CrossRef]
- EMBRAPA. Sistema Brasileiro de Classificação de Solos, 3rd ed.; Embrapa Solos Rio de Janeiro: Brasília, DF, Brazil, 2013; 306p. [Google Scholar]
- Duarte, M.M.; de Cássia Tomasi, J.; Helm, C.V.; Amano, E.; Lazzarotto, M.; de Godoy, R.C.B.; Nogueira, A.C.; Wendling, I. Caffeinated and decaffeinated mate tea: Effect of toasting on bioactive compounds and consumer acceptance. Rev. Bras. Cienc. Agrar. 2020, 15, 1–10. [Google Scholar] [CrossRef]
- Helm, C.V.; Ruiz, H.Z.; Hansel, F.A.; Stuepp, C.A.; Wendling, I. Efeito do Solvente na Extração de Teobromina e Cafeína em Progênies de Erva-Mate. Embrapa Florestas. 2015. Available online: https://www.embrapa.br/busca-de-publicacoes/-/publicacao/1038782/efeito-do-solvente-na-extracao-de-teobromina-e-cafeina-em-progenies-de-erva-mate (accessed on 26 October 2023).
- Rakocevic, M.; Maia, A.H.N.; Duarte, M.M.; Wendling, I. Secondary sexual dimorphism in biomass production of Ilex paraguariensis progenies associated to their provenances and morphotypes. Exp. Agric. 2023, 59, e3. [Google Scholar] [CrossRef]
- ANVISA—Agência Nacional de Vigilância Sanitária. Resolução da Diretoria Colegiada—RDC nº166 de 24 de Julho de 2017—Dispõe Sobre a Validação de Métodos Analíticos e dá Outras Providências. 2017; pp. 1–21. Available online: http://antigo.anvisa.gov.br/documents/10181/2721567/RDC_166_2017_COMP.pdf/d5fb92b3 (accessed on 6 November 2023).
- Magnusson, B.; Örnemark, U. Eurachem Guide: The Fitness for Purpose of Analytical Methods—A Laboratory Guide to Method Validation and Related Topics, 2nd ed. 2014. Available online: https://www.eurachem.org/images/stories/Guides/pdf/MV_guide_2nd_ed_EN.pdf (accessed on 9 November 2016).
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of AOAC International, 20th ed.; AOAC International: Rockville, MD, USA, 2016. [Google Scholar]
- Annicchiarico, P. Efeitos principais aditivos e análise de interação multiplicativa (AMMI) da interação genótipo-localização em ensaios de variedades repetidos ao longo dos anos. Theor. Appl. Genet. 1997, 94, 1072–1077. [Google Scholar] [CrossRef]
- Gabriel, K.R. Biometrika Trust The Biplot Graphic Display of Matrices with Application to Principal Component Analysis. Biometrika 1971, 58, 453–467. [Google Scholar] [CrossRef]
- Duarte, J.B.; Vencovsky, R. Interação genótipo x ambiente: Uma introdução à análise “AMMI”. Soc. Bras. Genét. 1999, 199, 60. [Google Scholar]
- Kassambara, A. Practical Guide to Cluster Analysis in R, 1st ed.; STHDA: Marseille, France, 2017. [Google Scholar]
- Cornelius, P.L.; Seyedsadr, M.; Crossa, J. Using the shifted multiplicative model to search for “separability” in crop cultivar trials. Theor. Appl. Genet. 1992, 84, 161–172. [Google Scholar] [CrossRef]
- Brasil (2005). Resolução de Diretoria Colegiada-RDC No. 277, de 22 de Setembro de 2005. Available online: https://bvsms.saude.gov.br/bvs/saudelegis/anvisa/2005/res0277_22_09_2005.html (accessed on 26 November 2023).
- Schuhli, G.S.; Penteado Junior, J.F.; Wendling, I. Descritores Mínimos em Cultivares de Espécies Florestais: Uma Contribuição para Erva-Mate; Embrapa Florestas. Documentos, 333; Embrapa Florestas: Colombo, Brazil, 2019; 22p, Available online: http://www.infoteca.cnptia.embrapa.br/infoteca/handle/doc/1117327 (accessed on 26 October 2023).
- Heckman, M.A.; Weil, J.; de Mejia, E.G. Caffeine (1, 3, 7-trimethylxanthine) in foods: A comprehensive review on consumption, functionality, safety, and regulatory matters. J. Food Sci. 2010, 75, 77–87. [Google Scholar] [CrossRef]
- Athayde, M.L.; Coelho, G.C.; Schenkel, P. Caffeine and theobromine in epicuticular wax of Ilex paraguariensis A. St.-Hil. Phytochemistry 2000, 55, 853–857. [Google Scholar] [CrossRef]
- Scherer, R.; Urfer, P.; Mayol, M.R.; Belingheri, L.D.; Marx, F.; Janssens, M.J.J. Inheritance studies of caffeine and theobromine content of Mate (Ilex paraguariensis) in Misiones, Argentina. Euphytica 2002, 126, 203–210. [Google Scholar] [CrossRef]
- Mohanpuria, P.; Kumar, V.; Yadav, S.K. Tea Caffeine Metabolism, Functions, and Reduction Strategies. Food Sci. Biotechnol. 2010, 19, 275–287. [Google Scholar] [CrossRef]
- Cardozo, E.L.; Ferrarese-Filho, O.; Filho, L.C.; Ferrarese, M.D.L.L.; Donaduzzi, C.M.; Sturion, J.A. Methylxanthines and phenolic compounds in mate (Ilex paraguariensis St. Hil.) progenies grown in Brazil. J. Food Compos. Anal. 2007, 20, 553–558. [Google Scholar] [CrossRef]
- Tfouni, S.A.V.; Camara, M.M.; Kamikata, K.; Gomes, F.M.L.; Furlani, R.P.Z. Caffeine in teas: Levels, transference to infusion and estimated intake. Food Sci. Technol. 2018, 38, 661–666. [Google Scholar] [CrossRef]
- Nakamura, K.L.; Cardozo Junior, L.; Donaduzzi, C.M.; Schuster, I. Genetic variation of phytochemical compounds in progenies of Ilex paraguariensis St. Hil. Crop Breed. Appl. Biotechnol. 2009, 9, 116–123. [Google Scholar] [CrossRef]
- Cardozo Junior, E.L.; Donaduzzi, M.C.; Ferrarese-Filho, O.; Friedrich, J.C.; Gonela, A.; Sturion, J.A. Quantitative genetic analysis of methylxanthines and phenolic compounds in mate progenies. Pesq. Agropecu. Bras. 2010, 45, 171–177. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, L.; Lu, M.; Zhang, J.; Han, J.; Deng, W.W.; Zhang, Z.Z. Caffeine Content and Related Gene Expression: Novel Insight into Caffeine Metabolism in Camellia Plants Containing Low, Normal, and High Caffeine Concentrations. J. Agric. Food Chem. 2019, 67, 3400–3411. [Google Scholar] [CrossRef] [PubMed]
- Ashihara, H.; Monteiro, A.M.; Gillies, F.M.; Crozier, A. Biosynthesis of Caffeine in Leaves of Coffee. Plant Physiol. 1996, 111, 747–753. [Google Scholar] [CrossRef]
- Da Croce, D.M. Características físico-químicas de extratos de erva-mate (Ilex paraguariensis St. Hil) no estado de Santa Catarina. Ciênc. Florest. 2002, 12, 107–113. [Google Scholar] [CrossRef][Green Version]
- Gugliucci, A.; Bastos, D.H.M. Chlorogenic acid protects paraoxonase 1 activity in high density lipoprotein from inactivation caused by physiological concentrations of hypochlorite. Fitoterapia 2009, 80, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Valerga, J.; Reta, M.; Lanari, M.C. Polyphenol input to the antioxidant activity of yerba mate (Ilex paraguariensis) extracts. Food Sci. Technol. 2012, 45, 28–35. [Google Scholar] [CrossRef]
- Sturion, J.Á.; Correa, G.; Resende, M.D.V.; Cardozo Junior, E.L.; Donaduzzi, C.M. Controle Genético dos Teores de Polifenóis Totais, Taninos e Cafeína em Progênies de Erva-Mate (Ilex paraguariensis St. Hil.) Cultivadas em Três Classes de Solos. Boletim de Pesquisa e Desenvolvimento (Embrapa Florestas). 2004. Available online: https://www.embrapa.br/busca-de-publicacoes/-/publicacao/287300/controle-genetico-dos-teores-de-polifenois-totais-taninos-e-cafeina-em-progenies-de-erva-mate-ilex-paraguariensis-st-hil-cultivadas-em-tres-classes-de-solos (accessed on 26 November 2023).
- Da Silveira, T.F.F.; Meinhart, A.D.; de Souza, T.C.L.L.; Teixeira Filho, J.; Godoy, H.T. Phenolic compounds from yerba mate based beverages—A multivariate optimization. Food Chem. 2016, 190, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Berté, K.A.S.; Beux, M.R.; Spada, P.K.; Salvador, M.; Hoffmann-Ribani, R. Chemical composition and antioxidant activity of yerba-mate (Ilex paraguariensis A.St.-Hil., Aquifoliaceae) extract as obtained by spray drying. J. Agric. Food Chem. 2011, 59, 5523–5527. [Google Scholar] [CrossRef]
- Frizon, C.; Perussello, C.; Sturion, J.; Hoffmann-Ribani, R. Novel Beverages of Yerba-Mate and Soy: Bioactive Compounds and Functional Properties. Beverages 2018, 4, 21. [Google Scholar] [CrossRef]
- Vieira, L.M.; de Almeida Maggioni, R.; de Cássia Tomasi, J.; Gomes, E.M.; Wendling, I.; Helm, C.V.; Zuffellato-Ribas, K.C. Vegetative propagation, chemical composition and antioxidant activity of yerba mate genotypes. Plant Genet. Resour. 2021, 19, 112–121. [Google Scholar] [CrossRef]
- Rakocevic, M.; Maia, A.D.H.N.; de Liz, M.V.; Imoski, R.; Helm, C.V.; Cardozo Junior, E.L.; Wendling, I. Stability of Leaf Yerba Mate (Ilex paraguariensis) Metabolite Concentrations over the Time from the Prism of Secondary Sexual Dimorphism. Plants 2023, 12, 2199. [Google Scholar] [CrossRef] [PubMed]

| Variation sources | GL | SQ | F (Pr > F) | ||||
| Model | 56 | 119.84 | |||||
| Genotype (G) | (53) | (119.03) | 37.87 (<0.0001) | ||||
| Year (A) | (3) | (0.81) | 4.57 (0.0042) | ||||
| Axes | High Value | Variance (%) | Explained Cumulative | GxA | 159 | 9.43 | |
| 1 | 5.50 | 58.35 | 58.35 | IPCA1 | (55) | (5.50) | 3.04 (<0.0001) |
| 2 | 2.14 | 22.74 | 81.10 | IPCA2 | (53) | (2.15) | 1.23 (0.1491) |
| 3 | 1.78 | 18.90 | 100.00 | IPCA3 | (51) | (1.78) | 1.06 (0.3731) |
| Mean Error | 290 | 9.57 | |||||
| Adjusted Total | 215 | 129.27 | |||||
| Genotypes | Means | Chi-Square | Pr > ChiSq | Genotypes | Means | Chi-Square | Pr > ChiSq |
|---|---|---|---|---|---|---|---|
| EC16 | 0.0396 | 0.17 | 0.6780 | EC44 | 0.0413 | 0.05 | 0.8203 |
| EC17 | 0.0542 | 0.79 | 0.3752 | EC16 | 0.0396 | 0.17 | 0.6780 |
| EC18 | 0.0674 | 2.71 | 0.0996 | EC44 | 0.0413 | 0.79 | 0.3752 |
| EC19 | 0.0348 | 1.32 | 0.2501 | EC45 | 2.3290 | 30.50 | <0.0001 |
| EC20 | 0.5961 | 27.19 | <0.0001 | EC47 | 0.1208 | 11.35 | 0.0008 |
| EC21 | 0.4373 | 25.61 | <0.0001 | EC48 | 1.7543 | 30.13 | <0.0001 |
| EC22 | 0.0374 | 0.43 | 0.5128 | EC49 | 2.2004 | 30.44 | <0.0001 |
| EC23 | 0.3752 | 24.52 | <0.0001 | EC50 | 0.0605 | 1.70 | 0.1925 |
| EC24 | 1.0741 | 29.17 | <0.0001 | EC51 | 0.0839 | 5.47 | 0.0193 |
| EC25 | 1.5091 | 29.88 | <0.0001 | EC52 | 0.0889 | 6.80 | 0.0091 |
| EC26 | 1.5221 | 29.90 | <0.0001 | EC53 | 2.3579 | 30.52 | <0.0001 |
| EC27 | 1.5769 | 29.96 | <0.0001 | EC65 | 0.3324 | 23.33 | <0.0001 |
| EC28 | 1.0096 | 27.96 | <0.0001 | EC66 | 0.4116 | 25.15 | <0.0001 |
| EC29 | 0.0850 | 5.89 | 0.0152 | EC67 | 0.0479 | 0.15 | 0.7018 |
| EC30 | 0.0374 | 0.38 | 0.5362 | EC68 | 0.7601 | 28.15 | <0.0001 |
| EC31 | 1.1940 | 29.42 | <0.0001 | EC69 | 0.8194 | 28.29 | <0.0001 |
| EC32 | 0.8286 | 28.44 | <0.0001 | EC70 | 1.5931 | 29.98 | <0.0001 |
| EC33 | 1.0823 | 29.19 | <0.0001 | EC71 | 2.1910 | 30.45 | <0.0001 |
| EC34 | 1.1340 | 29.30 | <0.0001 | EC72 | 1.2243 | 29.47 | <0.0001 |
| EC35 | 0.2751 | 22.21 | <0.0001 | EC73 | 1.6523 | 30.04 | <0.0001 |
| EC36 | 1.1766 | 29.38 | <0.0001 | EC74 | 1.1718 | 29.01 | <0.0001 |
| EC37 | 1.9442 | 30.28 | <0.0001 | EC76 | 0.0906 | 7.09 | 0.0078 |
| EC38 | 1.6107 | 29.99 | <0.0001 | EC77 | 0.6200 | 27.19 | <0.0001 |
| EC39 | 0.7686 | 28.19 | <0.0001 | EC78 | 0.0799 | 5.00 | 0.0253 |
| EC40 | 1.7908 | 30.16 | <0.0001 | EC79 | 2.3846 | 30.51 | <0.0001 |
| EC41 | 1.4447 | 29.80 | <0.0001 | EC80 | 0.7048 | 27.29 | <0.0001 |
| EC42 | 1.1004 | 29.23 | <0.0001 | EC81 | 0.0576 | 1.18 | 0.2770 |
| EC43 | 1.3957 | 29.74 | <0.0001 | EC82 | 0.0438 | 2.80 | 0.0946 |
| Variation sources | GL | SQ | F (Pr > F) | ||||
| Model | 57 | 19.86 | |||||
| Genotype (G) | (54) | (19.38) | 12.71 (<0.0001) | ||||
| Year (A) | (3) | (0.48) | 5.70 (0.0010) | ||||
| Axes | High Value | Variance (%) | Explained Cumulative | GxA | 162 | 4.57 | |
| 1 | 2.73 | 59.78 | 59.78 | IPCA1 | (56) | (2.73) | 3.11 (<0.0001) |
| 2 | 1.25 | 27.35 | 87.13 | IPCA2 | (54) | (1.25) | 1.47 (0.0238) |
| 3 | 0.59 | 12.87 | 100.00 | IPCA3 | (52) | (0.59) | 0.72 (0.9237) |
| Mean Error | 295 | 4.63 | |||||
| Adjusted Total | 219 | 24.43 | |||||
| Genotypes | Means | Chi-Square | Pr > ChiSq | Genotypes | Means | Chi-Square | Pr > ChiSq |
|---|---|---|---|---|---|---|---|
| EC16 | 0.3026 | 3.01 | 0.0829 | EC44 | 0.4760 | 5.52 | 0.0188 |
| EC17 | 0.2586 | 2.41 | 0.1204 | EC45 | 0.0161 | 0.26 | 0.6073 |
| EC18 | 0.4678 | 5.00 | 0.0253 | EC47 | 0.0627 | 0.48 | 0.4885 |
| EC19 | 0.3431 | 3.62 | 0.0570 | EC48 | 0.0076 | 2.37 | 0.1240 |
| EC20 | 0.0447 | 5.73 | 0.0167 | EC49 | 0.0898 | 1.44 | 0.2300 |
| EC21 | 0.0862 | 0.13 | 0.7216 | EC50 | 0.0543 | 0.35 | 0.5519 |
| EC22 | 0.0294 | 0.66 | 0.4177 | EC51 | 0.1211 | 4.58 | 0.0323 |
| EC23 | 0.0688 | 5.76 | 0.0164 | EC52 | 0.6187 | 6.47 | 0.0110 |
| EC24 | 0.1058 | 6.02 | 0.0141 | EC53 | 0.0173 | 0.00 | 0.9627 |
| EC25 | 0.3101 | 0.02 | 0.8859 | EC65 | 0.0237 | 1.81 | 0.1786 |
| EC26 | 0.0200 | 1.61 | 0.2045 | EC66 | 0.5809 | 3.59 | 0.0582 |
| EC27 | 0.0303 | 0.09 | 0.7673 | EC67 | 0.0045 | 4.99 | 0.0255 |
| EC28 | 0.0112 | 3.38 | 0.0661 | EC68 | 0.1182 | 8.03 | 0.0046 |
| EC29 | 0.0373 | 2.14 | 0.1431 | EC69 | 0.2487 | 1.92 | 0.1657 |
| EC30 | 0.0004 | 7.20 | 0.0073 | EC70 | 0.0162 | 6.18 | 0.0129 |
| EC31 | 1.7719 | 5.65 | 0.0174 | EC71 | 0.1529 | 5.22 | 0.0224 |
| EC32 | 0.0493 | 6.62 | 0.0101 | EC72 | 0.0796 | 2.19 | 0.1390 |
| EC33 | 0.0081 | 4.61 | 0.0317 | EC73 | 0.0581 | 0.47 | 0.4911 |
| EC34 | 0.0739 | 0.00 | 1.0000 | EC74 | 0.1612 | 1.15 | 0.2838 |
| EC35 | 0.0199 | 7.56 | 0.0060 | EC75 | 0.4280 | 0.01 | 0.9351 |
| EC36 | 0.0230 | 3.14 | 0.0766 | EC76 | 0.3440 | 5.87 | 0.0154 |
| EC37 | 0.5567 | 6.63 | 0.0101 | EC77 | 0.0090 | 3.85 | 0.0499 |
| EC38 | 0.0997 | 0.81 | 0.3695 | EC78 | 0.5312 | 5.58 | 0.0181 |
| EC39 | 0.1649 | 1.28 | 0.2572 | EC79 | 0.5640 | 0.29 | 0.5904 |
| EC40 | 0.0709 | 0.24 | 0.6236 | EC80 | 0.1005 | 3.04 | 0.0814 |
| EC41 | 0.9768 | 5.01 | 0.0251 | EC81 | 0.2100 | 1.31 | 0.2528 |
| EC42 | 0.1173 | 6.80 | 0.0091 | EC82 | 0.1230 | 4.58 | 0.0323 |
| EC43 | 0.0602 | 0.88 | 0.3491 |
| Variation sources | GL | SQ | F (Pr > F) | ||||
| Model | 57 | 95.58 | |||||
| Genotype (G) | (54) | (54.76) | 1.38 (0.0642) | ||||
| Year (A) | (3) | (40.82) | 18.50 (<0.0001) | ||||
| Axes | High Value | Variance (%) | Explained Cumulative | GxA | 162 | 118.50 | |
| 1 | 54.03 | 45.60 | 45.60 | IPCA1 | (56) | (54.03) | 2.37 (<0.0001) |
| 2 | 50.41 | 42.54 | 88.13 | IPCA2 | (54) | (50.41) | 2.29 (<0.0001) |
| 3 | 14.06 | 11.87 | 100.00 | IPCA3 | (52) | (14.06) | 0.66 (0.9625) |
| Mean Error | 295 | 120.12 | |||||
| Adjusted Total | 219 | 214.08 | |||||
| Genotypes | Means | Chi-Square | Pr > ChiSq | Genotypes | Means | Chi-Square | Pr > ChiSq |
|---|---|---|---|---|---|---|---|
| EC16 | 9.0128 | 0.00 | 0.9479 | EC44 | 8.1318 | 0.46 | 0.4999 |
| EC17 | 8.1033 | 1.05 | 0.3049 | EC45 | 8.6207 | 0.04 | 0.8496 |
| EC18 | 7.5834 | 2.91 | 0.0882 | EC47 | 8.1529 | 0.41 | 0.5245 |
| EC19 | 9.2024 | 1.33 | 0.2481 | EC48 | 8.2046 | 0.29 | 0.5871 |
| EC20 | 8.6407 | 0.28 | 0.5950 | EC49 | 8.5733 | 0.01 | 0.9140 |
| EC21 | 7.5239 | 3.31 | 0.0687 | EC50 | 8.6158 | 0.03 | 0.8563 |
| EC22 | 8.7405 | 0.16 | 0.6936 | EC51 | 8.3449 | 0.09 | 0.7704 |
| EC23 | 8.7693 | 0.20 | 0.6580 | EC52 | 8.8932 | 0.08 | 0.7752 |
| EC24 | 9.1320 | 0.41 | 0.5245 | EC53 | 8.5883 | 0.02 | 0.8935 |
| EC25 | 8.4304 | 0.26 | 0.6081 | EC65 | 8.2999 | 0.00 | 0.9828 |
| EC26 | 8.0816 | 0.59 | 0.4435 | EC66 | 8.2536 | 0.16 | 0.6896 |
| EC27 | 8.2580 | 0.55 | 0.4582 | EC67 | 8.7437 | 0.22 | 0.6425 |
| EC28 | 8.1072 | 1.07 | 0.3010 | EC68 | 8.7821 | 1.85 | 0.1739 |
| EC29 | 7.8740 | 1.32 | 0.2498 | EC69 | 7.7633 | 2.43 | 0.1194 |
| EC30 | 8.0060 | 0.82 | 0.3653 | EC70 | 7.4366 | 0.68 | 0.4084 |
| EC31 | 8.0287 | 0.75 | 0.3879 | EC71 | 9.0001 | 0.00 | 0.9637 |
| EC32 | 8.2664 | 0.19 | 0.6659 | EC72 | 8.7127 | 0.01 | 0.9114 |
| EC33 | 8.7675 | 0.19 | 0.6603 | EC73 | 8.6298 | 3.58 | 0.0586 |
| EC34 | 9.4237 | 2.27 | 0.1323 | EC74 | 7.4875 | 0.00 | 0.9658 |
| EC35 | 8.6304 | 0.04 | 0.8365 | EC75 | 8.2633 | 0.02 | 0.8873 |
| EC36 | 8.4143 | 0.03 | 0.8656 | EC76 | 8.3533 | 2.67 | 0.1023 |
| EC37 | 8.5419 | 0.00 | 0.9572 | EC77 | 7.6202 | 0.25 | 0.6157 |
| EC38 | 8.3793 | 0.05 | 0.8172 | EC78 | 8.1388 | 1.79 | 0.1806 |
| EC39 | 8.4205 | 0.03 | 0.8742 | EC79 | 7.7742 | 0.87 | 0.3509 |
| EC40 | 8.3520 | 0.08 | 0.7800 | EC80 | 7.8665 | 2.45 | 0.1177 |
| EC41 | 7.9771 | 0.92 | 0.3377 | EC81 | 7.0278 | 0.66 | 0.4170 |
| EC42 | 7.4232 | 4.07 | 0.0436 | EC82 | 8.9908 | 0.00 | 0.9828 |
| EC43 | 7.5776 | 2.50 | 0.1138 |
| Variation sources | GL | SQ | F (Pr > F) | ||||
| Model | 56 | 776.84 | |||||
| Genotype (G) | (53) | (354.64) | 2.68 (<0.0001) | ||||
| Year (A) | (3) | (422.20) | 56.31 (<0.0001) | ||||
| Axes | High Value | Variance (%) | Explained Cumulative | GxA | 159 | 397.41 | |
| 1 | 195.96 | 49.31 | 49.31 | IPCA1 | (55) | (195.96) | 3.04 (<0.0001) |
| 2 | 132.31 | 33.29 | 82.60 | IPCA2 | (53) | (132.31) | 1.23 (0.1491) |
| 3 | 69.14 | 17.40 | 100.00 | IPCA3 | (51) | (69.14) | 1.06 (0.3731) |
| Mean Error | 290 | 402.90 | |||||
| Adjusted Total | 215 | 1174.25 | |||||
| Genotypes | Means | Chi-Square | Pr > ChiSq | Genotypes | Means | Chi-Square | Pr > ChiSq |
|---|---|---|---|---|---|---|---|
| EC16 | 13.3475 | 0.03 | 0.8544 | EC43 | 14.7075 | 2.78 | 0.0952 |
| EC17 | 10.3950 | 8.91 | 0.0028 | EC44 | 12.1425 | 1.05 | 0.3046 |
| EC18 | 14.0125 | 0.65 | 0.4209 | EC45 | 16.2075 | 6.97 | 0.0083 |
| EC19 | 11.6125 | 2.54 | 0.1108 | EC47 | 11.3775 | 3.44 | 0.0638 |
| EC20 | 12.9750 | 0.24 | 0.6277 | EC48 | 13.8200 | 0.39 | 0.5298 |
| EC21 | 12.8350 | 0.10 | 0.7508 | EC49 | 15.2000 | 3.38 | 0.0662 |
| EC22 | 11.7525 | 2.08 | 0.1492 | EC50 | 11.1350 | 4.52 | 0.0335 |
| EC23 | 11.3125 | 3.71 | 0.0541 | EC51 | 14.4250 | 1.38 | 0.2402 |
| EC24 | 13.0875 | 0.55 | 0.4578 | EC52 | 13.5150 | 0.41 | 0.5238 |
| EC25 | 12.8925 | 0.01 | 0.9131 | EC53 | 16.5800 | 8.54 | 0.0035 |
| EC26 | 14.0875 | 0.76 | 0.3827 | EC65 | 12.7725 | 0.34 | 0.5608 |
| EC27 | 14.6025 | 1.77 | 0.1836 | EC66 | 12.6300 | 0.27 | 0.6005 |
| EC28 | 12.5550 | 0.34 | 0.5599 | EC67 | 11.6725 | 1.24 | 0.2663 |
| EC29 | 13.7550 | 0.74 | 0.3882 | EC68 | 14.1300 | 0.83 | 0.3621 |
| EC30 | 12.6575 | 0.47 | 0.4944 | EC69 | 14.1000 | 0.78 | 0.3766 |
| EC31 | 14.5175 | 1.58 | 0.2092 | EC70 | 12.9625 | 0.02 | 0.8908 |
| EC32 | 13.8325 | 0.41 | 0.5223 | EC71 | 13.9100 | 0.51 | 0.4770 |
| EC33 | 13.3525 | 0.04 | 0.8507 | EC72 | 13.7300 | 0.30 | 0.5859 |
| EC34 | 12.7325 | 0.05 | 0.8258 | EC73 | 16.0675 | 4.76 | 0.0290 |
| EC35 | 14.3075 | 1.15 | 0.2844 | EC74 | 14.0200 | 0.27 | 0.6009 |
| EC36 | 14.2100 | 0.97 | 0.3255 | EC76 | 13.0775 | 0.01 | 0.9378 |
| EC37 | 14.2125 | 0.97 | 0.3244 | EC77 | 12.7725 | 0.09 | 0.7675 |
| EC38 | 14.7925 | 2.23 | 0.1352 | EC78 | 13.3575 | 0.04 | 0.8469 |
| EC39 | 13.5625 | 0.15 | 0.6981 | EC79 | 12.6975 | 0.01 | 0.9114 |
| EC40 | 15.4900 | 4.31 | 0.0380 | EC80 | 12.5275 | 0.67 | 0.4140 |
| EC41 | 13.0575 | 0.01 | 0.9222 | EC81 | 11.5050 | 2.93 | 0.0868 |
| EC42 | 14.3575 | 1.24 | 0.2649 | EC82 | 13.1575 | 0.24 | 0.6277 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benedito, D.C.D.; Stuepp, C.A.; Helm, C.V.; Liz, M.V.d.; Miranda, A.C.d.; Imoski, R.; Lavoranti, O.J.; Wendling, I. Bioactive Compounds Concentrations and Stability in Leaves of Ilex paraguariensis Genotypes. Forests 2023, 14, 2411. https://doi.org/10.3390/f14122411
Benedito DCD, Stuepp CA, Helm CV, Liz MVd, Miranda ACd, Imoski R, Lavoranti OJ, Wendling I. Bioactive Compounds Concentrations and Stability in Leaves of Ilex paraguariensis Genotypes. Forests. 2023; 14(12):2411. https://doi.org/10.3390/f14122411
Chicago/Turabian StyleBenedito, Débora Caroline Defensor, Carlos André Stuepp, Cristiane Vieira Helm, Marcus Vinicius de Liz, Amanda Coelho de Miranda, Rafaela Imoski, Osmir José Lavoranti, and Ivar Wendling. 2023. "Bioactive Compounds Concentrations and Stability in Leaves of Ilex paraguariensis Genotypes" Forests 14, no. 12: 2411. https://doi.org/10.3390/f14122411
APA StyleBenedito, D. C. D., Stuepp, C. A., Helm, C. V., Liz, M. V. d., Miranda, A. C. d., Imoski, R., Lavoranti, O. J., & Wendling, I. (2023). Bioactive Compounds Concentrations and Stability in Leaves of Ilex paraguariensis Genotypes. Forests, 14(12), 2411. https://doi.org/10.3390/f14122411






