Root Traits and Soil Bacterial Composition Explain the Rhizosphere Effects along a Chronosequence of Rubber Plantations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site Information
2.2. Soil and Root Samples Collection
2.3. Soil Physicochemical Properties and SOM Decomposition
2.4. Soil DNA Extraction, Sequencing, and Bioinformatics Analysis
2.5. Root Morphological, Chemical, and Physiological Traits
2.6. Data Analysis
3. Results
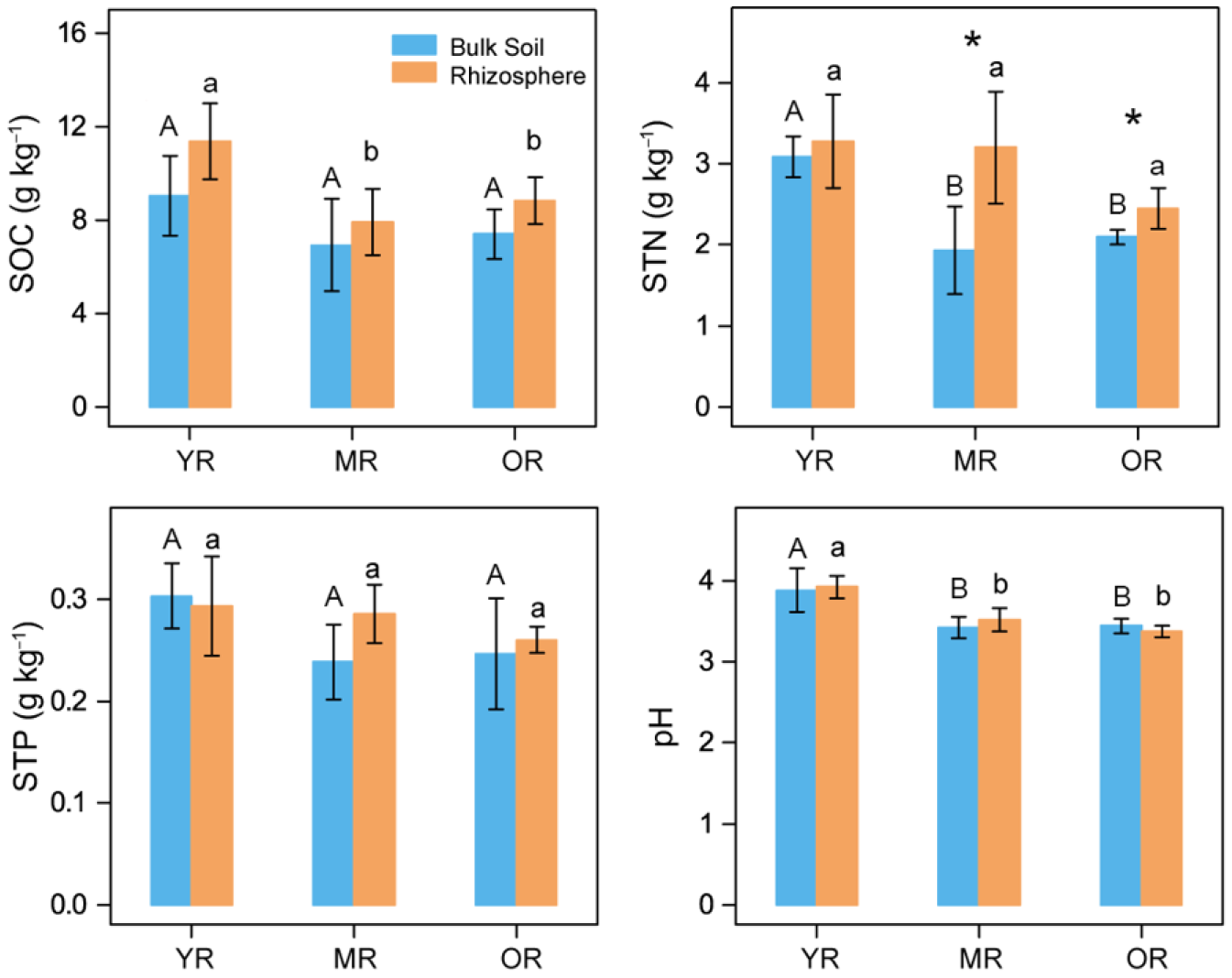
3.1. Soil Physicochemical Properties and Root Functional Traits
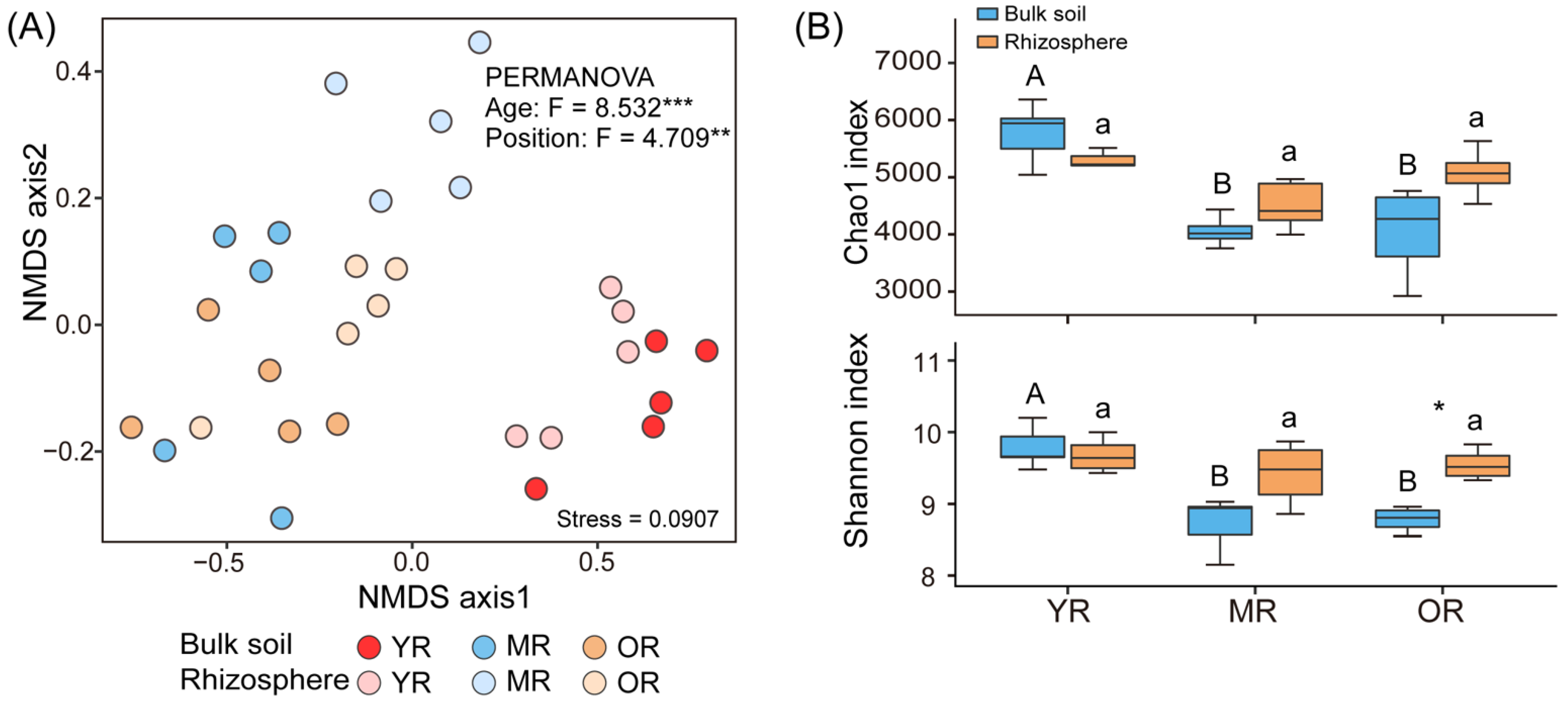
3.2. Soil Microbial Composition and Diversity
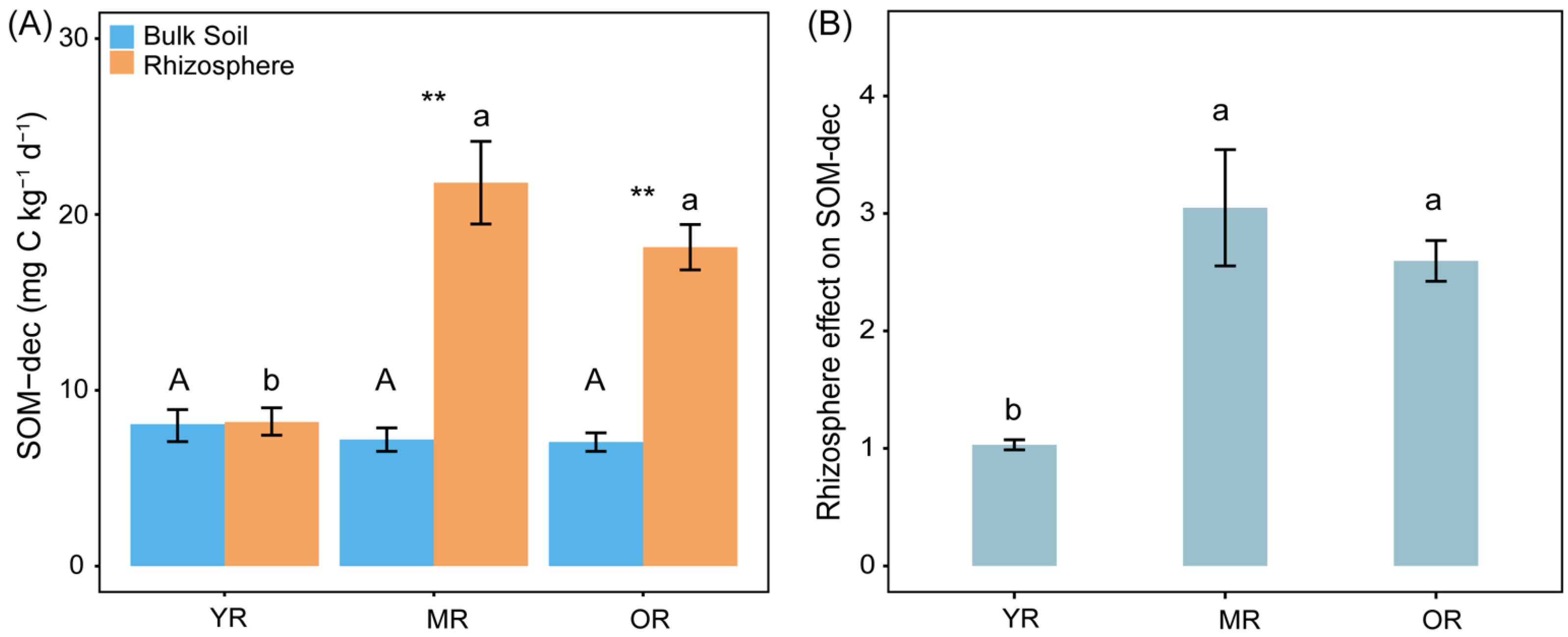
3.3. SOM Decomposition and RE on SOM Decomposition
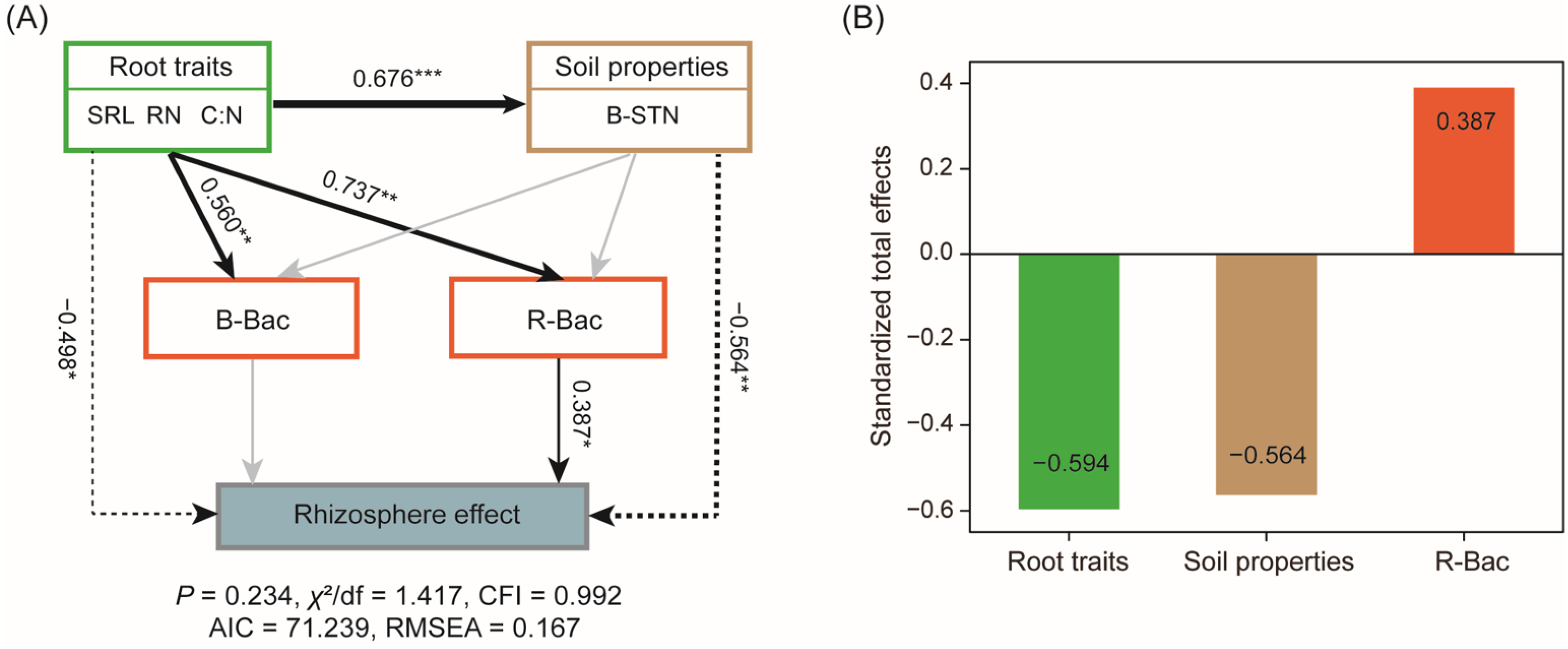
3.4. Dominant Determinants of RE on SOM Decomposition
4. Discussion
4.1. The RE on SOM Decomposition Increased with an Age Chronosequence of Rubber Plantations
4.2. Root Functional Traits Mediate RE on SOM Decomposition along a Chronosequence of Rubber Plantations
4.3. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Baudron, F.; Giller, K.E. Agriculture and nature: Trouble and strife? Biol. Conserv. 2014, 170, 232–245. [Google Scholar] [CrossRef]
- Gibbs, H.K.; Ruesch, A.S.; Achard, F.; Clayton, M.K.; Holmgren, P.; Ramankutty, N.; Foley, J.A. Tropical forests were the primary sources of new agricultural land in the 1980s and 1990s. Proc. Natl. Acad. Sci. USA 2010, 107, 16732–16737. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Production/Crops: Oil, Palm Fruit. 2018. Available online: http://faostat.fao.org (accessed on 10 December 2021).
- Lan, G.Y.; Wu, Z.X.; Yang, C.; Sun, R.; Chen, B.Q.; Zhang, X. Tropical rainforest conversion into rubber plantations results in changes in soil fungal composition, but underling mechanisms of community assembly remain unchanged. Geoderma 2020, 375, 114505. [Google Scholar] [CrossRef]
- Warren-Thomas, E.; Dolman, P.M.; Edwards, D.P. Increasing demand for natural rubber necessitates a robust sustainability initiative to mitigate impacts on tropical biodiversity. Conserv. Lett. 2015, 8, 230–241. [Google Scholar] [CrossRef]
- Guillaume, T.; Kotowska, M.M.; Hertel, D.; Knohl, A.; Krashevska, V.; Murtilaksono, K.; Scheu, S.; Kuzyakov, Y. Carbon costs and benefits of Indonesian rainforest conversion to plantations. Nat. Commun. 2018, 9, 2388. [Google Scholar] [CrossRef]
- Don, A.; Schumacher, J.; Freibauer, A. Impact of tropical land-use change on soil organic carbon stocks-a meta-analysis. Glob. Chang. Biol. 2011, 17, 1658–1670. [Google Scholar] [CrossRef]
- de Blécourt, M.; Hänsel, V.M.; Brumme, R.; Corre, M.D.; Veldkamp, E. Soil redistribution by terracing alleviates soil organic carbon losses caused by forest conversion to rubber plantation. For. Ecol. Manag. 2014, 313, 26–33. [Google Scholar] [CrossRef]
- Chen, C.F.; Liu, W.J.; Wu, J.N.; Jiang, X.J.; Zhu, X.A. Can intercropping with the cash crop help improve the soil physico-chemical properties of rubber plantations? Geoderma 2019, 335, 149–160. [Google Scholar] [CrossRef]
- Wauters, J.B.; Coudert, S.; Grallien, E.; Jonard, M.; Ponette, Q. Carbon stock in rubber tree plantations in Western Ghana and MatoGrosso (Brazil). For. Ecol. Manag. 2008, 255, 2347–2361. [Google Scholar] [CrossRef]
- Kurmi, B.; Nath, A.J.; Lal, R.; Das, A.K. Water stable aggregates and the associated active and recalcitrant carbon in soil under rubber plantation. Sci. Total Environ. 2020, 703, 135498. [Google Scholar] [CrossRef]
- Liu, C.G.; Jin, Y.Q.; Liu, C.A.; Tang, J.W.; Wang, Q.W.; Xu, M.X. Phosphorous fractions in soils of rubber-based agroforestry systems: Influence of season, management and stand age. Sci. Total Environ. 2018, 616–617, 1576–1588. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.C.; Ma, Y.X.; Cao, K.F.; Li, H.M.; Shen, J.X.; Liu, W.J.; Di, L.; Mei, C.C. Temporal changes of ecosystem carbon stocks in rubber plantations in Xishuangbanna, Southwest China. Pedosphere 2017, 27, 737–746. [Google Scholar] [CrossRef]
- Panklang, P.; Thoumazeau, A.; Chiarawipa, R.; Sdoodee, S.; Sebag, D.; Gay, F.; Thaler, P.; Brauman, A. Rubber, rubber and rubber: How 75 years of successive rubber plantations rotations affect topsoil quality. Land Degrad. Dev. 2022, 33, 1159–1169. [Google Scholar] [CrossRef]
- Finzi, A.C.; Abramoff, R.Z.; Spiller, K.S.; Brzostek, E.R.; Darby, B.A.; Kramer, M.A.; Phillips, R.P. Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Glob. Chang. Biol. 2015, 21, 2082–2094. [Google Scholar] [CrossRef]
- Jiang, Z.H.; Liu, Y.Z.; Yang, J.P.; Brookes, P.C.; Gunina, A. Rhizosphere priming regulates soil organic carbon and nitrogen mineralization: The significance of abiotic mechanisms. Geoderma 2021, 385, 114877. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Z.J.; Tang, M.; Zhu, B. Greater variations of rhizosphere effects within mycorrhizal group than between mycorrhizal group in a temperate forest. Soil Biol. Biochem. 2018, 126, 237–246. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Mommer, L.; De Vries, F.T. Going underground: Root traits as drivers of ecosystem processes. Trends Ecol. Evol. 2014, 29, 692–699. [Google Scholar] [CrossRef]
- Carrillo, Y.; Bell, C.; Koyama, A.; Canarini, A.; Boot, C.M.; Wallenstein, M.; Pendall, E. Plant traits, stoichiometry and microbes as drivers of decomposition in the rhizosphere in a temperate grassland. J. Ecol. 2017, 105, 1750–1765. [Google Scholar] [CrossRef]
- Orwin, K.H.; Buckland, S.M.; Johnson, D.; Turner, B.L.; Smart, S.; Oakley, S.; Bardgett, R.D. Linkages of plant traits to soil properties and the functioning of temperate grassland. J. Ecol. 2010, 98, 1074–1083. [Google Scholar] [CrossRef]
- Reich, P.B. The world-wide ‘fast–slow’ plant economics spectrum: A traits manifesto. J. Ecol. 2014, 102, 275–301. [Google Scholar] [CrossRef]
- Sun, L.J.; Ataka, M.; Kominami, Y.; Yoshimura, K. Relationship between fine-root exudation and respiration of two Quercus species in a Japanese temperate forest. Tree Physiol. 2017, 37, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- de Graaff, M.A.; Six, J.; Jastrow, J.D.; Schadt, C.W.; Wullschleger, S.D. Variation in root architecture among switchgrass cultivars impacts root decomposition rates. Soil Biol. Biochem. 2013, 58, 198–206. [Google Scholar] [CrossRef]
- Ontl, T.A.; Cambardella, C.A.; Schulte, L.A.; Kolka, R.K. Factors influencing soil aggregation and particulate organic matter responses to bioenergy crops across a topographic gradient. Geoderma 2015, 255, 1–11. [Google Scholar] [CrossRef]
- Sun, L.J.; Ataka, M.; Han, M.G.; Han, Y.F.; Gan, D.Y.; Xu, T.L.; Guo, Y.P.; Zhu, B. Root exudation as a major competitive fine-root functional trait of 18 coexisting species in a subtropical forest. New Phytol. 2021, 229, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, T.; Chenu, C.; Genet, P.; Barot, S.; Perveen, N.; Mougin, C.; Fontaine, S. Contribution of exudates, arbuscular mycorrhizal fungi and litter depositions to the rhizosphere priming effect induced by grassland species. Soil Biol. Biochem. 2015, 80, 146–155. [Google Scholar] [CrossRef]
- Han, M.G.; Sun, L.J.; Gan, D.Y.; Fu, L.C.; Zhu, B. Root functional traits are key determinants of the rhizosphere effect on soil organic matter decomposition across 14 temperate hardwood species. Soil Biol. Biochem. 2020, 151, 108019. [Google Scholar] [CrossRef]
- Cheng, W.; Kuzyakov, Y. Root effects on soil organic matter decomposition. In Roots and Soil Management: Interactions between Roots and the Soil; Agronomy Monograph No. 48; Zobel, R.W., Wright, S.F., Eds.; American Society of Agronomy, Crop Science Society of America, Soil Science Society of America: Madison, WI, USA, 2005; pp. 119–143. [Google Scholar]
- Baldrian, P.; Kolařík, M.; Štursová, M.; Kopecký, J.; Valášková, V.; Větrovský, T.; Žifčáková, L.; Šnajdr, J.; Rídl, J.; Vlček, Č.; et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012, 6, 248–258. [Google Scholar] [CrossRef]
- Chen, R.R.; Senbayram, M.; Blagodatsky, S.; Myachina, O.; Dittert, K.; Lin, X.G.; Blagodatskaya, E.; Kuzyakov, Y. Soil C and N availability determine the priming effect: Microbial N mining and stoichiometric decomposition theories. Glob. Chang. Biol. 2014, 20, 2356–2367. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Hill, P.W.; Jones, D.L. Root exudate components change litter decomposition in a simulated rhizosphere depending on temperature. Plant Soil 2007, 290, 293–305. [Google Scholar] [CrossRef]
- Ai, C.; Liang, G.Q.; Sun, J.W.; Wang, X.B.; Zhou, W. Responses of extracellular enzyme activities and microbial community in both the rhizosphere and bulk soil to long-term fertilization practices in a fluvo-aquic soil. Geoderma 2012, 173, 330–338. [Google Scholar] [CrossRef]
- Spitzer, C.M.; Lindahl, B.; Wardle, D.A.; Sundqvist, M.K.; Gundale, M.J.; Fanin, N.; Kardol, P. Root trait–microbial relationships across tundra plant species. New Phytol. 2021, 229, 1508–1520. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.T.; Han, F.; Ning, P.; Li, H.B.; Rengel, Z. Root traits and soil nutrient and carbon availability drive soil microbial diversity and composition in a northern temperate forest. Plant Soil 2022, 479, 281–299. [Google Scholar] [CrossRef]
- Meier, I.C.; Tückmantel, T.; Heitkötter, J.; Müller, K.; Preusser, S.; Wrobel, T.J.; Kandeler, E.; Marschner, B.; Leuschner, C. Root exudation of mature beech forests across a nutrient availability gradient: The role of root morphology and fungal activity. New Phytol. 2020, 226, 583–594. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Guebel, D.V.; Nudel, B.C.; Giulietti, A.M. A simple and rapid micro-Kjeldahl method for total nitrogen analysis. Biotechnol. Tech. 1991, 5, 427–430. [Google Scholar] [CrossRef]
- Bao, S. Soil and Agricultural Chemistry Analysis; Agriculture Publication: Beijing, China, 2000; pp. 25–103. [Google Scholar]
- Zheng, J.F.; Zhang, X.H.; Li, L.Q.; Zhang, P.J.; Pan, G.X. Effect of long-term fertilization on C mineralization and production of CH4 and CO2 under anaerobic incubation from bulk samples and particle size fractions of a typical paddy soil. Agric. Ecosyst. Environ. 2007, 120, 129–138. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-SyedMohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, 141–145. [Google Scholar] [CrossRef]
- Phillips, R.P.; Erlitz, Y.; Bier, R.; Bernhardt, E.S. New approach for capturing soluble root exudates in forest soils. Funct. Ecol. 2008, 22, 990–999. [Google Scholar] [CrossRef]
- Meyer, N.; Welp, G.; Rodionov, A.; Borchard, N.; Martius, C.; Amelung, W. Nitrogen and phosphorus supply controls soil organic carbon mineralization in tropical topsoil and subsoil. Soil Biol. Biochem. 2018, 119, 152–161. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Sahrawat, K.L. Organic matter accumulation in submerged soils. Adv. Agron. 2004, 81, 169–201. [Google Scholar]
- De Deyn, G.B.; Cornelissen, J.H.C.; Bardgett, R.D. Plant functional traits and soil carbon sequestration in contrasting biomes. Ecol. Lett. 2008, 11, 516–531. [Google Scholar] [CrossRef] [PubMed]
- Ghafoor, A.; Poeplau, C.; Kätterer, T. Fate of straw-and root-derived carbon in a Swedish agricultural soil. Biol. Fertil. Soils 2017, 53, 257–267. [Google Scholar] [CrossRef]
- Keller, A.B.; Brzostek, E.R.; Craig, M.E.; Fisher, J.B.; Phillips, R.P. Root-derived inputs are major contributors to soil carbon in temperate forests, but vary by mycorrhizal type. Ecol. lett. 2021, 24, 626–635. [Google Scholar] [CrossRef]
- Sokol, N.W.; Bradford, M.A. Microbial formation of stable soil carbon is more efficient from belowground than aboveground input. Nat. Geosci. 2019, 12, 46–53. [Google Scholar] [CrossRef]
- Villarino, S.H.; Pinto, P.; Jackson, R.B.; Piñeiro, G. Plant rhizodeposition: A key factor for soil organic matter formation in stable fractions. Sci. Adv. 2021, 7, eabd3176. [Google Scholar] [CrossRef] [PubMed]
- Cotrufo, M.F.; Wallenstein, M.D.; Boot, C.M.; Denef, K.; Paul, E.B. The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Glob. Chang. Biol. 2013, 19, 988–995. [Google Scholar] [CrossRef]
- Pausch, J.; Kuzyakov, Y. Carbon input by roots into the soil: Quantification of rhizodeposition from root to ecosystem scale. Glob. Chang. Biol. 2018, 24, 1–12. [Google Scholar] [CrossRef]
- Hobbie, S.E. Contrasting effects of substrate and fertilizer nitrogen on the early stages of litter decomposition. Ecosystems 2005, 8, 644–656. [Google Scholar] [CrossRef]
- Manzoni, S.; Jackson, R.B.; Trofymow, J.A.; Porporato, A. The global stoichiometry of litter nitrogen mineralization. Science 2008, 321, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Bardgett, R.D.; Freeman, C.; Ostle, N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008, 2, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Kramer, C.; Gleixner, G. Soil organic matter in soil depth profiles: Distinct carbon preferences of microbial groups during carbon transformation. Soil Biol. Biochem. 2008, 40, 425–433. [Google Scholar] [CrossRef]
- Henneron, L.; Cros, C.; Picon-Cochard, C.; Rahimian, V.; Fontaine, S. Plant economic strategies of grassland species control soil carbon dynamics through rhizodeposition. J. Ecol. 2020, 108, 528–545. [Google Scholar] [CrossRef]
- Shahbaz, M.; Kuzyakov, Y.; Sanaullah, M.; Heitkamp, F.; Zelenev, V.; Kumar, A.; Blagodatskaya, E. Microbial decomposition of soil organic matter is mediated by quality and quantity of crop residues: Mechanisms and thresholds. Biol. Fertil. Soils 2017, 53, 287–301. [Google Scholar] [CrossRef]
- Poirier, V.; Roumet, C.; Munson, A.D. The root of the matter: Linking root traits and soil organic matter stabilization processes. Soil Biol. Biochem. 2018, 120, 246–259. [Google Scholar] [CrossRef]
- Garcia, L.; Damour, G.; Gary, C.; Follain, S.; Le Bissonnais, Y.; Metay, A. Trait-based approach for agroecology: Contribution of service crop root traits to explain soil aggregate stability in vineyards. Plant Soil 2019, 435, 1–14. [Google Scholar] [CrossRef]
- de Vries, F.T.; Manning, P.; Tallowin, J.R.B.; Mortimer, S.R.; Pilgrim, E.S.; Harrison, K.A.; Hobbs, P.J.; Quirk, H.; Shipley, B.; Cornelissen, J.H.C.; et al. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol. Lett. 2012, 15, 1230–1239. [Google Scholar] [CrossRef]
- Chao, L.; Liu, Y.; Zhang, W.; Wang, Q.K.; Guan, X.; Yang, Q.P.; Chen, L.C.; Zhang, J.B.; Hu, B.Q.; Liu, Z.F.; et al. Root functional traits determine the magnitude of the rhizosphere priming effect among eight tree species. Oikos 2023, 2023, e09638. [Google Scholar] [CrossRef]
- Yin, L.M.; Xiao, W.; Dijkstra, F.A.; Zhu, B.; Wang, P.; Cheng, W.X. Linking absorptive roots and their functional traits with rhizosphere priming of tree species. Soil Biol. Biochem. 2020, 150, 107997. [Google Scholar] [CrossRef]
- Fanin, N.; Kardol, P.; Farrell, M.; Nilsson, M.; Gundale, M.J.; Wardle, D.A. The ratio of Gram-positive to Gram-negative bacterial PLFA markers as an indicator of carbon availability in organic soils. Soil Biol. Biochem. 2019, 128, 111–114. [Google Scholar] [CrossRef]
- Zhou, G.X.; Qiu, X.W.; Chen, L.; Zhang, C.Z.; Ma, D.H.; Zhang, J.B. Succession of organics metabolic function of bacterial community in response to addition of earthworm casts and zeolite in maize straw composting. Bioresour. Technol. 2019, 280, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Chen, P.F.; He, C.; Jiang, Z.H.; Lan, R.; Yang, J.P. Soil microbial community structure dynamics shape the rhizosphere priming effect patterns in the paddy soil. Sci. Total Environ. 2023, 857, 159459. [Google Scholar] [CrossRef]
- Monteiro, P.H.R.; Kaschuk, G.; Winagraski, E.; Auer, C.G.; Higa, A.R. Rhizobial inoculation in black wattle plantation (Acacia mearnsii De Wild.) in production systems of southern Brazil. Braz. J. Microbiol. 2019, 50, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y. Sources of CO2 efflux from soil and review of partitioning methods. Soil Biol. Biochem. 2006, 38, 425–448. [Google Scholar] [CrossRef]
- Phillips, R.P.; Fahey, T.J. Tree species and mycorrhizal associations influence the magnitude of rhizosphere effects. Ecology 2006, 87, 1302–1313. [Google Scholar] [CrossRef]
- Gan, D.Y.; Zeng, H.; Zhu, B. The rhizosphere effect on soil gross nitrogen mineralization: A meta-analysis. Soil Ecol. Lett. 2022, 4, 144–154. [Google Scholar] [CrossRef]
- Kong, C.H.; Zhang, S.Z.; Li, Y.H.; Xia, Z.C.; Yang, X.F.; Meiners, S.J.; Wang, P. Plant neighbor detection and allelochemical response are driven by root-secreted signaling chemicals. Nat. Commun. 2018, 9, 3867. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqu’e, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef]

| Traits | YR | MR | OR | |
|---|---|---|---|---|
| Morphology traits | Root diameter (mm) | 0.816 ± 0.054 a | 0.782 ± 0.020 a | 0.743 ± 0.057 a |
| SRL (m g−1) | 4.058 ± 1.104 c | 9.389 ± 1.576 b | 12.395 ± 2.119 a | |
| RTD (g cm−3) | 0.225 ± 0.022 b | 0.313 ± 0.038 a | 0.356 ±0.017 a | |
| SRA (cm2 g−1) | 219.792 ± 10.994 a | 166.525 ± 25.506 b | 152.858 ± 15.754 b | |
| Chemical traits | RC (g kg−1) | 489.865 ± 33.713 a | 427.421 ± 45.974 a | 427.133 ± 57.750 a |
| RN (g kg−1) | 14.248 ± 1.582 b | 20.036 ± 2.743 a | 24.352 ± 2.394 a | |
| Root C:N | 34.638 ± 3.044 a | 21.653 ± 3.373 b | 17.528 ± 1.404 b | |
| Physiological traits | Root exudation (mg C g−1 h−1) | 0.043 ± 0.008 c | 0.087 ± 0.012 b | 0.122 ± 0.029 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Deng, D.; Yang, M.; Sun, Y. Root Traits and Soil Bacterial Composition Explain the Rhizosphere Effects along a Chronosequence of Rubber Plantations. Forests 2023, 14, 2209. https://doi.org/10.3390/f14112209
Liu G, Deng D, Yang M, Sun Y. Root Traits and Soil Bacterial Composition Explain the Rhizosphere Effects along a Chronosequence of Rubber Plantations. Forests. 2023; 14(11):2209. https://doi.org/10.3390/f14112209
Chicago/Turabian StyleLiu, Guoyin, Danting Deng, Meiqiu Yang, and Yanfei Sun. 2023. "Root Traits and Soil Bacterial Composition Explain the Rhizosphere Effects along a Chronosequence of Rubber Plantations" Forests 14, no. 11: 2209. https://doi.org/10.3390/f14112209
APA StyleLiu, G., Deng, D., Yang, M., & Sun, Y. (2023). Root Traits and Soil Bacterial Composition Explain the Rhizosphere Effects along a Chronosequence of Rubber Plantations. Forests, 14(11), 2209. https://doi.org/10.3390/f14112209





