Transcriptomic and Non-Targeted Metabolomic Analyses Reveal Changes in Metabolic Networks during Leaf Coloration in Cyclocarya paliurus (Batalin) Iljinsk
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Materials
2.2. Determination of Metabolites
2.3. Transcriptome Analysis
2.4. Data Analysis
3. Results
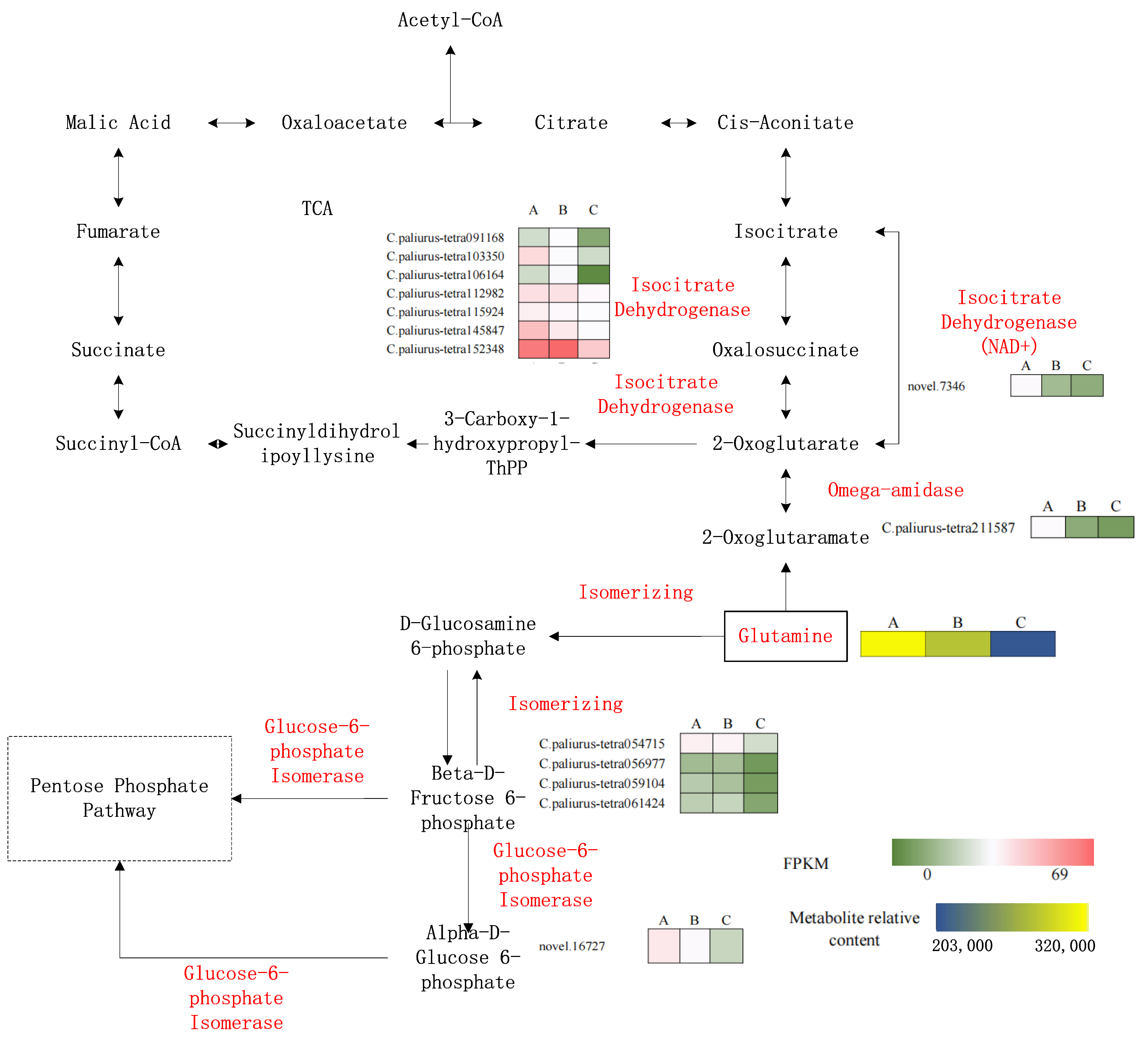
3.1. Metabolism and Transcriptome Association Analysis
3.2. N Metabolism
3.3. Relationships between Metabolites in Specific Pathways and DEGs
3.4. Acetyl-CoA Biosynthesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Cao, Y.N.; Fang, S.Z.; Wang, T.L.; Yin, Z.Q.; Shang, X.L.; Yang, W.X.; Fu, X.X. Antidiabetic effect of Cyclocarya paliurus leaves depends on the contents of antihyperglycemic flavonoids and antihyperlipidemic triterpenoids. Molecules 2018, 23, 1042. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.Q.; Shang, X.L.; Fang, S.Z.; Zhang, X.T.; Fu, X.X. Genome assembly of two diploid and one auto-tetraploid Cyclocarya paliurus genomes. Sci. Data 2021, 12, 146. [Google Scholar] [CrossRef]
- Sun, C.; Zhong, W.; Fu, X.; Shang, X.; Fang, S. A study on growth and aboveground biomass production of juvenile Cyclocarya paliurus plantations. J. Nanjing For. Univ. 2022, 46, 138–144. [Google Scholar] [CrossRef]
- Yang, W.X.; Zhuang, J.Q.; Ding, S.Y.; Zhang, M.; Tian, Y.; Wan, S.Y.; Fang, S.Z. Study on cutting cultivation technology and rooting mechanism of Cyclocarya paliurus. Ecol. Chem. Eng. 2022, 29, 379–389. [Google Scholar] [CrossRef]
- Fang, S. A review on the development history and the resource silviculture of Cyclocarya paliurus industry. J. Nanjing For. Univ. 2022, 46, 115–126. [Google Scholar] [CrossRef]
- Qu, Y.Q.; Shang, X.L.; Zeng, Z.Y.; Yu, Y.H.; Bian, G.L.; Wang, W.L.; Liu, L.; Tian, L.; Zhang, S.C.; Wang, Q.; et al. Whole-genome duplication reshaped adaptive evolution in a relict plant species, Cyclocarya paliurus. Dev. Reprod. Biol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, B.; Chen, X.; Mao, X.; Fu, X. Squalene epoxidase (SE) gene related to triterpenoid biosynthesis assists to select elite genotypes in medicinal plant: Cyclocarya paliurus (Batal.) Iljinskaja. Plant Physiol. Biochem. 2023, 199, 107726. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Zhang, Z.; Fang, S.Z. Physiological response and molecular regulatory mechanism reveal a positive role of nitric oxide and hydrogen sulfide applications in salt tolerance of Cyclocarya paliurus. Front. Plant Sci. 2023, 14, 1211162. [Google Scholar] [CrossRef]
- Deng, B.; Li, Y.Y.; Xu, D.D.; Ye, Q.Q.; Liu, G.H. Nitrogen availability alters flavonoid accumulation in Cyclocarya paliurus via the effects on the internal carbon/nitrogen balance. Sci. Rep. 2019, 9, 2370. [Google Scholar] [CrossRef]
- Qin, J.; Yue, X.L.; Fang, S.Z.; Qian, M.Y.; Zhou, S.T.; Shang, X.L.; Yang, W.X. Nitrogen addition modifies the relative gene expression level and accumulation of carbon-based bioactive substances in Cyclocarya paliurus. Plant Physiol. Biochem. 2022, 188, 70–80. [Google Scholar] [CrossRef]
- Sun, C.; Shang, X.; Ding, H.; Cao, Y.; Fang, S. Natural variations in flavonoids and triterpenoids of Cyclocarya paliurus leaves. J. For Res. 2020, 32, 805–814. [Google Scholar] [CrossRef]
- Sun, C.; Shang, X.; Fang, S.; Yang, W.; Cao, Y.; Ding, H.; Li, X. Association Analysis between Genotype and Environment: Differentiation between Cyclocarya paliurus Resources That Accumulate Triterpenoids. Front. Plant Sci. 2022, 13, 945897. [Google Scholar] [CrossRef]
- Ruan, J.; Haerdter, R.; Gerendás, J. Impact of nitrogen supply on carbon/nitrogen allocation: A case study on amino acids and catechins in green tea [Camellia sinensis (L.) O. Kuntze] plants. J. Plant Biol. 2010, 12, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Lin, H.; Liu, Z.; Liu, Z. Metabolomics analysis of C. sinensis with respect to harvesting time. Food Res. Int. 2020, 128, 108814. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, M.; Fang, H.; Zhang, Q.; Ruan, J. Accumulation of Amino Acids and Flavonoids in Young Tea Shoots Is Highly Correlated With Carbon and Nitrogen Metabolism in Roots and Mature Leaves. Front. Plant Sci. 2021, 12, 756433. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Liang, X.; Li, K.; Dai, P.; Li, J.; Liang, B.; Sun, C.; Lin, X. Metabolomics analysis reveals potential mechanisms of phenolic accumulation in lettuce (Lactuca sativa L.) induced by low nitrogen supply. Plant Physiol. Biochem. 2021, 158, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, B.; Huang, H.J.; Huang, W.J.; Zhang, Z.R.; Wang, Q.N.; Luo, H.L.; An, B. Metabolomic and transcriptomic analyses provide insights into metabolic networks during cashew fruit development and ripening. Food Chem. 2022, 404, 134765. [Google Scholar] [CrossRef] [PubMed]
- Boex-Fontvieille, E.R.A.; Gauthier, P.P.G.; Gilard, F.; Hodges, M.; Tcherkez, G.G.B. A new anaplerotic respiratory pathway involving lysine biosynthesis in isocitrate dehydrogenase-deficient Arabidopsis mutants. New Phytol. 2013, 199, 673–682. [Google Scholar] [CrossRef]
- Sun, C.; Fang, S.; Shang, X. Triterpenoids biosynthesis regulation for leaf coloring of wheel wingnut (Cyclocarya paliurus). Forests 2021, 12, 1733. [Google Scholar] [CrossRef]
- Sun, C.; Fang, S.; Shang, X. Flavonoid biosynthesis regulation for leaf coloring of Cyclocarya paliurus. Acta Physiol. Plant. Vol. 2023, 45, 91. [Google Scholar] [CrossRef]
- Lyubetskaya, A.V.; Rubanov, L.I.; Gelfand, M.S. Use of the flux model of amino acid metabolism of Escherichia coli. Biochemistry 2006, 71, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Chen, H.; Wang, J.; Zheng, Y.; Li, J. Joint transcriptomic and metabolic analysis of flavonoids in Cyclocarya paliurus leaves. ACS Omega 2021, 6, 9028–9038. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.K.; Lin, W.D.; Zhu, J.X.; Li, J.M. Amino acids profiling and transcriptomic data integration demonstrates the dynamic regulation of amino acids synthesis in the leaves of Cyclocarya paliurus. PeerJ 2022, 10, e13689. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.L.; Liu, X.F.; Fang, S.Z. Influence of nitrogen and phosphorus additions on parameters of photosynthesis and chlorophyll fluorescence in Cyclocarya paliurus seedlings. Photosynthetica 2023, 61, 318–327. [Google Scholar] [CrossRef]
- Qin, J.; Yue, X.L.; Ling, Y.; Zhou, Y.S.; Li, N.; Shang, X.L.; Fang, S.Z. Nitrogen form and ratio impact phenolic accumulation and related gene expression in Cyclocarya paliurus. Trees 2021, 35, 685–696. [Google Scholar] [CrossRef]
- Lian, D.H.; Li, L.; Liu, X.; Zhong, X.; Wang, H.Z.; Zhou, S.; Gu, L. Time-scale dynamics of proteome predicts the central carbon metabolism involved in triterpenoid accumulation responsive to nitrogen limitation in Ganoderma lucidum. Fungal Biol. 2021, 125, 294–304. [Google Scholar] [CrossRef]
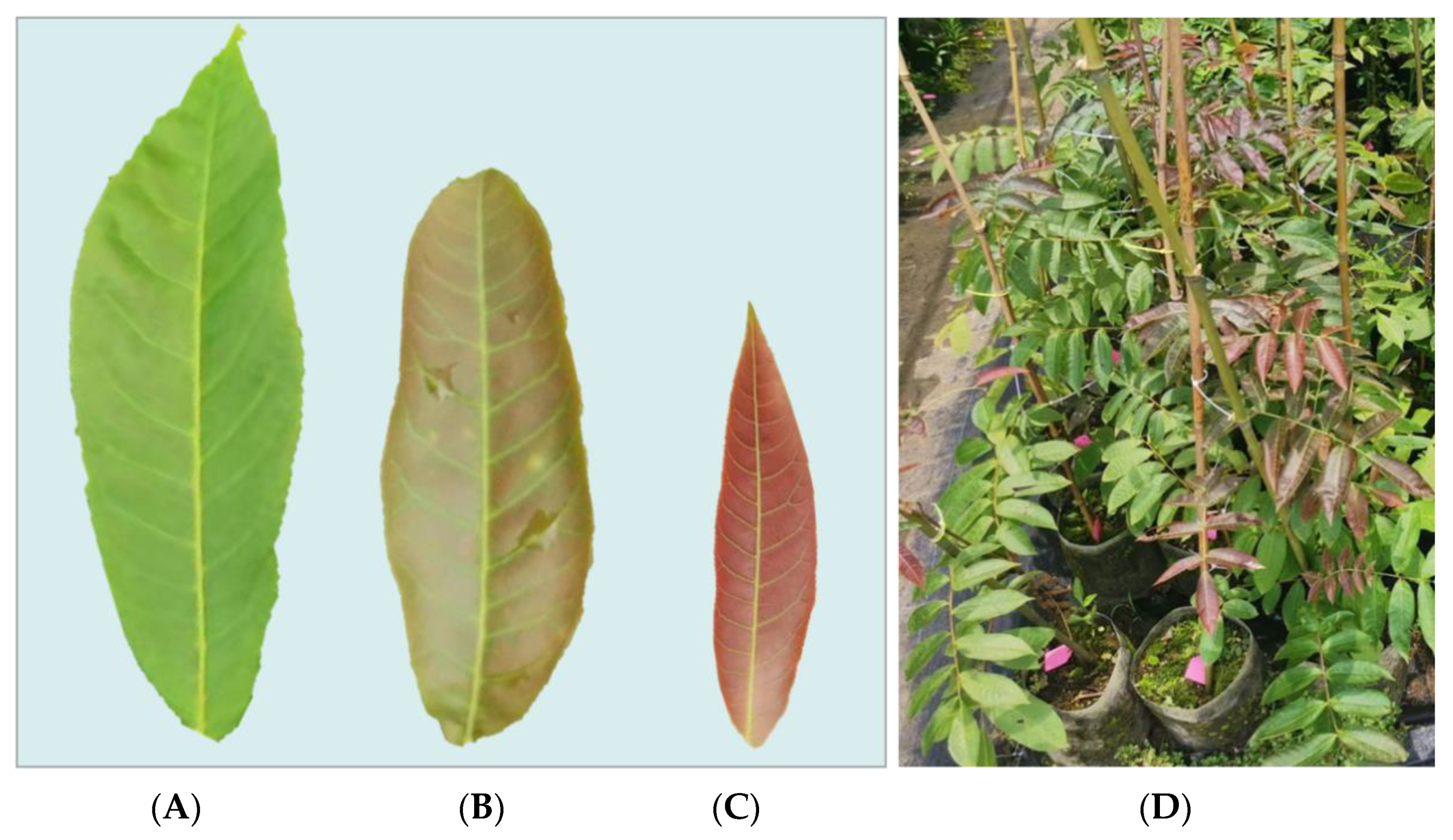
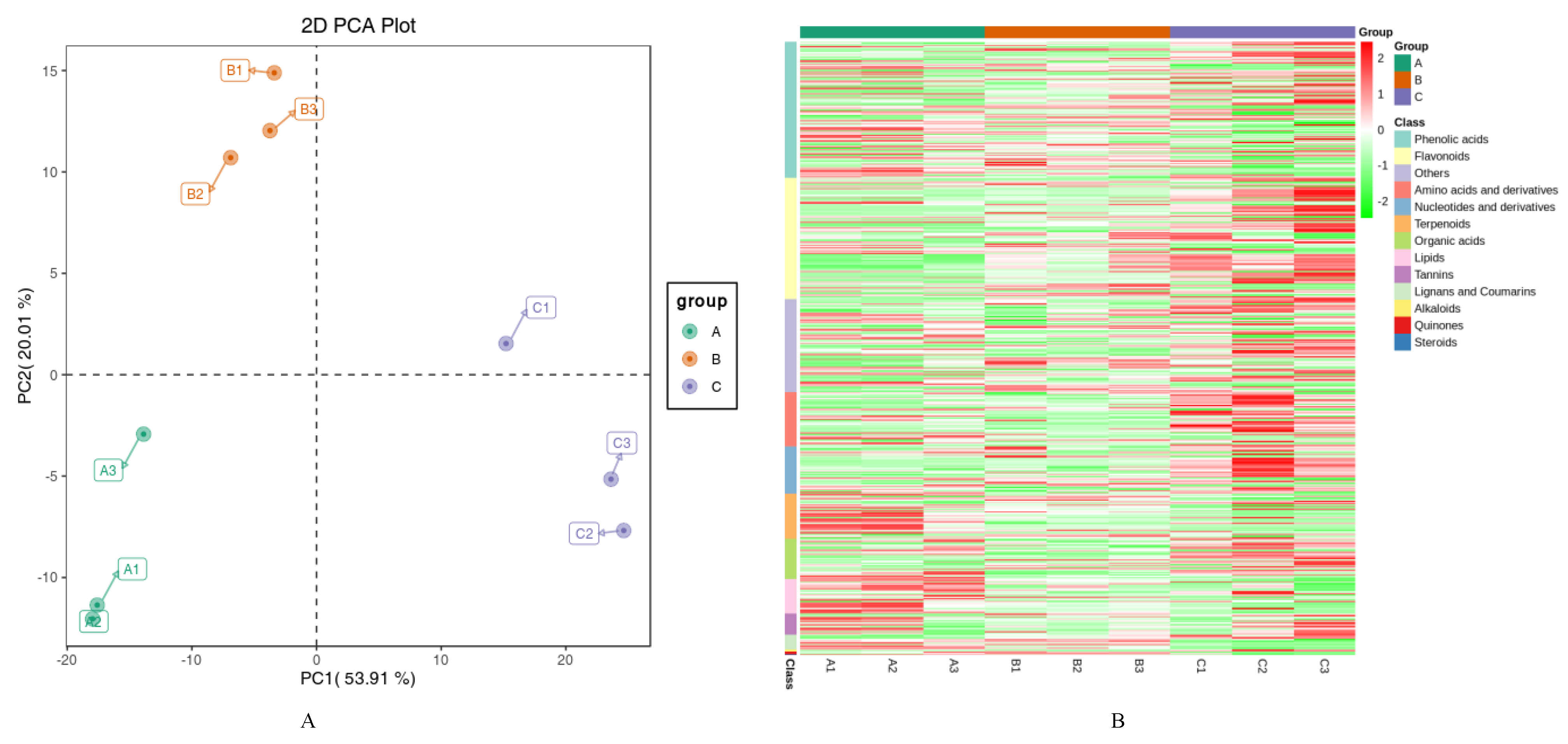
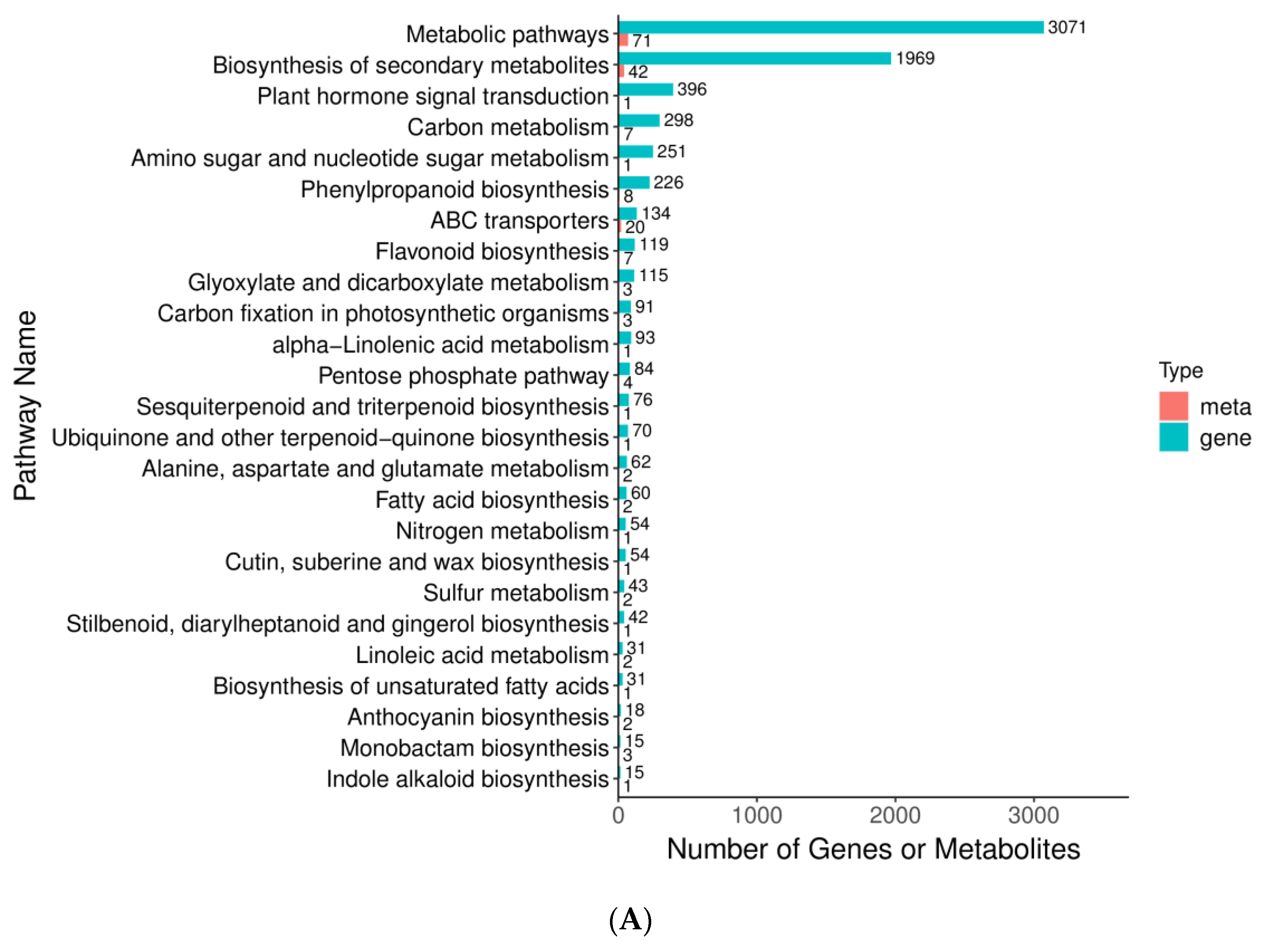
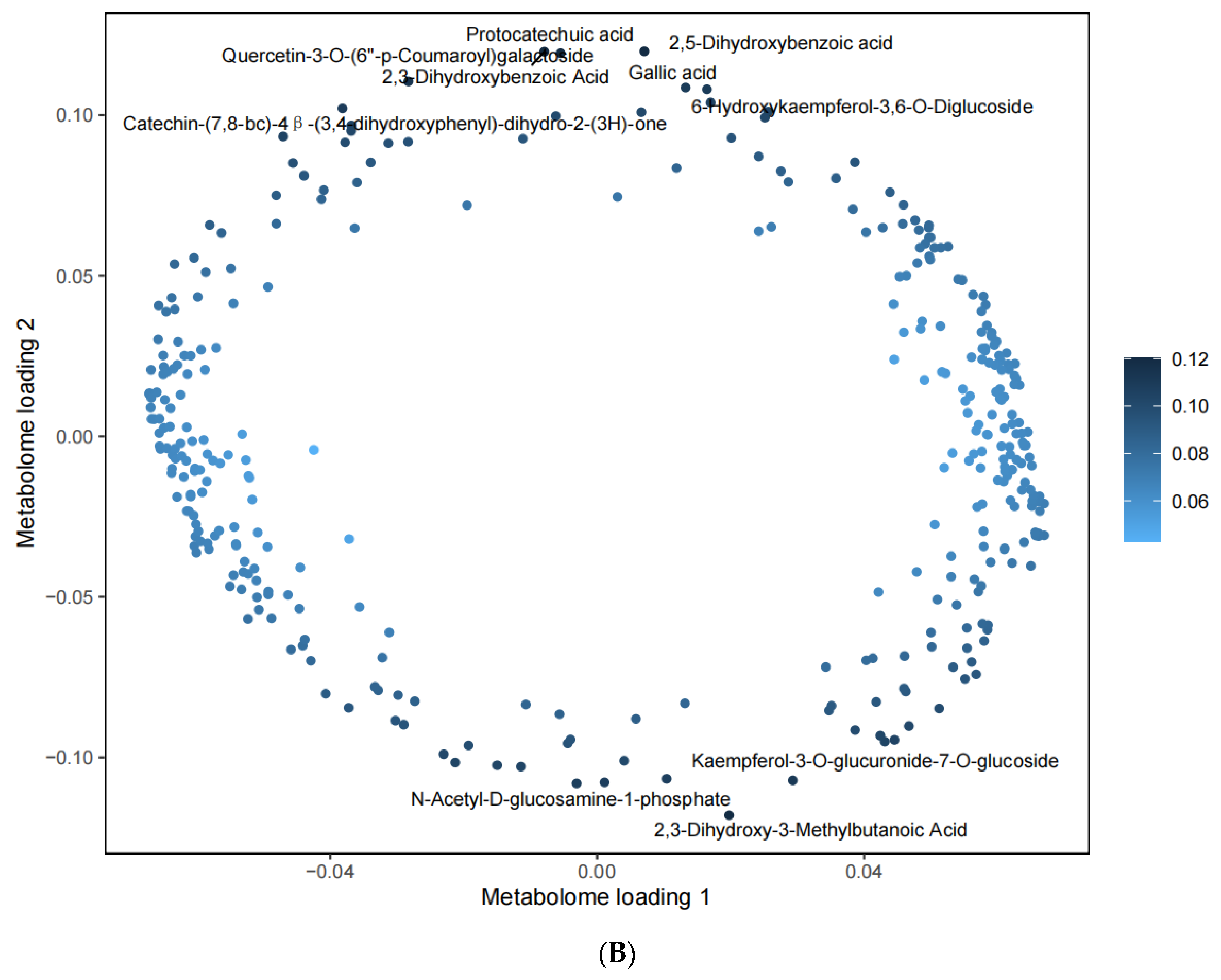


| Metabolite | Stage | Class | ||
|---|---|---|---|---|
| A | B | C | ||
| L-Glutamine | 3.05 × 106 ± 1.45 × 105 b | 3.17 × 106 ± 2.03 × 105 b | 4.94 × 106 ± 6.38 × 105 a | Amino acids and derivatives |
| N-Acetyl-L-Glutamine | 3.05 × 106 ± 1.45 × 105 a | 3.17 × 106 ± 2.03 × 105 a | 4.94 × 106 ± 6.38 × 105 b | Amino acids and derivatives |
| L-Phenylalanine | 1.15 × 107 ± 7.09 × 105 a | 8.26 × 106 ± 5.12 × 105 a | 2.43 × 107 ± 4.36 × 106 b | Amino acids and derivatives |
| L-Aspartic Acid | 3.13 × 105 ± 1.55 × 104 a | 3.79 × 105 ± 3.03 × 104 a | 1.14 × 106 ± 2.67 × 105 b | Amino acids and derivatives |
| L-Lysine | 3.32 × 105 ± 1.00 × 104 b | 2.95 × 105 ± 8.33 × 103 b | 2.08 × 105 ± 1.82 × 104 a | Amino acids and derivatives |
| L-Histidine | 4.14 × 104 ± 2.10 × 103 b | 2.68 × 104 ± 2.65 × 102 a | 2.69 × 104 ± 3.61 × 103 a | Amino acids and derivatives |
| L-Arginine | 9.39 × 104 ± 3.73 × 103 a | 1.13 × 105 ± 5.69 × 103 a | 1.59 × 106 ± 5.41 × 105 b | Amino acids and derivatives |
| N-Acetyl-L-glutamic acid | 7.48 × 104 ± 9.68 × 102 c | 4.60 × 104 ± 2.67 × 103 b | 3.48 × 104 ± 4.51 × 103 a | Amino acids and derivatives |
| L-Tryptophan | 7.91 × 105 ± 1.08 × 105 b | 2.87 × 105 ± 2.29 × 104 a | 1.15 × 105 ± 2.60 × 104 a | Amino acids and derivatives |
| Syringin | 2.21 × 105 ± 2.19 × 103 b | 2.76 × 105 ± 8.62 × 103 c | 1.45 × 105 ± 1.15 × 104 a | Phenolic acids |
| Coniferaldehyde | 4.18 × 104 ± 3.20 × 103 b | 3.70 × 104 ± 3.70 × 103 b | 2.56 × 104 ± 9.81 × 102 a | Phenolic acids |
| Coniferyl alcohol | 3.44 × 105 ± 6.41 × 104 b | 3.33 × 105 ± 1.88 × 104 b | 1.33 × 105 ± 4.04 × 103 a | Phenolic acids |
| Cyanidin-3-O-glucoside | 2.87 × 105 ± 7.56 × 104 a | 3.08 × 106 ± 2.48 × 105 b | 8.07 × 106 ± 6.13 × 105 c | Anthocyanins |
| Delphinidin-3-O-glucoside | 2.11 × 106 ± 5.92 × 105 a | 1.87 × 107 ± 2.81 × 106 b | 8.85 × 107 ± 6.02 × 106 c | Anthocyanins |
| Luteolin | 9.30 × 103 ± 2.48 × 103 a | 1.24 × 104 ± 1.86 × 102 a | 6.54 × 104 ± 1.51 × 104 b | Flavonoid |
| Kaempferol-3-O-rutinoside | 1.20 × 105 ± 3.51 × 103 c | 7.18 × 104 ± 1.61 × 104 b | 4.28 × 103 ± 4.77 × 102 a | Flavonoid |
| Quercetin | 3.31 × 105 ± 4.80 × 104 a | 5.06 × 105 ± 2.80 × 104 a | 9.68 × 105 ± 1.37 × 105 b | Flavonols |
| Quercetin-3-O-xyloside (Reynoutrin) | 3.70 × 106 ± 1.76 × 105 a | 9.12 × 106 ± 5.47 × 105 b | 1.27 × 107 ± 1.56 × 106 c | Flavonols |
| Quercetin-3-O-glucoside (Isoquercitrin) | 7.87 × 106 ± 5.29 × 105 a | 9.95 × 106 ± 6.76 × 105 a b | 1.20 × 107 ± 7.75 × 105 b | Flavonols |
| Quercetin-3-O-rutinoside (Rutin) | 2.62 × 104 ± 5.27 × 103 a | 3.44 × 104 ± 4.00 × 103 a | 1.70 × 105 ± 5.18 × 104 b | Flavonols |
| Kaempferol-3,7-O-dirhamnoside (Kaempferitrin) | 1.71 × 105 ± 3.60 × 104 a | 1.58 × 105 ± 4.98 × 103 a | 1.98 × 105 ± 6.18 × 104 a | Flavonols |
| Apigenin-6-C-glucoside (Isovitexin) | 4.88 × 105 ± 9.50 × 103 b | 1.69 × 105 ± 4.53 × 104 a | 1.30 × 105 ± 3.20 × 104 a | Flavonoid carbonoside |
| D-Erythrose-4-phosphate | 2.71 × 104 ± 2.11 × 103 a | 2.67 × 104 ± 2.43 × 103 a | 5.10 × 104 ± 8.61 × 103 b | Saccharides and Alcohols |
| Ribulose-5-phosphate | 3.20 × 104 ± 4.16 × 102 b | 1.38 × 104 ± 1.18 × 103 a | 1.89 × 104 ± 3.21 × 103 a | Saccharides and Alcohols |
| Shikimic acid | 1.06 × 105 ± 5.09 × 103 a | 1.01 × 105 ± 1.43 × 104 a | 2.21 × 105 ± 2.14 × 104 b | Organic acids |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Fang, S.; Shang, X. Transcriptomic and Non-Targeted Metabolomic Analyses Reveal Changes in Metabolic Networks during Leaf Coloration in Cyclocarya paliurus (Batalin) Iljinsk. Forests 2023, 14, 1948. https://doi.org/10.3390/f14101948
Sun C, Fang S, Shang X. Transcriptomic and Non-Targeted Metabolomic Analyses Reveal Changes in Metabolic Networks during Leaf Coloration in Cyclocarya paliurus (Batalin) Iljinsk. Forests. 2023; 14(10):1948. https://doi.org/10.3390/f14101948
Chicago/Turabian StyleSun, Caowen, Shengzuo Fang, and Xulan Shang. 2023. "Transcriptomic and Non-Targeted Metabolomic Analyses Reveal Changes in Metabolic Networks during Leaf Coloration in Cyclocarya paliurus (Batalin) Iljinsk" Forests 14, no. 10: 1948. https://doi.org/10.3390/f14101948
APA StyleSun, C., Fang, S., & Shang, X. (2023). Transcriptomic and Non-Targeted Metabolomic Analyses Reveal Changes in Metabolic Networks during Leaf Coloration in Cyclocarya paliurus (Batalin) Iljinsk. Forests, 14(10), 1948. https://doi.org/10.3390/f14101948







