The Jujube TCP Transcription Factor ZjTCP16 Regulates Plant Growth and Cell Size by Affecting the Expression of Genes Involved in Plant Morphogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Cloning, Sequence Analysis and Subcellular Localization of ZjTCP16
2.3. qRT-PCR Assay
2.4. Generation of ZjTCP16-OE Lines of Arabidopsis and Z. Jujuba
2.5. Generation of zjtcp16 CRISPR Mutant Jujube Shoots
2.6. Phenotypic Analysis and Microscopic Examination
2.7. Y1H and Y2H Assay
2.8. BiFC Analysis
3. Results
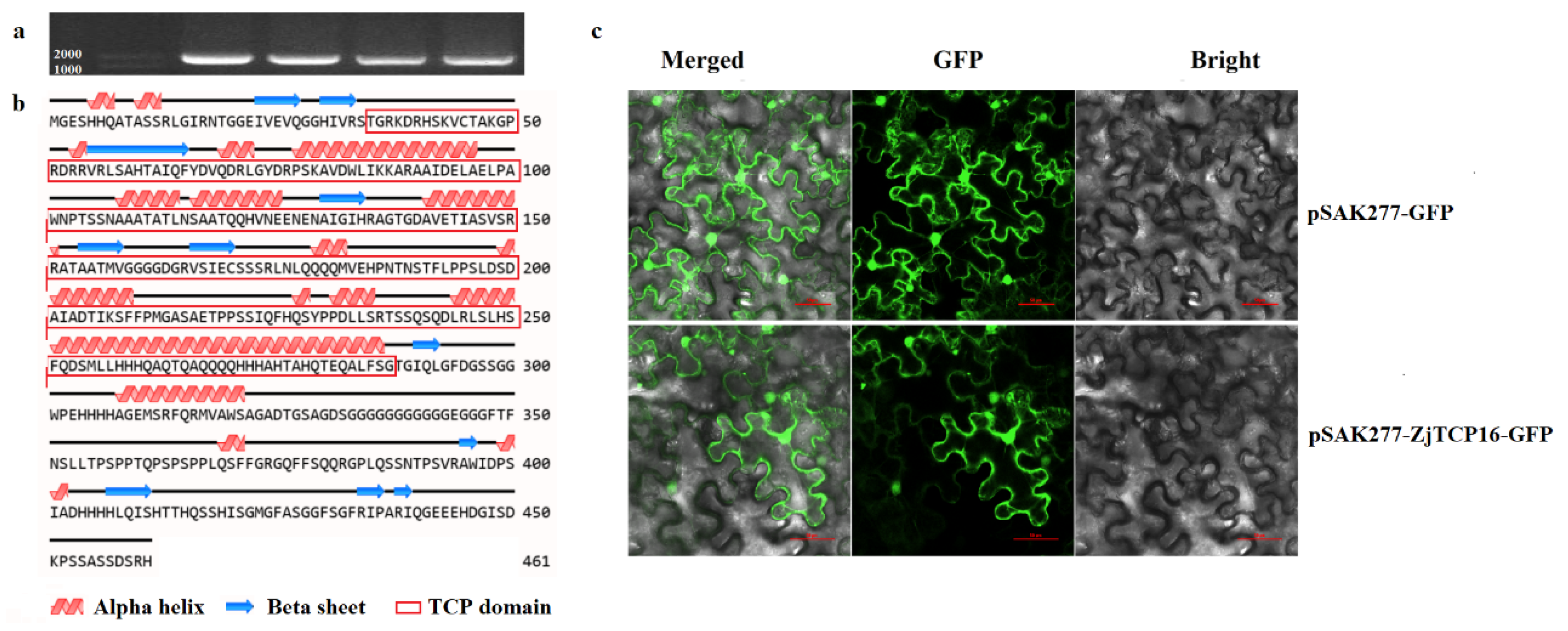
3.1. Sequence and Protein Structure Analysis and Subcellular Localization of ZjTCP16
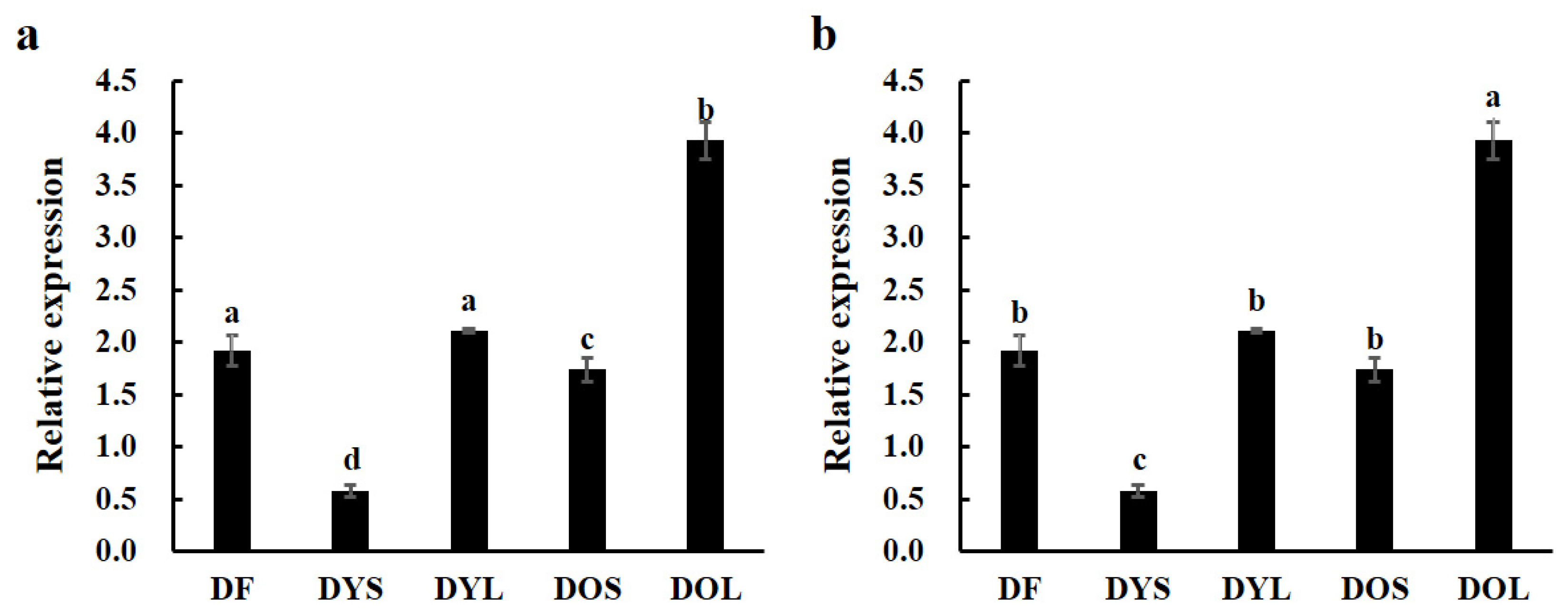
3.2. Expression of ZjTCP16 in Different Tissues of JWB-Positive and -Negative Jujube Trees
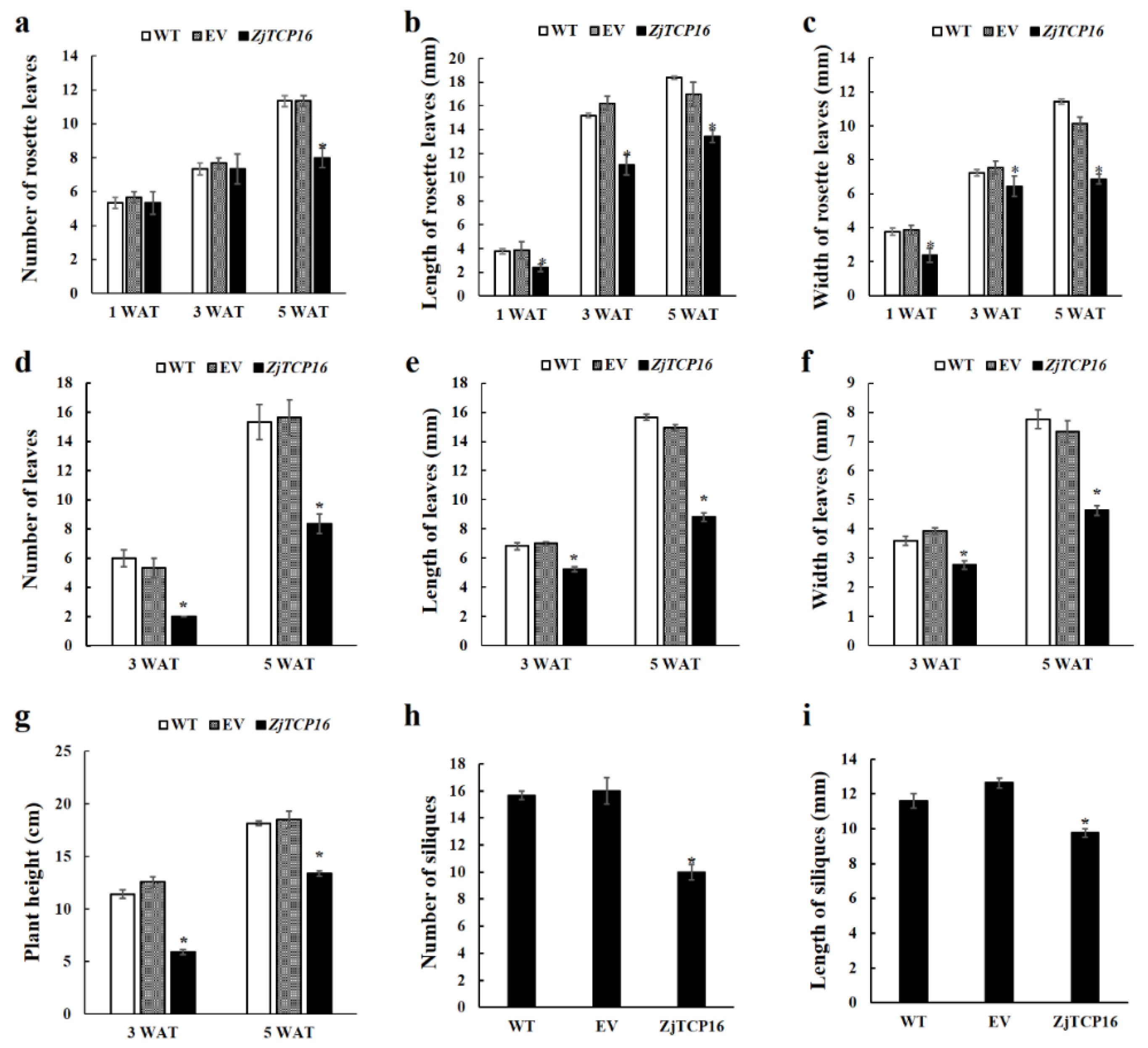
3.3. Overexpression of ZjTCP16 Resulted in Dwarfism and Smaller Leaves in Arabidopsis
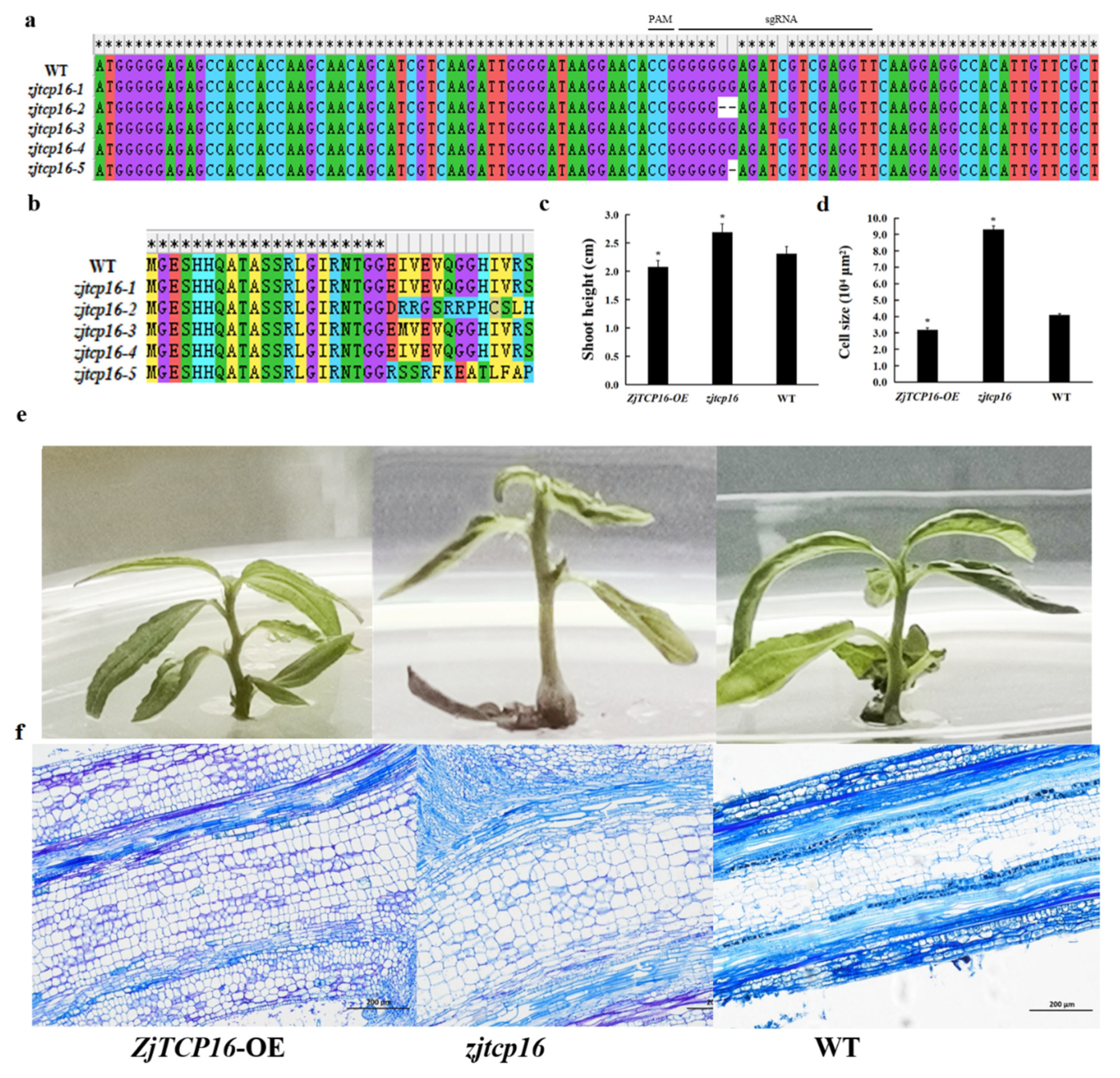
3.4. Alteration of ZjTCP16 Expression Regulated Jujube Growth and Cell Size in Z. Jujube Shoots
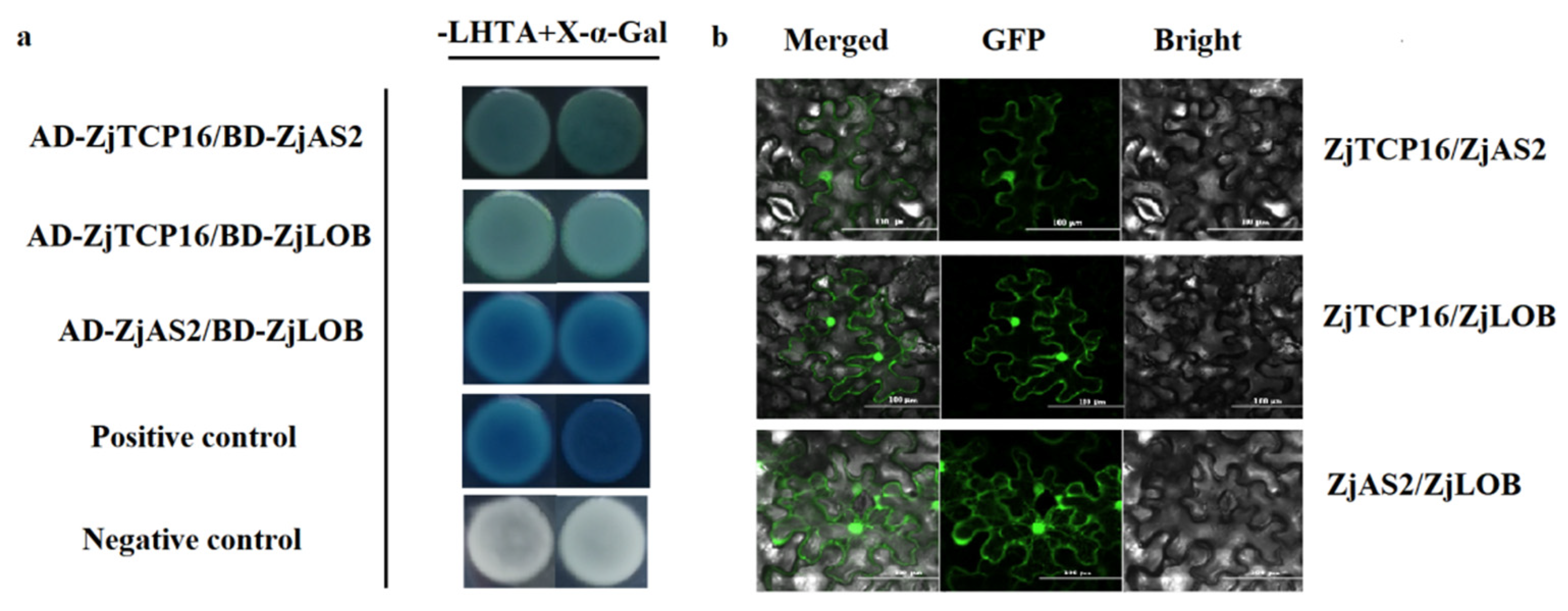
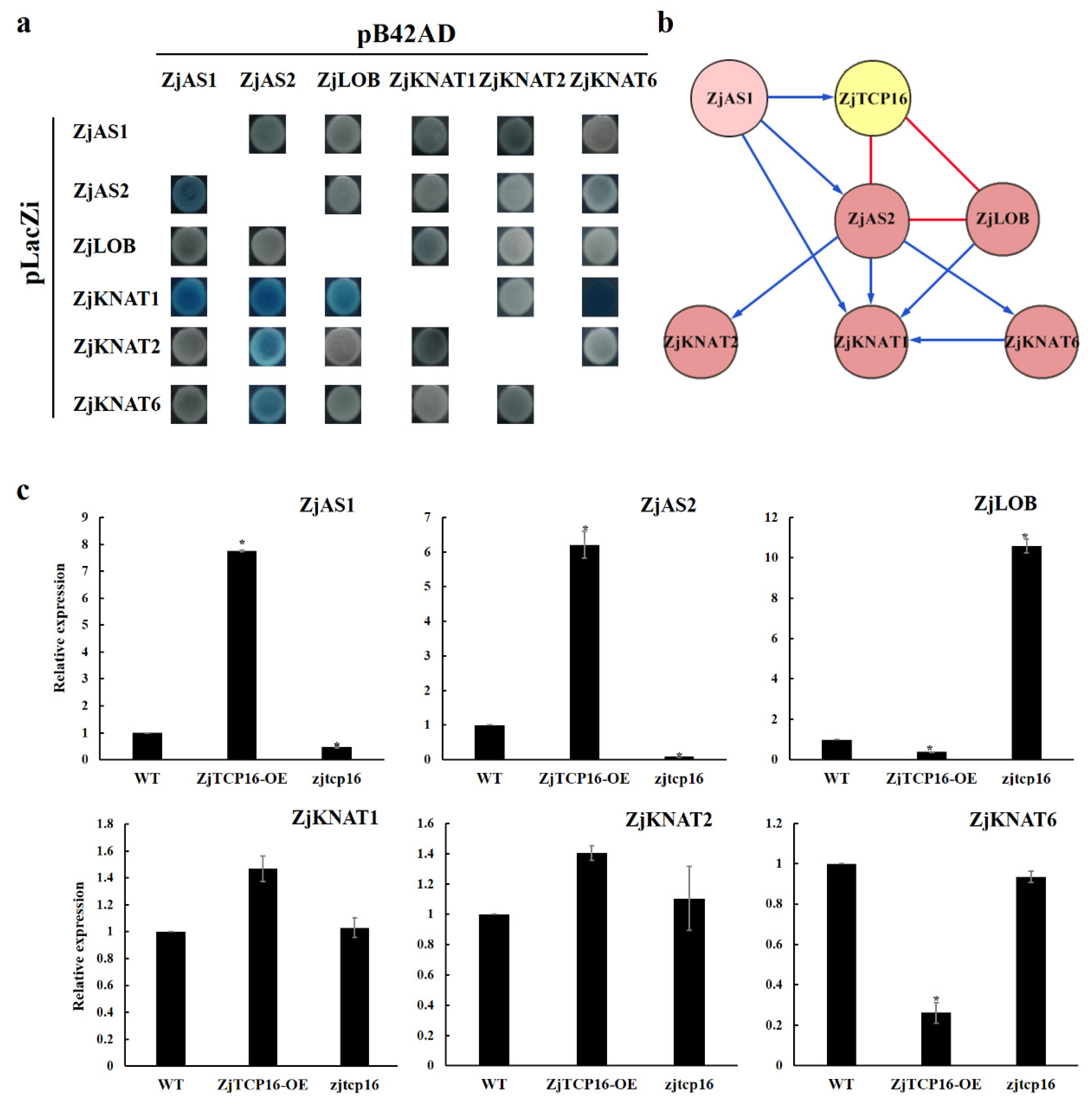
3.5. ZjTCP16 Interacted with ZjAS2 and ZjLOB
3.6. ZjTCP16 Altered the Expression Levels of Genes Involved in Plant Morphogenesis
4. Discussion
4.1. ZjTCP16 Induced Dwarf and Smaller Leaf Phenotypes by Changing the Expression of Morphogenesis-Related Genes
4.2. Mutation of ZjTCP16 by CRISPR/Cas9-Mediated Gene Editing
4.3. The Expression of ZjTCP16 in Response to JWB Phytoplasma
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yao, S. Past, Present, and Future of Jujubes-Chinese dates in the United States. HortScience A Publ. Am. Soc. Hortic. Sci. 2013, 48, 672–680. [Google Scholar] [CrossRef]
- Liu, M.; Wang, J.; Wang, L.; Liu, P.; Zhao, J.; Zhao, Z.; Yao, S.; Stănică, F.; Liu, Z.; Wang, L.; et al. The historical and current research progress on jujube–a superfruit for the future. Hortic. Res. 2020, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.Y.; Sawayanagi, T.; Kakizawa, S.; Nishigawa, H.; Wei, W.; Oshima, K.; Miyata, S.I.; Ugaki, M.; Hibi, T.; Namba, S. ‘Candidatus phytoplasma Ziziphi’, a novel phytoplasma taxon associated with jujube witches’-broom disease. Int. J. Syst. Evol. Microbiol. 2003, 53 Pt 4, 1037–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Wang, H.; Chen, P.; Fu, B.; Zhang, M.; Li, J.; Zheng, X.; Tan, B.; Feng, J. Combination of iTRAQ proteomics and RNA-seq transcriptomics reveals multiple levels of regulation in phytoplasma-infected Ziziphus jujuba Mill. Hortic. Res. 2017, 4, 17080. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Ye, X.; Li, J.; Tan, B.; Chen, P.; Cheng, J.; Wang, W.; Zheng, X.; Feng, J. Transcriptome profiling analysis revealed co-regulation of multiple pathways in jujube during infection by ‘Candidatus Phytoplasma ziziphi’. Gene 2018, 665, 82–95. [Google Scholar] [CrossRef]
- Chen, P.; Li, J.; Ye, X.; Tan, B.; Zheng, X.; Cheng, J.; Wang, W.; Wang, H.; Gu, L.; Feng, J. Genome-wide identification of ziziphus jujuba TCP transcription factors and their expression in response to infection with jujube witches’ broom phytoplasma. Acta Physiol. Plant. 2019, 41, 86. [Google Scholar] [CrossRef]
- Wang, H.; Ye, X.; Li, J.; Tan, B.; Chen, P.; Jiang, Y.; Cheng, J.; Wang, W.; Zheng, X.; Feng, J. Combination of iTRAQ proteomics and RNA-seq transcriptomics reveals jasmonate-related-metabolisms central regulation during the process of jujube witches’ broom recovery by tetracycline treatment. Sci. Hortic. 2019, 243, 197–206. [Google Scholar] [CrossRef]
- Li, J.D.; Chen, L.C.; Chen, P.; Li, Q.C.; Yang, Q.Q.; Zhang, Y.; Tan, B.; Ye, X.; Zheng, X.B.; Feng, J.C. Genome-wide identification and expression of the lipoxygenase gene family in jujube (Ziziphus jujuba) in response to phytoplasma infection. J. Plant Biochem. Biotechnol. 2021, 31, 139–153. [Google Scholar] [CrossRef]
- Sugio, A.; Kingdom, H.N.; MacLean, A.M.; Grieve, V.M.; Hogenhout, S.A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, E1254–E1263. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Yang, H.; Yin, Z.; Liu, W.; Sun, L.; Wu, Y. Phytoplasma effector SWP1 induces witches’ broom symptom by destabilizing the TCP transcription factor BRANCHED1. Mol. Plant Pathol. 2018, 19, 2623–2634. [Google Scholar] [CrossRef]
- Zhou, J.; Ma, F.; Yao, Y.; Deng, M.; Chen, M.; Zhang, S.; Li, Y.; Yang, J.; Zhang, N.; Huang, J.; et al. Jujube witches’ broom phytoplasma effectors SJP1 and SJP2 induce lateral bud outgrowth by repressing the ZjBRC1-controlled auxin efflux channel. Plant Cell Environ. 2021, 44, 3257–3272. [Google Scholar] [CrossRef]
- Chen, P.; Chen, L.; Ye, X.; Tan, B.; Zheng, X.; Cheng, J.; Wang, W.; Yang, Q.; Zhang, Y.; Li, J.; et al. Phytoplasma effector Zaofeng6 induces shoot proliferation by decreasing the expression of ZjTCP7 in Ziziphus jujuba. Hortic. Res. 2022, 9, uhab032. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.C.; Yu, X.M.; Shang, X.L.; Li, J.D.; Wu, Y.X. Factors influencing efficiency of shoot regeneration in Ziziphus jujuba Mill. ‘Huizao’. Plant Cell Tissue Organ Cult. 2010, 101, 111–117. [Google Scholar] [CrossRef]
- Gundersen, D.E.; Lee, I.M. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol. Mediterr. 1996, 35, 144–151. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 2010, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, M.; Shi, M.; Wang, Y.; Huang, Q.; Yuan, T.; Wang, Q.; Wang, C.; Zhou, W.; Kai, G. The biosynthesis of phenolic acids is positively regulated by the JA-responsive transcription factor ERF115 in Salvia miltiorrhiza. J. Exp. Bot. 2019, 70, 243–254. [Google Scholar] [CrossRef]
- Yan, P.; Zeng, Y.; Shen, W.; Tuo, D.; Li, X.; Zhou, P. Nimble Cloning: A Simple, Versatile, and Efficient System for Standardized Molecular Cloning. Front. Bioeng. Biotechnol. 2020, 7, 460. [Google Scholar] [CrossRef]
- Lopez, J.A.; Sun, Y.; Blair, P.B.; Mukhtar, M.S. TCP three-way handshake: Linking developmental processes with plant immunity. Trends Plant Sci. 2015, 20, 238–245. [Google Scholar] [CrossRef]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- Carrara, S.; Dornelas, M.C. TCP genes and the orchestration of plant architecture. Trop. Plant Biol. 2020, 14, 1–10. [Google Scholar] [CrossRef]
- Sarvepalli, K.; Nath, U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. Cell Mol. Biol. 2011, 67, 595–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Li, B.; Shen, W.H.; Huang, H.; Dong, A. TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 2012, 71, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Challa, K.R.; Aggarwal, P.; Nath, U. Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in arabidopsis. Plant Cell 2016, 28, 2117–2130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wu, S.; Xu, J.; Sui, C.; Wei, J. Application of CRISPR/Cas9 in plant biology. Acta Pharm. Sin. 2017, 7, 292–302. [Google Scholar] [CrossRef]
- Ramirez-Torres, F.; Ghogare, R.; Stowe, E.; Cerdá-Bennasser, P.; Lobato-Gómez, M.; Williamson-Benavides, B.A.; Giron-Calva, P.S.; Hewitt, S.; Christou, P.; Dhingra, A. Genome editing in fruit, ornamental, and industrial crops. Transgenic Res. 2021, 30, 499–528. [Google Scholar] [CrossRef]
- Ma, X.; Zhu, Q.; Chen, Y.; Liu, Y.G. CRISPR/Cas9 Platforms for Genome Editing in Plants: Developments and Applications. Mol. Plant 2016, 9, 961–974. [Google Scholar] [CrossRef] [Green Version]
- Rao, M.J.; Wang, L. CRISPR/Cas9 technology for improving agronomic traits and future prospective in agriculture. Planta 2021, 254, 68. [Google Scholar] [CrossRef]
- Muhr, M.; Paulat, M.; Awwanah, M.; Brinkkötter, M.; Teichmann, T. CRISPR/Cas9-mediated knockout of Populus BRANCHED1 and BRANCHED2 orthologs reveals a major function in bud outgrowth control. Tree Physiol. 2018, 38, 1588–1597. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, S.; Garner, C.M.; Gassmann, W. New clues in the nucleus: Transcriptional reprogramming in effector-triggered immunity. Front. Plant Sci. 2013, 4, 364. [Google Scholar] [CrossRef] [Green Version]
- Mukhtar, M.S.; Carvunis, A.R.; Dreze, M.; Epple, P.; Steinbrenner, J.; Moore, J.; Tasan, M.; Galli, M.; Hao, T.; Nishimura, M.T.; et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011, 333, 596–601. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Li, Q.; Gu, L.; Chen, P.; Zhang, Y.; Li, Y.; Chen, Y.; Ye, X.; Tan, B.; Zheng, X.; et al. The Jujube TCP Transcription Factor ZjTCP16 Regulates Plant Growth and Cell Size by Affecting the Expression of Genes Involved in Plant Morphogenesis. Forests 2022, 13, 723. https://doi.org/10.3390/f13050723
Yang Q, Li Q, Gu L, Chen P, Zhang Y, Li Y, Chen Y, Ye X, Tan B, Zheng X, et al. The Jujube TCP Transcription Factor ZjTCP16 Regulates Plant Growth and Cell Size by Affecting the Expression of Genes Involved in Plant Morphogenesis. Forests. 2022; 13(5):723. https://doi.org/10.3390/f13050723
Chicago/Turabian StyleYang, Qiqi, Qicheng Li, Liyuan Gu, Peng Chen, Yu Zhang, Yonghua Li, Yun Chen, Xia Ye, Bin Tan, Xianbo Zheng, and et al. 2022. "The Jujube TCP Transcription Factor ZjTCP16 Regulates Plant Growth and Cell Size by Affecting the Expression of Genes Involved in Plant Morphogenesis" Forests 13, no. 5: 723. https://doi.org/10.3390/f13050723
APA StyleYang, Q., Li, Q., Gu, L., Chen, P., Zhang, Y., Li, Y., Chen, Y., Ye, X., Tan, B., Zheng, X., Li, J., & Feng, J. (2022). The Jujube TCP Transcription Factor ZjTCP16 Regulates Plant Growth and Cell Size by Affecting the Expression of Genes Involved in Plant Morphogenesis. Forests, 13(5), 723. https://doi.org/10.3390/f13050723





