Chloroplast Microsatellite-Based High-Resolution Melting Analysis for Authentication and Discrimination of Ilex Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
2.2. Comparison of Chloroplast Genomes and Identification of Microsatellite Loci
2.3. Quantitative PCR and HRM Analysis of Candidate DNA Barcodes
3. Results
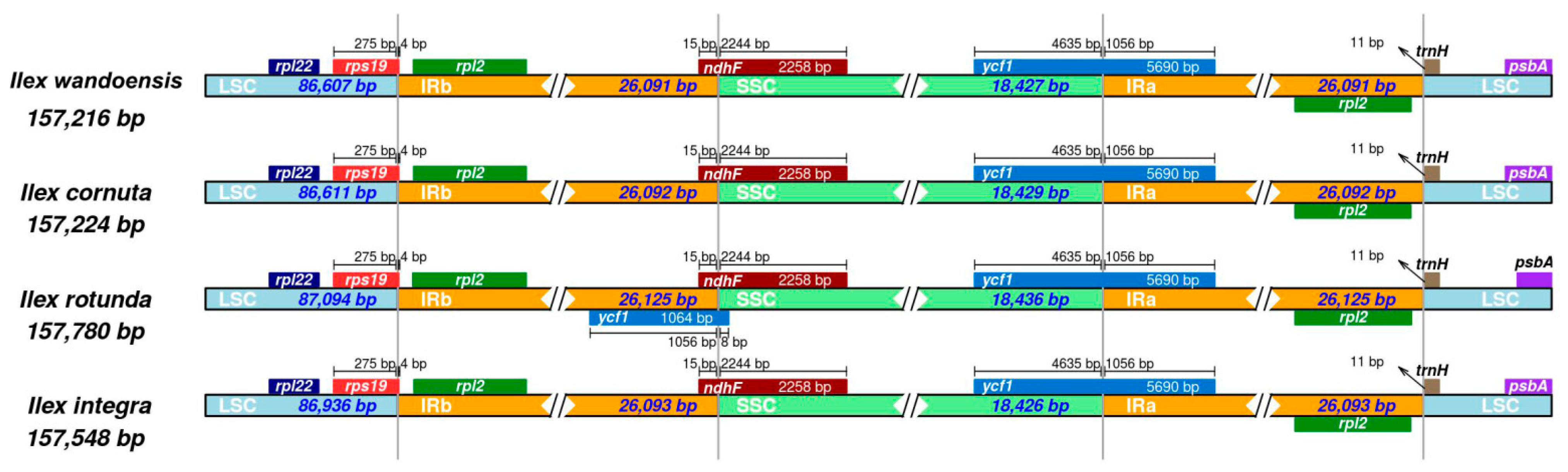
3.1. Comparative Analysis of the Chloroplast Genome of Four Ilex Species
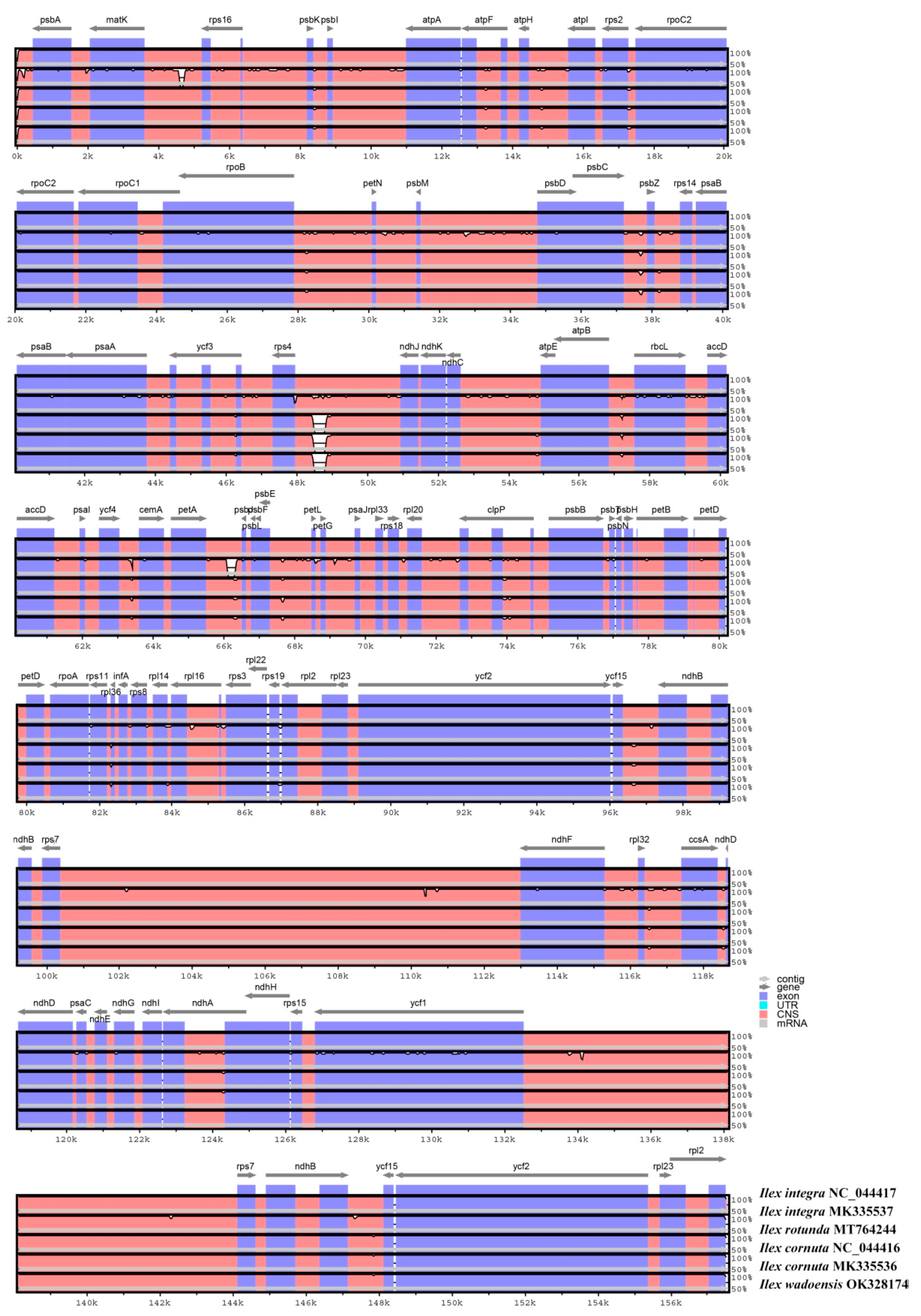
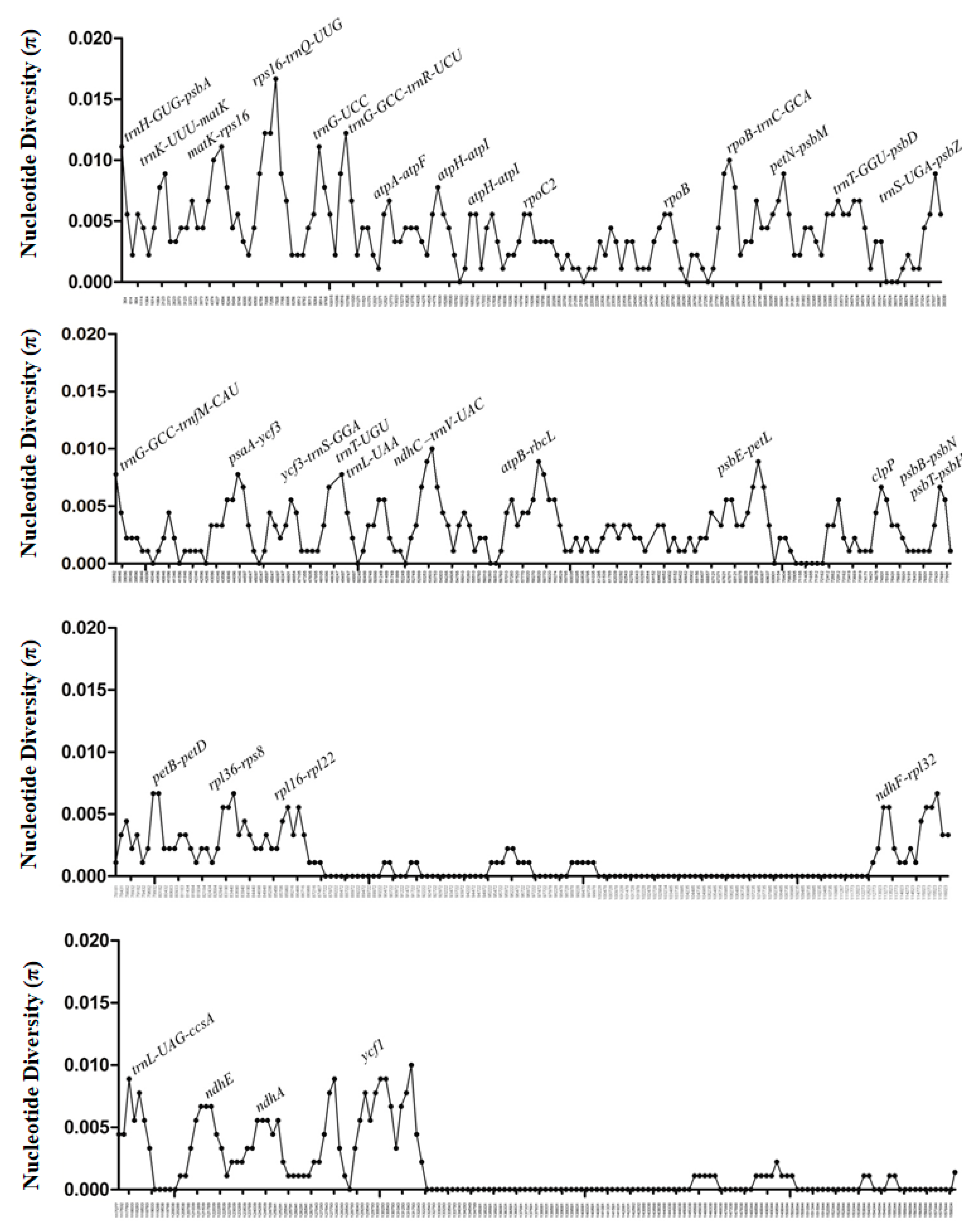
3.2. Divergence Hotspots in the Four Ilex Cp Genomes
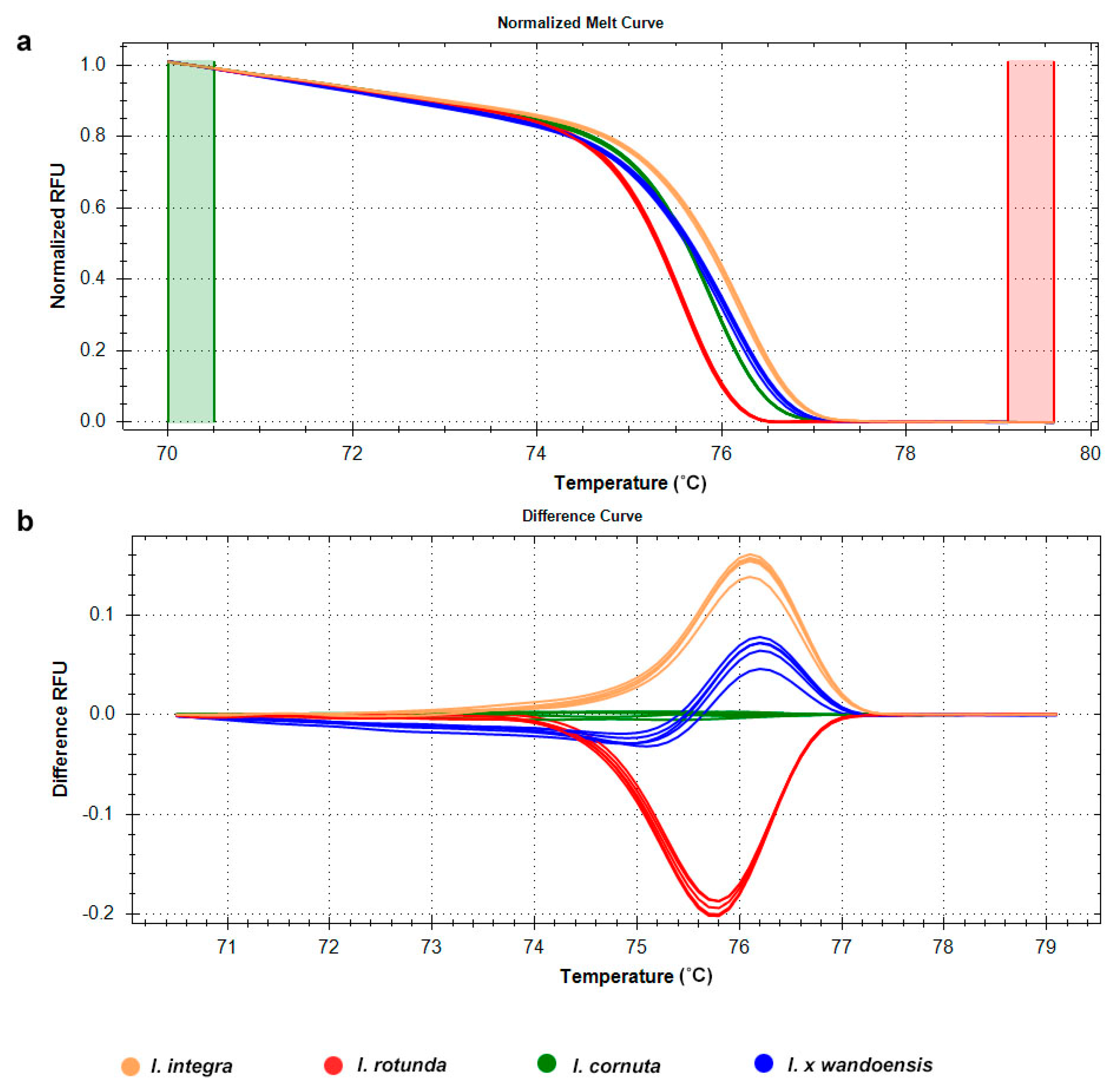
3.3. Microsatellite Genotyping of the Four Ilex Species Using HRM Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gan, R.-Y.; Zhang, D.; Wang, M.; Corke, H. Health Benefits of Bioactive Compounds from the Genus Ilex, a Source of Traditional Caffeinated Beverages. Nutrients 2018, 10, 1682. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.H.; Corrêa, R.C.; Barros, L.; Calhelha, R.C.; Santos-Buelga, C.; Peralta, R.M.; Bracht, A.; Matsushita, M.; Ferreira, I.C. Phytochemicals and bioactive properties of Ilex paraguariensis: An in-vitro comparative study between the whole plant, leaves and stems. Food Res. Int. 2015, 78, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Chaudhuri, P.K. Structural characteristics, bioavailability and cardioprotective potential of saponins. Integr. Med. Res. 2018, 7, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.A.; Connolly, J.D. Triterpenoids. Nat. Prod. Rep. 2020, 37, 962–998. [Google Scholar] [CrossRef]
- Correa, V.G.; de Sá-Nakanishi, A.B.; Gonçalves, G.d.A.; Barros, L.; Ferreira, I.C.F.R.; Bracht, A.; Peralta, R.M. Yerba mate aqueous extract improves the oxidative and inflammatory states of rats with adjuvant-induced arthritis. Food Funct. 2019, 10, 5682–5696. [Google Scholar] [CrossRef] [PubMed]
- Panza, V.P.; Brunetta, H.S.; de Oliveira, M.V.; Nunes, E.A.; da Silva, E.L. Effect of mate tea (Ilex paraguariensis) on the expression of the leukocyte NADPH oxidase subunit p47phox and on circulating inflammatory cytokines in healthy men: A pilot study. Int. J. Food Sci. Nutr. 2019, 70, 212–221. [Google Scholar] [CrossRef]
- Cahuê, F.; Nascimento, J.H.M.; Barcellos, L.; Salerno, V.P. Ilex paraguariensis, exercise and cardioprotection: A retrospective analysis. J. Funct. Foods 2019, 53, 105–108. [Google Scholar] [CrossRef]
- Rocha, D.S.; Casagrande, L.; Model, J.F.A.; Dos Santos, J.T.; Hoefel, A.L.; Kucharski, L.C. Effect of yerba mate (Ilex paraguariensis) extract on the metabolism of diabetic rats. Biomed. Pharmacother. 2018, 105, 370–376. [Google Scholar] [CrossRef]
- Luís, Â.F.S.; Domingues, F.D.C.; Amaral, L.M.J.P. The anti-obesity potential of Ilex paraguariensis: Results from a meta-analysis. Braz. J. Pharm. Sci. 2019, 55, e17615. [Google Scholar] [CrossRef]
- Zhu, F.; Cai, Y.Z.; Sun, M.; Ke, J.; Lu, D.; Corke, H. Comparison of major phenolic constituents and in vitro antioxidant activity of diverse Kudingcha genotypes from Ilex kudingcha, Ilex cornuta, and Ligustrum robustum. J. Agric. Food Chem. 2009, 57, 6082–6089. [Google Scholar] [CrossRef]
- Chen, G.; Han, Y.; Feng, Y.; Wang, A.; Li, X.; Deng, S.; Zhang, L.; Xiao, J.; Li, Y.; Li, N. Extract of Ilex rotunda Thunb alleviates experimental colitis-associated cancer via suppressing inflammation-induced miR-31-5p/YAP overexpression. Phytomedicine 2019, 62, 152941. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shen, Y.-J.; Tu, Q.-B.; Zhao, Y.-R.; Guo, H.; Wang, J.; Zhang, L.; Shi, H.-W.; Sun, Y. Pedunculoside, a novel triterpene saponin extracted from Ilex rotunda, ameliorates high-fat diet induced hyperlipidemia in rats. Biomed. Pharmacother. 2018, 101, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, K.; Harada, A. Character of warm-temperate Quercus forests in Asia. In Warm-Temperate Deciduous Forests around the Northern Hemisphere; Springer: Cham, Switzerland, 2015; pp. 27–80. [Google Scholar]
- Jin, E.J.; Yoon, J.H.; Choi, M.S.; Sung, C.H. Seasonal Changes in the Absorption of Particulate Matter and the Fine Structure of Street Trees in the Southern Areas, Korea: With a Reference to Quercus myrsinifolia, Quercus glauca, Quercus salicina, Camellia japonica, and Prunus× yedoensis. J. Korean Soc. For. Sci. 2021, 110, 129–140. [Google Scholar]
- Joung, Y.H.; Picton, D.; Park, J.O.; Roh, M.S. Molecular evidence for the interspecific hybrid origin of Ilex× wandoensis. Hortic. Environ. Biotechnol. 2011, 52, 516–523. [Google Scholar] [CrossRef]
- Ahn, Y.H.; Choi, C.H. The Ecological Characteristics of Native Habitat of Korean Native Wando Holly (llex X wandoensis). J. Environ. Sci. Int. 2007, 16, 1011–1018. [Google Scholar]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA barcoding: From gene to genome. Biol. Rev. 2015, 90, 157–166. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Graham, S.; Little, D.P. Choosing and Using a Plant DNA Barcode. PLoS ONE 2011, 6, e19254. [Google Scholar]
- Krawczyk, K.; Nobis, M.; Myszczyński, K.; Klichowska, E.; Sawicki, J. Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa). Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Wu, L.; Wu, M.; Cui, N.; Xiang, L.; Li, Y.; Li, X.; Chen, S. Plant super-barcode: A case study on genome-based identification for closely related species of Fritillaria. Chin. Med. 2021, 16, 1–11. [Google Scholar] [CrossRef]
- Yu, J.; Wu, X.; Liu, C.; Newmaster, S.; Ragupathy, S.; Kress, W.J. Progress in the use of DNA barcodes in the identification and classification of medicinal plants. Ecotoxicol. Environ. Saf. 2021, 208, 111691. [Google Scholar] [CrossRef]
- Song, M.; Li, J.; Xiong, C.; Liu, H.; Liang, J. Applying high-resolution melting (HRM) technology to identify five commonly used Artemisia species. Sci. Rep. 2016, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gesto-Borroto, R.; Medina-Jiménez, K.; Lorence, A.; Villarreal, M.L. Application of DNA Barcoding for Quality Control of Herbal Drugs and Their Phytopharmaceuticals. Rev. Bras. Farm. 2021, 31, 127–141. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Sudhir, K.K.C.; Satish, S.C.; Mohan, S.M.R.; Manisha, D.S.; Deepak, K.S.; Ruchi, B.S.; Amita, S.; Bipin, R.; Ashok, K. Chemical constituents and biological significance of the genus Ilex (Aquifoliaceae). Nat. Prod. J. 2012, 2, 212–224. [Google Scholar]
- Yi, F.; Zhao, X.L.; Peng, Y.; Xiao, P.G. Genus llex L.: Phytochemistry, ethnopharmacology, and pharmacology. Chin. Herb. Med. 2016, 8, 209–230. [Google Scholar] [CrossRef]
- Parveen, I.; Gafner, S.; Techen, N.; Murch, S.J.; Khan, I.A. DNA Barcoding for the Identification of Botanicals in Herbal Medicine and Dietary Supplements: Strengths and Limitations. Planta Medica 2016, 82, 1225–1235. [Google Scholar] [CrossRef]
- Dong, W.; Liu, H.; Xu, C.; Zuo, Y.; Chen, Z.; Zhou, S. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: A case study on ginsengs. BMC Genet. 2014, 15, 138. [Google Scholar] [CrossRef]
- Chen, Q.; Wu, X.; Zhang, D. Comparison of the abilities of universal, super, and specific DNA barcodes to discriminate among the original species of Fritillariae cirrhosae bulbus and its adulterants. PLoS ONE 2020, 15, e0229181. [Google Scholar] [CrossRef]
- Yik, M.; Kong, B.; Siu, T.-Y.; Lau, D.; Cao, H.; Shaw, P.-C. Differentiation of Hedyotis diffusa and Common Adulterants Based on Chloroplast Genome Sequencing and DNA Barcoding Markers. Plants 2021, 10, 161. [Google Scholar] [CrossRef] [PubMed]


| No. | Scientific Name | Common Name | Collection Site | Specimen Code |
|---|---|---|---|---|
| 1 | Ilex integra Thunb. | Kamtangnamu | 34°21′37.8″ N126°39′51.2″ E | NIBRGR0000630040 |
| 2 | 34°21′37.5″ N 126°39′51.5″ E | JINR000003111 | ||
| 3 | 34°21′37.4″ N 126°39′52.2″ E | JINR000003112 | ||
| 4 | 34°21′37.3″ N 126°39′52.5″ E | JINR000003113 | ||
| 5 | 34°41′12.1″ N 126°53′47.6″ E | JINR000003114 | ||
| 6 | Ilex rotunda Thunb. | Meonnamu | 33°18′09.4″ N 126°18′47.9″ E | NIBRGR0000429868 |
| 7 | 34°41′11.8″ N 126°53′48.8″ E | JINR000003115 | ||
| 8 | 34°21′44.5″ N 126°39′43.9″ E | JINR000003116 | ||
| 9 | 34°21′44.4″ N 126°39′43.7″ E | JINR000003117 | ||
| 10 | 34°21′44.2″ N 126°39′44.0″ E | JINR000003118 | ||
| 11 | Ilex cornuta Lindl.andPaxton | Horanggasinamu | 33°20′12.7″ N 126°12′03.5″ E | NIBRGR0000429502 |
| 12 | 33°20′00.6″ N 126°10′40.6″ E | NIBRGR0000630049 | ||
| 13 | 33°18′55.3″ N 126°10′33.4″ E | NIBRGR0000630051 | ||
| 14 | 35°00′23.2″ N 126°49′21.5″ E | JINR000003119 | ||
| 15 | 35°00′23.3″ N 126°49′24.3″ E | JINR000003120 | ||
| 16 | Ilex x wandoensis C.F. Mill. and M. Kim | Wandohoranggasinamu | 34°55′38.1″ N 127°30′06.6″ E | JINR000003121 |
| 17 | 35°00′23.5″ N 126°49′20.1″ E | JINR000003122 | ||
| 18 | 35°00′23.5″ N 126°49′20.2″ E | JINR000003123 | ||
| 19 | 34°21′38.3″ N 126°39′51.4″ E | JINR000003124 | ||
| 20 | 34°21′38.1″ N 126°39′51.8″ E | JINR000003125 |
| Category | Gene Group | Gene Name | ||||
|---|---|---|---|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4.5 a | rrn5 a | rrn16 a | rrn23 a | |
| Transfer RNA genes | trnA-UGC * | trnC-GCA | trnD-GUC | trnE-UUC | trnF-GAA | |
| trnfM-CAU | trnG-GCC | trnG-UCC * | trnH-GUG | trnI-CAU a | ||
| trnI-GAU a,* | trnK-UUU * | trnL-CAA a | trnL-UAA * | trnL-UAG | ||
| trnM-CAU | trnN-GUU a | trnP-UGG | trnQ-UUG | trnR-ACG a | ||
| trnR-UCU | trnS-GCU | trnS-UGA | trnS-GGA | trnT-UGU | ||
| trnT-GGU | trnV-GAC a | trnV-UAC * | trnV-GAC | trnW-CCA | ||
| trendy-GUA | ||||||
| Small ribosome subunit | rps2 | rps3 | rps4 | rps7 a | rps8 | |
| rps11 | rps12A * | rps12B a,** | rps14 | rps15 | ||
| rps16 * | rps18 | rps19 | ||||
| Large ribosome subunit | rpl2 a,* | rpl14 | rpl16 * | rpl20 | rpl22 | |
| rpl23 a | rpl32 | rpl33 | rpl36 | |||
| RNA polymerase | rpoA | rpoB | rpoC1 * | rpoC2 | ||
| Translation initiation factor | infA | |||||
| Photosynthesis | Photosystem I subunit | psaA | psaB | psaC | psaI | psaJ |
| ycf3 ** | ycf4 | |||||
| Photosystem II subunit | psbA | psbB | psbC | psbD | psbE | |
| psbF | psbH | psbI | psbJ | psbK | ||
| psbL | psbM | psbT | psbZ | |||
| Cytochrome b/f complex | petA | petB* | petD * | petG | petL | |
| petN | ||||||
| ATP synthase | atpA | atpB | atpE | atpF* | atpH | |
| atpI | ||||||
| Rubisco large chain | rbcL | |||||
| NADH dehydrogenase | ndhA * | ndhB a,* | ndhC | ndhD | ndhE | |
| ndhF | ndhG | ndhH | ndhI | ndhJ | ||
| ndhK | ||||||
| Other genes | Maturase | matK | ||||
| Membrane protein | cemA | |||||
| Acetyl-CoA carboxylase | accD | |||||
| Cytochrome c biogenesis | ccsA | |||||
| ATP-dependent protease | clpP ** | |||||
| TIC complex component | ycf1 a | |||||
| Genes of unknown functions | Open reading frames | ycf2 | ycf15 | |||
| Locus | Region | Repeat Motif | Sequence (5’-3’) | Tm (°C) | Size (bp) |
|---|---|---|---|---|---|
| trnSUGA-psbZ | IGS | (T/A)(A)4(A/T/G/C)(A)n(T)2 (T/G)(C)2(A)n(C)n | TGCATGCCCATTTGTGAAA CATTTGATCCCTCTATCAGCCA | 60 | 170–180 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Oh, D.-R.; Kim, Y.-J.; Oh, K.-N.; Bae, D. Chloroplast Microsatellite-Based High-Resolution Melting Analysis for Authentication and Discrimination of Ilex Species. Forests 2022, 13, 1718. https://doi.org/10.3390/f13101718
Kim Y, Oh D-R, Kim Y-J, Oh K-N, Bae D. Chloroplast Microsatellite-Based High-Resolution Melting Analysis for Authentication and Discrimination of Ilex Species. Forests. 2022; 13(10):1718. https://doi.org/10.3390/f13101718
Chicago/Turabian StyleKim, Yonguk, Dool-Ri Oh, Yu-Jin Kim, Kyo-Nyeo Oh, and Donghyuk Bae. 2022. "Chloroplast Microsatellite-Based High-Resolution Melting Analysis for Authentication and Discrimination of Ilex Species" Forests 13, no. 10: 1718. https://doi.org/10.3390/f13101718
APA StyleKim, Y., Oh, D.-R., Kim, Y.-J., Oh, K.-N., & Bae, D. (2022). Chloroplast Microsatellite-Based High-Resolution Melting Analysis for Authentication and Discrimination of Ilex Species. Forests, 13(10), 1718. https://doi.org/10.3390/f13101718






