Abstract
ICE (inducer of CBF expression) is a positive regulator of cold signaling pathway in plants. Identification of ICE transcription factors is important for the sustainable development of the natural rubber planting industry in nontraditional regions where sudden cold waves often occur. In this study, five ICE genes were isolated from genome of rubber tree (Hevea brasiliensis Muell. Arg.) for analysing tolerance to cold stress. They shared an ICE-specific region in the highly conserved bHLH-ZIP domain and were localized in the nucleus. The HbICEs were different in transcript abundance and expression patterns in response to cold and drought stresses and among different rubber tree clones. Generally, the expression level of HbICEs was significantly higher in the cold-tolerant rubber tree clones than that in the cold-sensitive rubber tree clones. Overexpression of HbICE1, HbICE2, and HbICE4 significantly enhanced the cold tolerance of transgenic Arabidopsis and tobacco, which showed a significant increase in chlorophyll content and decrease in relative water content and conductivity at the early stage of cold stress in comparison with wild-type plants. Furthermore, overexpression of HbICE2 and HbICE4, but also HbICE1 enhanced drought tolerance in transgenic Arabidopsis. The cold tolerance of rubber tree clones is positively controlled by the expression level of HbICE1, HbICE2, and HbICE4.
1. Introduction
Plants have evolved efficient and sophisticated mechanisms involving altered physiological and biochemical processes to adapt to cold and drought stress. These changed physiological and biochemical processes are involved in changes of gene expression profiles. For example, inducer of CBF expressions (ICEs) are MYC-like basic helix-loop-helix (bHLH) transcription factors regulating plant responses to cold and drought stress. They contain some very similar motifs, such as acidic domain, Ser-rich domain, bHLH domain, zipper region, and dimerization domain [1,2,3]. Alignments of the bHLH domains of various plant ICE homologs exhibit a highly conserved 19 amino acids sequence (KMDRASILGDAI(D/E)YLKELL) that is specific to ICEs but not to other MYC-like proteins [3,4]. AtICE1 regulates the freezing tolerance of Arabidopsis [5] and enhances cold and drought tolerance in transgenic indica rice [6]. ZjICE1 in Zoysia japonica, PtrICE1 in trifoliate orange, and ICEs (TaICE1a and TaICE1d) in wheat are induced by drought and cold treatment [7,8,9]. The rice OsICE1 confers enhanced cold resistance of transgenic Arabidopsis [10]. Tomato SlICE1a, a MYC-type ICE1-like transcription factor, enhances cold, salt, and osmotic resistance in transgenic tobacco [11]. Ectopic expression of Vitis amurensis ICE1 and ICE2 improves tolerance to cold stresses in Arabidopsis [12]. Under cold stress, ICEs are activated and specifically bind to the cis-elements (CANNTG) that are present in the promoters of DREB1/CBF (dehydration-responsive element-binding protein 1/C-repeat binding factor), and they form the ICE-DREB1/CBF transcriptional regulatory cascade pathway [2,3,13]. The signal pathway ICE-DREB1/CBF plays a key role in the cold responses [14,15,16]. In Arabidopsis, the ICE-DREB1/CBF pathway is activated by cold stress, and CBF1, CBF2, and CBF3 transcriptional factors induce transcription of a large cluster of downstream COR (cold regulated) genes, leading to the improved cold resistance [14,15,17,18].
JAZs (jasmonate-ZIM domain) are repressors of the jasmonate acid signaling pathway that physically interact with the MYC-like protein to inhibit its transcriptional activity. For example, in Arabidopsis, overexpression of JAZ1 or JAZ4 inhibits the cold signaling pathway (ICE-DREB1/CBF) and cold stress tolerance [19]. The F-box protein COI1 (coronatine insensitive 1) acts as the jasmonate acid receptor that perceives the bioactive phytohormone JA-Ile (jasmonoyl-isoleucine) and recruits JAZ repressors for degradation, leading to activation of ICE, MYC, and DREB transcription factors [5]. JA serves as an upstream positive regulator of the cold signaling pathway through up-regulating ICE and DREB1/CBF to cope with cold stress in plants. SnRK2.6 (SNF1-related protein kinases), which is known as OPEN STOMATA 1 (OST1), serves as a positive regulator of the ABA signaling pathway [19]. It is reported that the SnRK2.6/OST1 physically interacts with ICE1 for its phosphorylation to enhance its transcriptional activity and stability under cold stress [20]. Therefore, ABA acts as a positive regulator of the ICE-DREB1/CBF cold signaling pathway to modulate cold stress tolerance in plants. SIZ1 (SAP and MIZ SUMO E3 ligase) is a SUMO E3 ligase which modulates tolerance to cold stresses in Arabidopsis [21]. SIZ1 physically interacts with ICE1 for its sumoylation to increase its stability under cold stress, which modulates the ICE-DREB1/CBF transcriptional regulatory cascade pathway and cold stress responses [22]. In addition, several key regulators, such as CAMTA3 (Calmodulin binding transcription activator 3) [23], HOS1 (High expression of osmotically responsive genes 1) [24], and COLD1 [25] also modulate the ICE-DREB1/CBF pathway and cold stress tolerance in plants.
Rubber tree is a typical tropical economic crop which is a significant source of natural rubber worldwide. Abiotic stresses such as cold and drought stresses adversely affect the natural rubber tree planting industry. The traditional rubber planting area of rubber tree has a dry season and wet seasons, and therefore the rubber tree faces drought stress. In order to satisfy the demand for natural rubber, rubber tree plantation has been extended from traditional rubber planting areas to non-traditional rubber planting areas; therefore, in addition to drought stress, the rubber tree also faces cold stress. The rubber tree plantation in China is in the northern edge of the world’s tropics and frequently suffers from both drought and sudden, extreme cold waves, which seriously threaten the sustainable development of natural rubber planting industry. Studying the resistance mechanism of the rubber tree has important practical value for improving the stress resistance of high-yield varieties by means of genetic engineering. In this study, five HbICE genes were identified and cloned from the rubber tree clone 93-114. Then, bioinformatic analyses of the ICEs’ evolutionary relationship and the structure of HbICE genes and encoded proteins were performed. The functional identification of HbICE genes was conducted by their differentially expression profiles in cold-tolerant and cold-sensitive rubber tree clones, subcellular localization, and overexpression in Arabidopsis and tobacco.
2. Materials and Methods
2.1. Plant Materials
Grafted plantlets of four cold-tolerant rubber tree clones (INA873, 93-114, Zhanshi327-13, and GT1) and four cold-sensitive rubber tree clones (Reken515, Reken501, Haiken1, and Reken514) were bred on the breeding base of Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences on Hainan Island, China. When the plantlets had formed one extension unit with completely matured leaves in the high temperature (25–35 °C) and humidity (70%–85%) shade of the breeding base, they were used for subsequent stress treatment.
2.2. Stress Treatments
To analyse gene expression in four cold-tolerant rubber tree clones and four cold-sensitive rubber tree clones, the treatment of the grafted plantlets was performed under cold stress in accordance with a published protocol [26]. Bark samples were collected from five plantlets of each rubber tree clone at 0 h, 4 h, 8 h, and 24 h of treatment and mixed as one sample. Triplicate was conducted at each time point. The control was conducted in the same manner while the plantlets remained at 28 °C.
For gene expression analysis in response to drought stimuli, plantlets of rubber tree clone 93-114 were placed into a conviron PGR15 growth chamber (Winnipeg, Canada) with a constant temperature of 28 °C, light of 16 h at 125 μmol·m−2·s−1, and relative humidity of 80%. Drought stress was performed by placing the bare-rooted plantlets in the chamber at 28 °C. Leaf samples were respectively collected from fifteen plantlets at 0 h, 2 h, 4 h, 8 h, 12 h, and 24 h under treatments. The five leaves from five plantlets at the same interval time were mixed as one sample. Experiments were performed independently in triplicate. As control, plantlets remained at 28 °C without any treatments.
2.3. DNA and RNA Isolation and cDNA Synthesis
Extraction of genomic DNA was performed using the DNA secure Plant Kit (Tiangen, Beijing, China) according to the manufacture’s protocol. Total RNA was extracted and remaining DNA was eliminated using the RNAprep Pure Plant Kit (Tiangen, Beijing, China). The concentration and quality assay of the RNA was performed in accordance with the described protocol [27]. First-strand cDNA was synthesised from 1 µg RNA using Fastking gDNA Dispelling RT SuperMix Fastking (Tiangen, Beijing, China).
2.4. Open Reading Frame (ORF) Cloning
Candidate HbICEs were cloned from the rubber tree DNA to analyse the gene structures. The full-length sequences of ICE ORFs were cloned using ICE-specific primers (Supplementary Table S1) by PCR and confirmed by sequencing. The amplification of ICE genes was performed according to the described protocol [27].
2.5. Bioinformatic Analysis
Multiple-sequence alignment of ICE proteins from Vitis vinifera, Populus trichocarpa, Arabidopsis thaliana, Jatropha curcas, Ricinus communis, and rubber tree was performed using the DNAMAN software (Lynnon Biosoft, QC, Canada). The phylogenetic tree was generated using the full-length amino acid sequences of ICE proteins from Populus trichocarpa, Camellia sinensis, Solanum lycopersicum, Arabidopsis, Plantain, Triticum aestivum, rice, and rubber tree by MEGA5.10 in accordance with the described protocol [27]. The theoretical iso-electric point and protein molecular weight of HbICE proteins were analyzed using Expasy software. The exon-intron structures of HbICE genes were analyzed by the GSDS 2.0 software, which aligned the open reading frames with the corresponding full-length sequences.
2.6. Quantitative Expression Analysis
Quantitative real-time PCR (qPCR) was carried out with 2 ×TB GreenTM Premix Ex TaqTM II (TliRNaseH Plus) (TaKaRa, Shiga, Japan) through the CFX384 Real-Time System (Bio-Rad, Herakles, CA, USA) according to the described protocol [26]. Each sample used three biological replicates, and each biological replicate used three technical replicates for qPCR analysis. The ICE-specific primers for qPCR and the primers of reference genes (HbACTIN7a, HbRH8, and HbUBC4) are all listed in Supplementary Table S1.
2.7. Subcellular Localization Analysis
Entire coding sequences of HbICE family members without stop codons were amplified with specific primers (Supplementary Table S1). The encoding sequences of HbICE1, HbICE2, HbICE3, HbICE4, and HbICE5 were constructed into the pCAMBIA1302 vector, respectively. HbICE1::GFP, HbICE2::GFP, HbICE3::GFP, HbICE4::GFP, and HbICE5::GFP recombinant plasmids and the empty pCAMBIA1302 vector were introduced into living onion epidermal cells by Agrobacterium GV3101 as previously described [26]. The localization of fusion proteins analysis was carried out using a laser confocal scanning microscope (Zeiss LSM 800, Germany) after 24 h incubation in the dark. The nucleus was stained by DAPI.
2.8. Genetic Transformation of Arabidopsis and Tobacco
The ORFs of HbICE genes were constructed into the pCAMBIA1302 vector using specific primers (Supplementary Table S1), respectively. The tobacco (Nicotiana benthamiana) leaf disks were utilised to introduce 35S::HbICE1, 35S::HbICE2, 35S::HbICE3, 35S::HbICE4, and 35S::HbICE5 into tobacco through Agrobacterium strain GV3101 as previously described [28]. The floral-dipping was used to introduce 35S::HbICE1, 35S::HbICE2, 35S::HbICE3, 35S::HbICE4, and 35S::HbICE5 into Arabidopsis thaliana (Columbia) through the Agrobacterium strain GV3101 as previously described [29]. Transformants screening were performed on MS medium with 25 mg liter–1 hygromycin, and resistant transformants were placed to seedling cup with nutrient soil. Transgenic T2 seedlings were further identified using genomic RT-PCR. Wild-type plants (WT) and transgenic T2 seedlings were grown in the chamber with a 12 h photoperiod and light intensity of 200 μmol photon m−2 s−1 at 24 °C.
2.9. Analysis of Cold Tolerance and Drought Tolerance
To evaluate freezing tolerance, 2-week-old Arabidopsis plants (WT and transgenic lines) and 4-week-old tobacco seedlings (WT and transgenic lines) grown under normal growth conditions were treated in a low-temperature chamber at −5 °C for 20 h and 15 h respectively, then transferred to the chamber at 23/21 °C (day/night) with a 12 h photoperiod and light intensity of 200 μmol photon m−2 s−1. The survival ratio was calculated when the plants were recovered at 14 days and 7 days under normal conditions, respectively. Wild-type Columbia Arabidopsis and tobacco were used as controls. To investigate the drought tolerance, 2-week-old WT and HbICEs-overexpressed Arabidopsis plants were not watered for 20 days, followed by normal watering in a chamber. Then, the survival ratio calculation of WT and transgenic plants was performed.
2.10. Physiological Parameter Determination
5-week-old Arabidopsis plants and 4-week-old tobacco plants were treated under −5 °C for 1 h, then the chlorophyll content (SPAD value) was measured with SPAD-502 Plus chlorophyll meter. After cold treatment, the leaves of seedlings were collected, and relative electrolyte leakage analysis was performed according to Verma et al. [6]. 0.2 g clean leaves from the same part of each strain were cut into 5 mm × 5 mm pieces and put into a centrifuge tube containing 30 mL ultrapure water. The transgenic Arabidopsis lines cultured in the seedling cup for 5 weeks were treated in a low-temperature incubator at −5 °C for 1 h. The chlorophyll content (SPAD), relative water content, and relative electrical conductivity of the leaves were measured. The fresh weight M1 of leaves in the same part of each strain was measured, and then the chopped leaves were put into a 50 mL centrifuge tube containing deionized water, soaked for 5–6 h, and the leaves absorbed water until saturated. Leaves were removed, and absorbent paper was used to absorb the residual water on the leaves, and then the weight M2 of the leaves was measured. Finally, the leaves were placed into an oven at 85 °C, and dried overnight to constant weight. The drying blade M3 was weighed. Relative water content = (M1 − M3)/(M2 − M3) × 100%. 0.2 g of clean leaves from the same part of each strain were cut into 5 mm × 5 mm pieces and put into a centrifuge tube containing 30 mL ultrapure water. They were placed in a vacuum dryer and vacuumed for 10 min, removed from the vacuum dryer, left at room temperature for 30 min, and oscillated once every 5 min. The prepared conductivity meter was used to measure the conductivity L1. The centrifuge tube was put into boiling water for 20 min, oscillated twice during the process, removed and cooled to room temperature, and then the conductivity L2 was measured. Relative conductivity = L1/L2 × 100%.
2.11. Statistical Analysis
Ct values are shown as mean ± standard deviation (S.D.). The calculation of relative HbICE expression was carried out using the 2−ΔΔCT method. The statistical significance of relative expression, physiological parameter, and survival ratio was analysed using SPSS software. p < 0.05 and <0.01 were considered significant and highly significant, respectively.
3. Results
3.1. Genome-Wide Identification of HbICEs
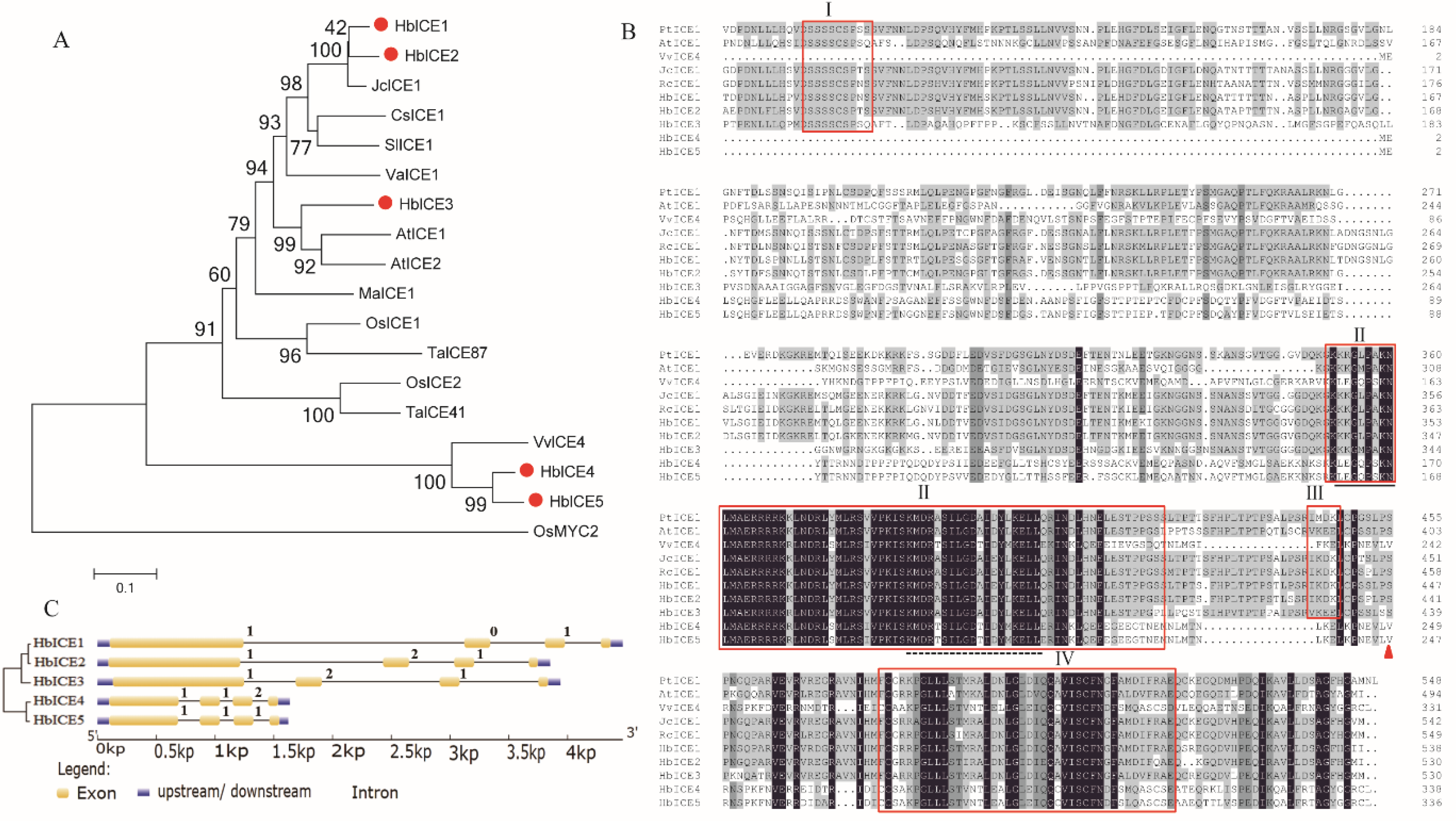
The full-length cDNAs of HbICE genes were cloned from rubber tree 93-114 and their names were shown on the basis of the phylogenetic relationship between rubber tree and other plant species. The critical features of HbICE nucleotide and protein sequences are shown in Supplementary Table S2. The coding sequences of HbICE genes exhibited the range of 1011 bp (HbICE5) to 1617 bp (HbICE1), the encoded proteins ranged from 336 to 538 amino acid residues, with corresponding molecular masses of 37.89–59.00 kDa, with the predicted isoelectric points of 4.75 (HbICE5) to 5.86 (HbICE2). To characterise the evolutionary relationship between rubber tree ICE proteins and other ICEs, a phylogenetic relationship tree was constructed on the basis of the amino acid sequence of ICEs from rubber tree and other plant species (Figure 1A). The results showed that HbICE1 and HbICE2 of rubber tree had close homology with JcICE1 of Jatropha curcas. The protein sequence of rubber tree HbICE3 was highly homologous to that of Arabidopsis AtICE1 and AtICE2. Rubber tree HbICE4 and HbICE5 had highly homology with Vitis vinifera VvICE4.
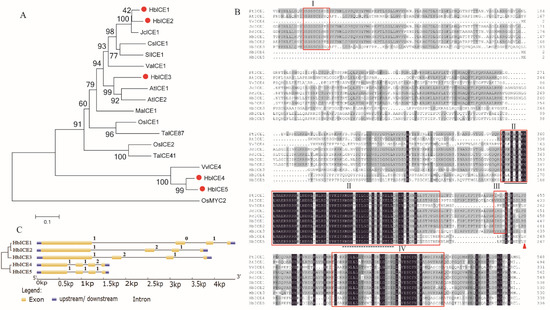
Figure 1.
Phylogenetic tree, alignment, and gene structure of five HbICEs. Phylogenetic tree was constructed according to the deduced amino acid sequences through MEGA5.10 program (A). GenBank accession numbers listed in parentheses: rubber tree (HbICE1, KY406162; HbICE2, KY406163; HbICE3, KY406164; HbICE4, KY406165; HbICE5, KY406166), rice (OsICE1, XP_015616517; OsICE2, KAF2954112; OsMYC2, XP_015614012), Arabidopsis (AtICE1, AT3G26744; AtICE2, AT1G12860), Populus trichocarpa (PtICE1, XP_002318166), Camellia sinensis (CsICE1, ACT90640), Solanum lycopersicum (SlICE2, AGG38826), Triticum aestivum (TaICE41, ACB69501; TaICE87, ACB69502), Vitis vinifera (VvICE1, AGP04217; VvICE4, AHM24956), and Jatropha curcas (JcICE1, NP_001306859). Multiple alignment of the deduced amino sequence of HbICEs with typical ICE proteins from other plants (B). Identical and similar residues are shaded black and gray, respectively. The conserved fragments, such as S-rich region (box I), bHLH-ZIP domain (box II), the putative SUMO conjugation motif (box III), and ACT-like domain (box IV), are specifically found in ICE proteins and asterisks indicate serine, a key residue for the protein stabilization of ICEs. The black line below the sequence represents the nuclear localization signal (NLS). The dotted line indicates a conserved region of ICEs. The origin and GenBank accession numbers are as follows: Vitis vinifera (VvICE4, AHM24956), Populus trichocarpa (PtICE1, XP_002318166), Arabidopsis (AtICE1, AT3G26744), Jatropha curcas (JcICE1, NP_001306859), Ricinus communis (RcICE1, XP_002511101). Gene structures of HbICEs (C).
Alignment of rubber tree HbICEs with other plant ICE proteins shown that HbICE proteins shared highly conservative regions in their C-terminal region (Figure 1B), such as the bHLH-ZIP domain and ACT-like domain. The bHLH-ZIP domain was a functional domain of HbICEs, which directly bound to cis-elements in the downstream gene promoter to regulate its transcription level. In the bHLH-ZIP domain of HbICEs, there was a conserved region composed of 19 amino acid residues (KMDRA/TSILGDA/TID/EYLKELL). In addition, there was a nuclear localization signal composed of 20 amino acid residues in the bHLH-ZIP domain, which guided the protein into the nucleus. The ACT-like domain was also present in the C terminus of HbICE proteins, which participated in the dimerization of several bHLH transcription factors. The conserved S-rich region (SSSSCS) was present in motif 9 of the HbICE1, HbICE2, and HbICE3 N-terminal region, but not in HbICE4 and HbICE5. The putative SUMO (sumoylation) conjugation motif and phosphorylation site were present in the C-terminal region of HbICE1, HbICE2, and HbICE3, but not of HbICE4 and HbICE5 (Figure 1B).
Exon-intron organization identification of the 5 HbICE revealed that all HbICE genes possessed four exons and three introns. The introns were inserted into the bHLH-ZIP domain and ACT-like domain. Most introns were phase-one introns, each of HbICE2, HbICE3, and HbICE4 genes had phase-two introns, and the HbICE1 gene had a phase-zero intron (Figure 1C).
3.2. Subcellular Localization of HbICE Members
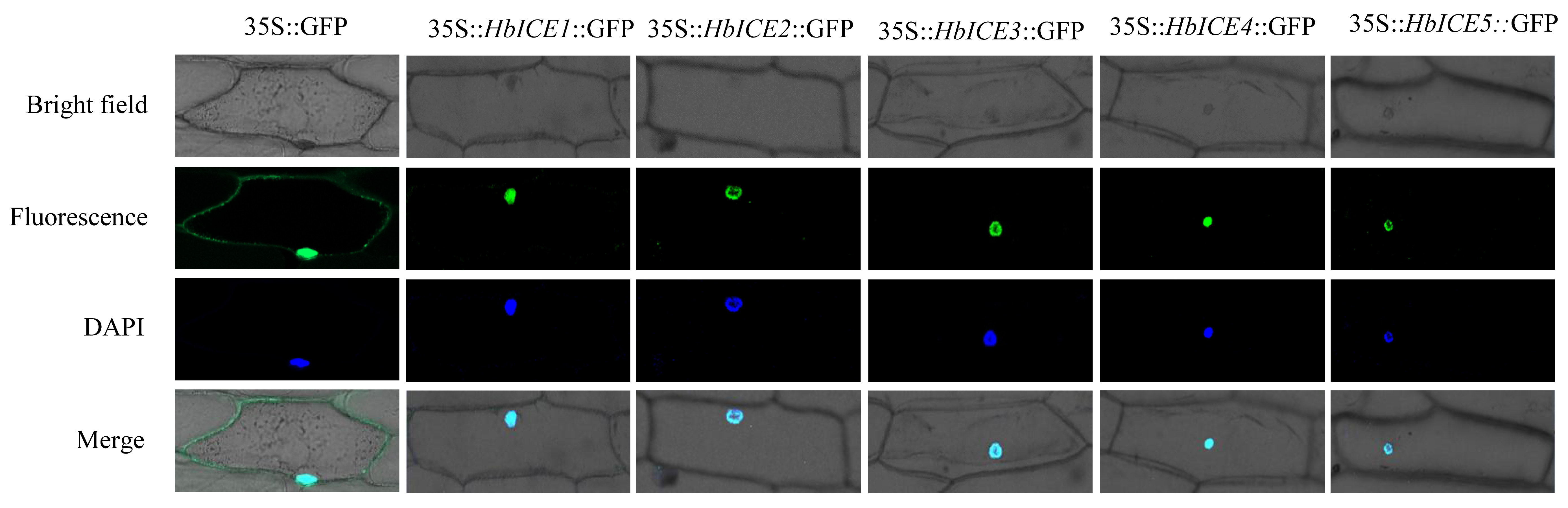
HbICE1, HbICE2, HbICE3, HbICE4, and HbICE5 were inserted into the pCAMBIA1302 vector respectively, and the HbICE proteins could be fused with GFP. The subcellular localizations of HbICE1, HbICE2, HbICE3, HbICE4, and HbICE5 were performed in living onion epidermal cells, respectively. The subcellular localization results showed that the onion epidermal cells containing the empty vector exhibited fluorescence in the whole cells, while the green fluorescence signal was specifically tested in the nucleus of the onion cells containing 35S::HbICE1::GFP, 35S::HbICE2::GFP, 35S::HbICE3::GFP, 35S::HbICE4::GFP, and 35S::HbICE5::GFP, respectively (Figure 2).
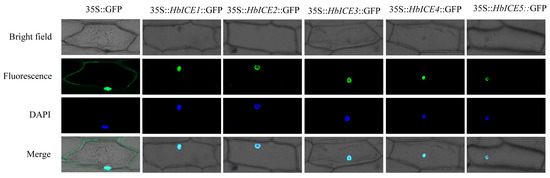
Figure 2.
Subcellular localization of HbICEs.
3.3. Expression Patterns of HbICEs in Two Rubber Tree Clones with Differential Cold Tolerance
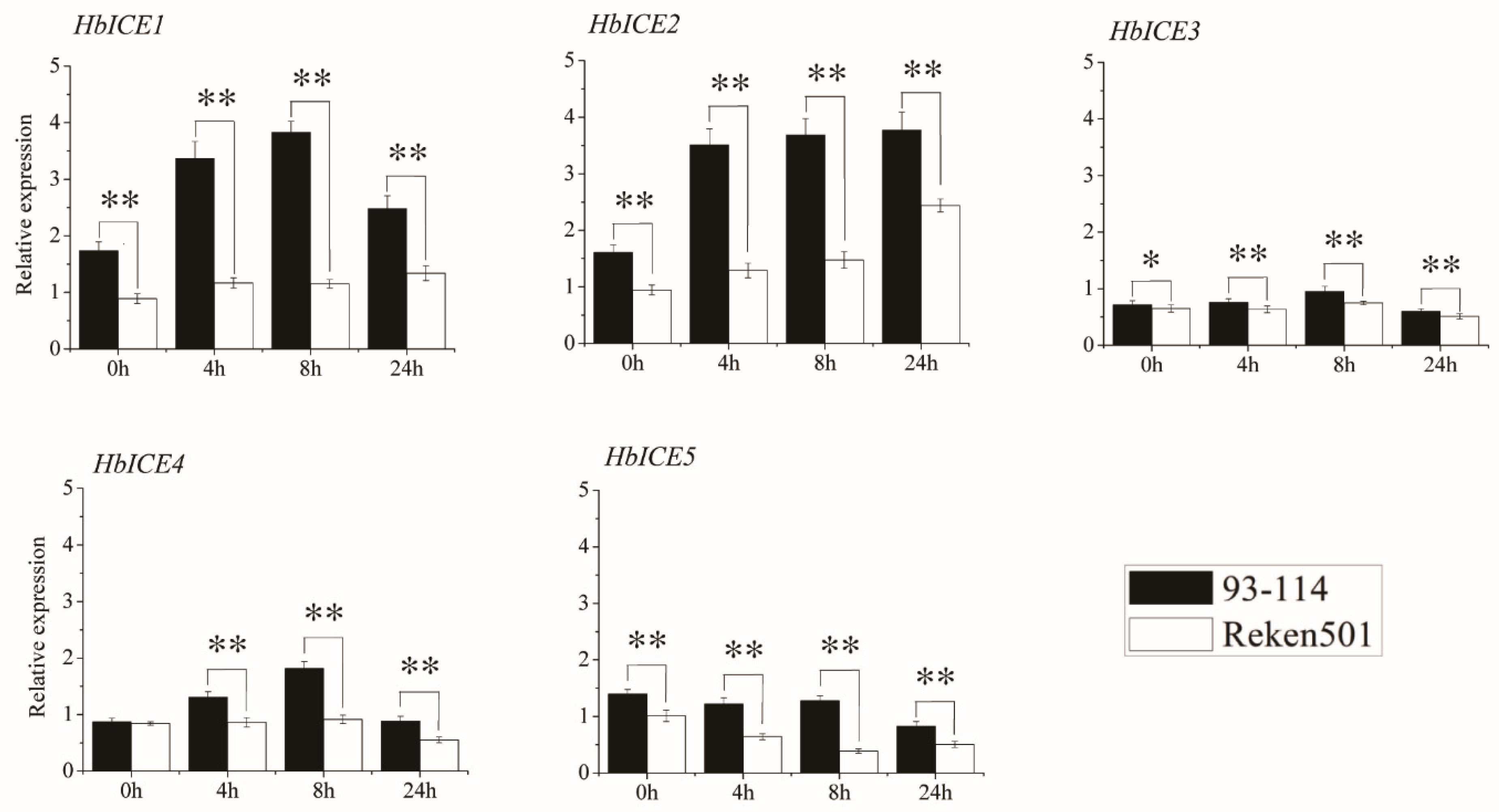
The rubber tree clone 93–114 was cold-tolerant while rubber tree clone Reken501 was cold-sensitive. The background expression levels of HbICE1, HbICE2, HbICE3, and HbICE5 in the bark tissues of 93-114 were significantly higher than those of Reken501, but the background expression of HbICE4 had no significant difference (Figure 3). Under cold stress, the expression levels of all the HbICEs in 93-114 were significantly higher than those in Reken501 at each time intervals (Figure 3). The expression levels of HbICE1 and HbICE2 in 93-114 were more than 2 times of those in Reken501 at 4 h and 8 h under cold treatment. The expression of HbICE4 was up-regulated in 93-114 and down-regulated in Reken501, and the expression level of HbICE4 in 93-114 was significantly higher than that in Reken501. The expression of HbICE5 was down regulated in 93-114 and Reken501, but the expression level in 93-114 was still higher than that in Reken501 (Figure 3).
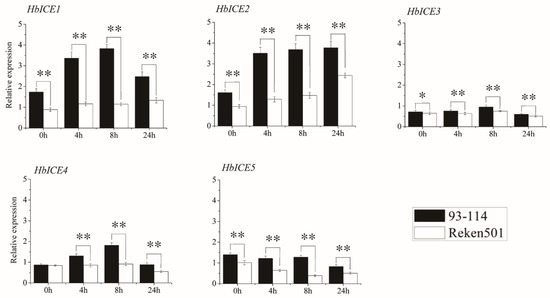
Figure 3.
Expression profiles of HbICE genes in the stem bark of Reken501 and 93-114 upon cold stress. The asterisks (**) and (*) respectively represented very significant difference (p < 0.01) and significant difference (p < 0.05).
3.4. Fuctional Identification of HbICE Genes in Arabidopsis Thaliana
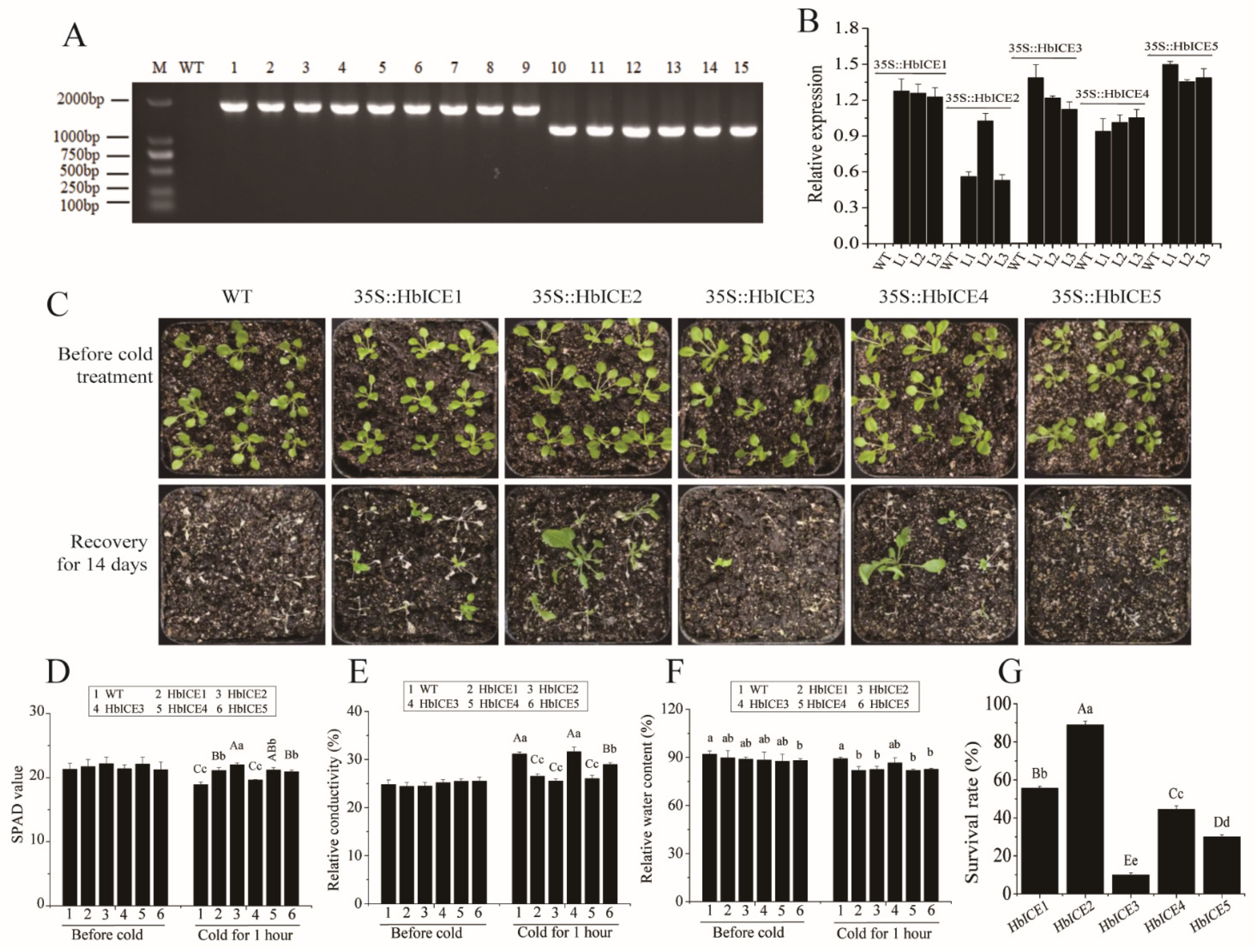
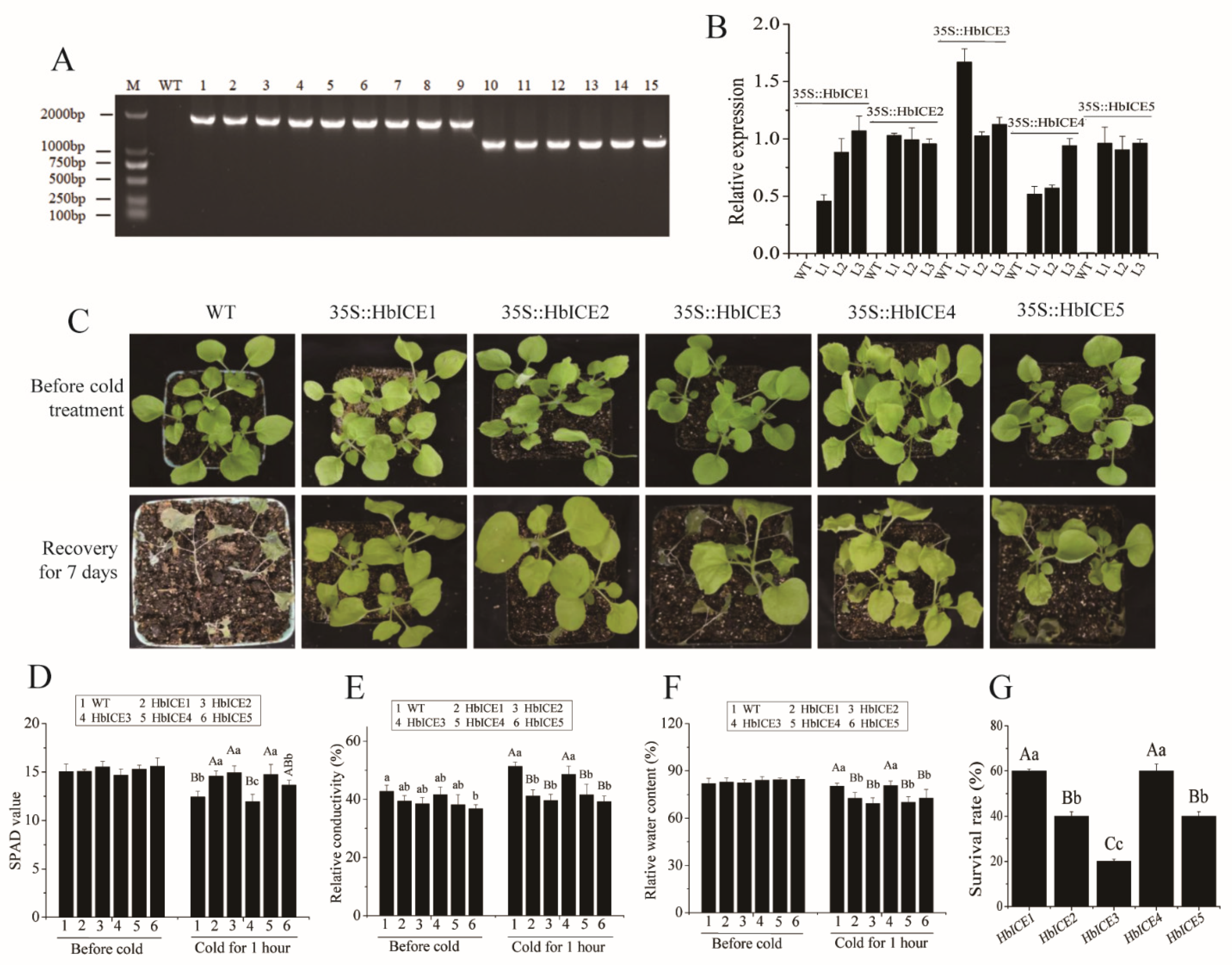
As the expression levels of HbICEs were significantly higher in 93-114 than those in Reken501 under cold stress, the effects of HbICEs overexpression on cold tolerance of Arabidopsis were analysed. At the DNA level, RT-PCR analysis demonstrated that HbICE1, HbICE2, HbICE3, HbICE4, and HbICE5 were successfully introduced into the Arabidopsis plants respectively (Figure 4A). At the RNA level, expression of the five genes was detected by qPCR in transgenic lines T2, but not in WT (Figure 4B). The survival ratio of the HbICEs-overexpressed plants was higher than that of WT (Figure 4C,G). Although cold stress generally reduced the SPAD value (a measure of leaf cellular chlorophyll content) of both WT and transgenic plants (Figure 4D), the SPAD value of wild-type and HbICE3-overexpressed plants was significantly reduced in comparison with the other HbICEs-overexpressed plants (Figure 4D). By contrast, the relative conductivity (Figure 4E) and water content (Figure 4F) of wild-type and HbICE3-overexpressed plants were significantly higher in comparison with the other HbICEs-overexpressed plants. The level of relative conductivity and water content was negatively related to the cold tolerance. The survival ratio was 55.56%, 88.89%, 10.00%, 44.44%, and 30.00% for the HbICE1-, HbICE2-, HbICE3-, HbICE4-, and HbICE5-overexpressed plants, respectively. The wild-type plants did not recover and showed 100% mortality when kept in the chamber with normal growth conditions for recovery (Figure 4G). The results showed that rubber tree HbICEs, especially HbICE1, HbICE2, and HbICE4, enhanced the cold endurance of transgenic Arabidopsis.
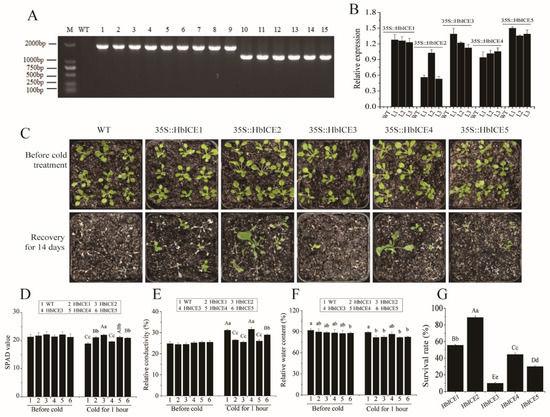
Figure 4.
Overexpression of HbICEs conferred enhanced cold tolerance in Arabidopsis. Identification of transgenic Arabidopsis lines overexpressing HbICEs by Genomic RT-PCR (A) and qPCR (B). M, marker; WT, wild-type; 1–3, transgenic lines overexpressing HbICE1; 4–6, transgenic lines overexpressing HbICE2; 7–9, transgenic lines overexpressing HbICE3; 10–12, transgenic lines overexpressing HbICE4; 13–15, transgenic lines overexpressing HbICE5. The phenotypes of WT and transgenic plants before and after cold treatment (C). SPAD value (D), relative conductivity (E), relative water content (F), and survival rate (G) in Arabidopsis WT and transgenic plants analyzed before and after cold treatment. The capital letters and lowercase letters respectively represent very significant difference (p < 0.01) and significant difference (p < 0.05). The same below.
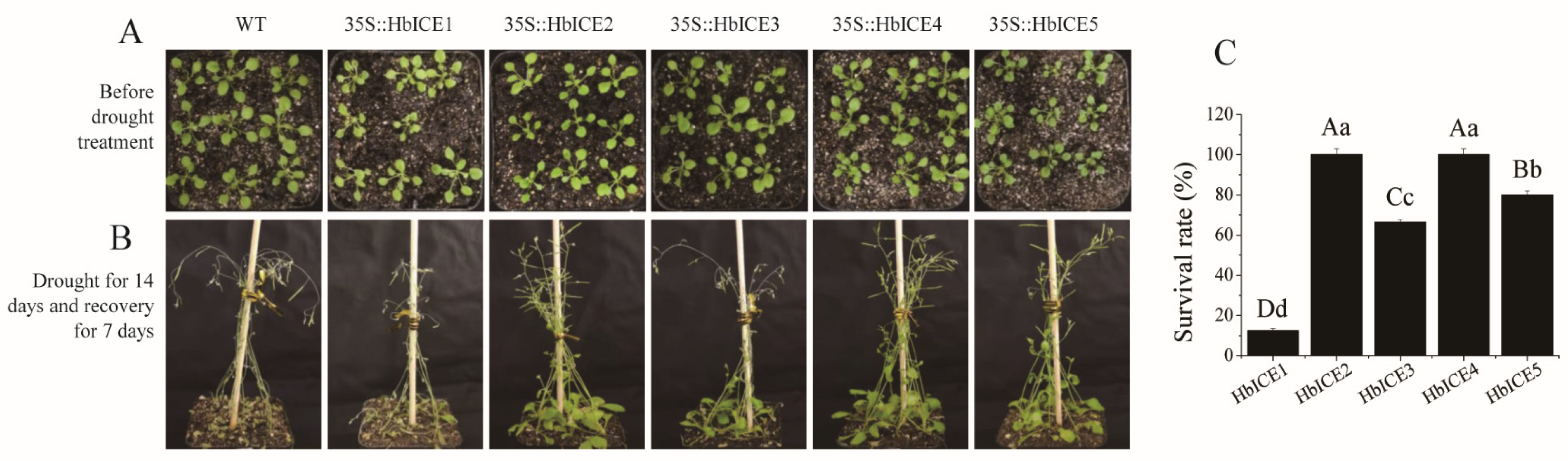
To identify the function of HbICEs in drought tolerance, 2-week-old wild type plants and HbICEs-overexpressed transgenic plants suffered drought through lack of watering for 14 days in the chamber and were then re-watered for recovery (Figure 5A). The WT and transgenic plants were growing normally until they encountered drought. Upon drought stress, all WT and transgenic plants stopped vegetative growth and initiated reproductive growth, started turning yellow, and wilted afterwards. Stems and leaves of some WT and transgenic seedlings were dried 14 days after being subjected to continuous drought. After re-watering, some of the HbICEs-overexpressed transgenic plants rejuvenated while WT did not recover and showed 100% mortality (Figure 5B). The survival ratio was 12.50%, 100.00%, 66.67%, 100.00%, and 80.00% for the HbICE1-, HbICE2-, HbICE3-, HbICE4-, and HbICE5-overexpressed transgenic plants, respectively (Figure 5C).
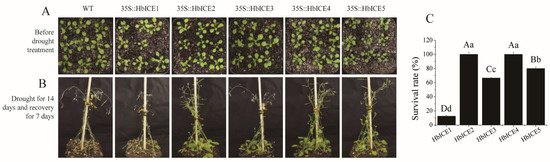
Figure 5.
Effect of overexpression of HbICEs on drought tolerance in Arabidopsis. (A,B) The phenotypes of WT and transgenic plants before drought treatment (A) and after drought followed by recovery (B). (C) Survival ratio of transgenic plants after drought treatment.
3.5. Fuctional Identification of HbICE Genes in Tobacco Plants
At the DNA and RNA levels, the results of RT-PCR and qPCR demonstrated that HbICE1, HbICE2, HbICE3, HbICE4, and HbICE5 were successfully introduced into the tobacco plants respectively (Figure 6A,B). Four-week old WT plants and transgenic plants were exposed to −5 °C freezing stress for 15 h in a chamber, then transferred to another chamber with normal growth conditions and recovered for one week. Freezing tolerance assays showed that HbICEs-overexpressed plants were more tolerant than the WT (Figure 6C). Although cold stress reduced the SPAD value and relative water content of both WT and HbICEs-overexpressed plants, the SPAD values of HbICE1-, HbICE2-, HbICE4-, and HbICE5-overexpressed plants were obviously higher than that of WT while it was the reverse for the relative water content (Figure 6D,F). The SPAD values and relative water content and relative conductivity of HbICE3-overexpressed plants were respectively similar to those of WT under cold stress (Figure 6D–F). However, the relative conductivity of HbICE1-, HbICE2-, HbICE4-, and HbICE5-overexpressed plants was significantly lower than that of WT upon cold stress (Figure 6E). The survival ratio of HbICE1-, HbICE2-, HbICE3-, HbICE4-, and HbICE5-overexpressed transgenic plants was 60.00%, 40.00%, 20.00%, 60.00%, and 40.00%, respectively while wild-type plants did not recover and showed 100% mortality (Figure 5G). These results were different to some extent from the results in Arabidopsis.
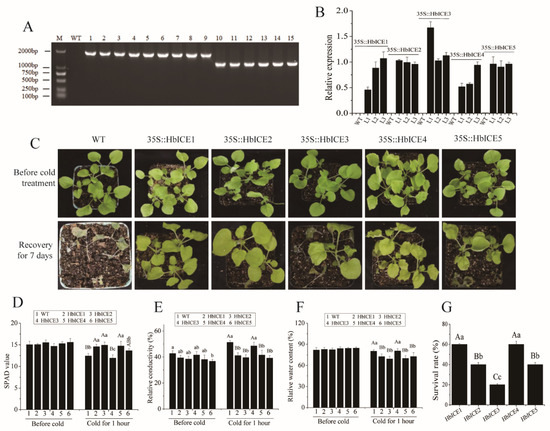
Figure 6.
Effect of overexpression of HbICEs on cold tolerance in tobacco. (A,B) Identification of HbICEs-overexpressed tobacco lines by genomic RT-PCR (A) and qPCR (B). M, marker; WT, wild-type; 1–3, HbICE1-overexpressed transgenic lines; 4–6, HbICE2- overexpressed transgenic lines; 7–9, HbICE3-overexpressed transgenic lines; 10–12, HbICE4-overexpressed transgenic lines; 13–15, HbICE5-overexpressed transgenic lines. (C) The phenotypes of WT and transgenic plants before and after cold treatment. (D–G) Analysis of the SPAD value (D), relative conductivity (E), relative water content (F), and survival rate (G) in tobacco WT and transgenic plants.
3.6. Expression Identification of HbICE Genes in Rubber Trees
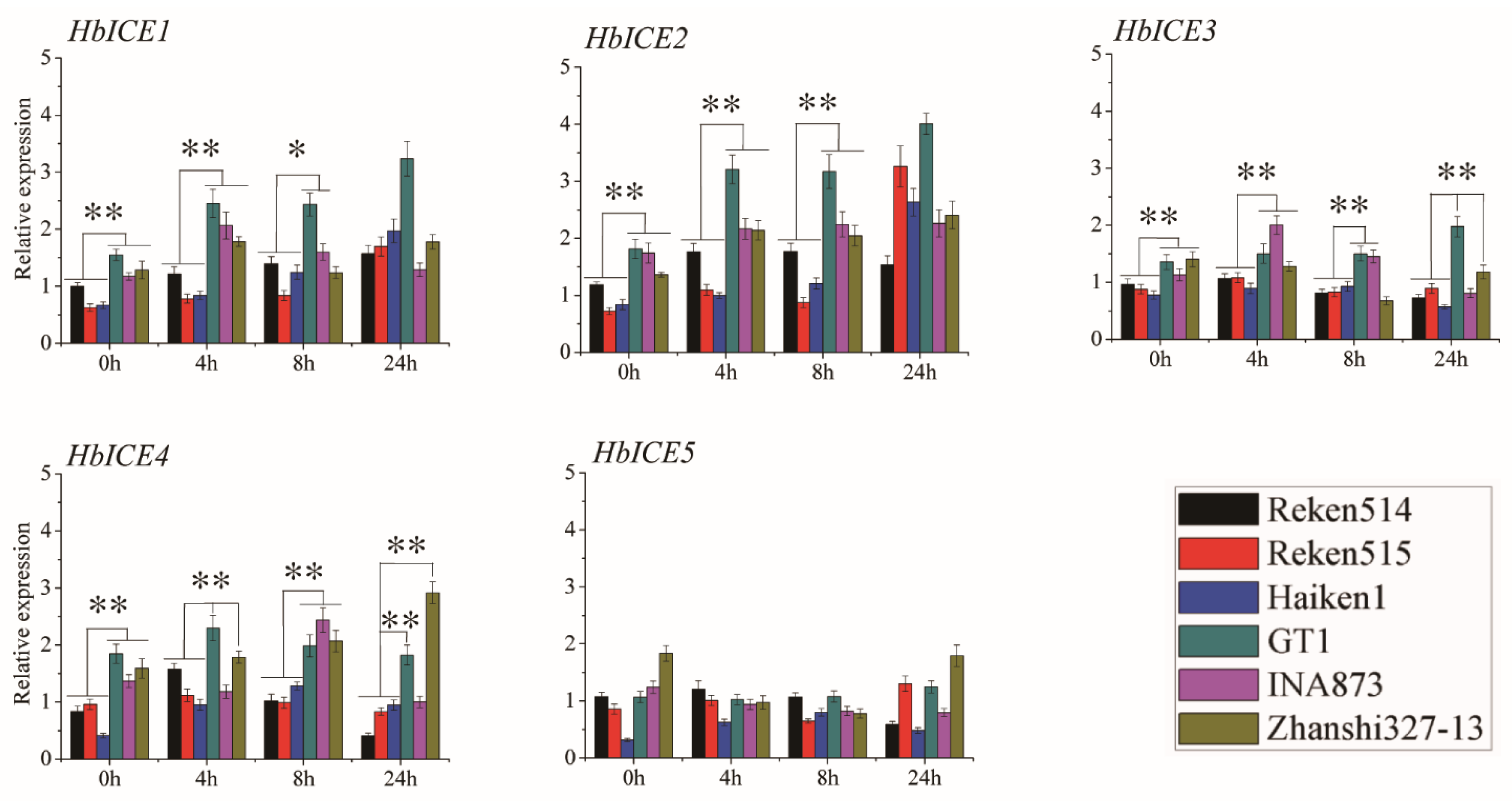
The expression pattern of HbICEs upon cold stress was comparatively analysed between three cold-tolerant rubber tree clones (Zhanshi327-13, INA873, GT1) and three cold-sensitive rubber tree clones (Reken514, Reken515, Haiken1) by qPCR. HbICE1, HbICE2, HbICE3, and HbICE4 were obviously up-regulated by cold stress in all the three cold-tolerant and three sensitive rubber tree clones. However, the expression levels of HbICE1, HbICE2, HbICE3, and HbICE4 in all the three cold-tolerant rubber tree clones were significantly higher at least at two time intervals than those in the three cold-sensitive rubber tree clones (Figure 7). The expression level of HbICE1 was significantly higher in background and at 4 h upon cold stress in the three cold-tolerant rubber tree clones as compared with all the three cold-sensitive rubber tree clones. The expression level of HbICE2 was significantly higher in background and during cold stress in three cold-tolerant rubber tree clones as compared to the three cold-sensitive rubber tree clones, except for no difference at 24 h between the cold-tolerant rubber tree clones and the cold-sensitive rubber tree clones. The expression level of HbICE3 was significantly higher in background and at 4 h upon cold stress in the three cold-tolerant rubber tree clones in comparison with all the three cold-sensitive rubber tree clones. The expression level of HbICE4 was significantly higher in background and at 8 h upon cold stress in the three cold-tolerant rubber tree clones in contrast to all the three cold-sensitive rubber tree clones, and it was significantly higher at 4 h and 24 h upon cold stress in cold-tolerant rubber tree clones GT1 and Zhanshi327-13 as compared with the three cold-sensitive rubber tree clones. There was no difference in the expression level of HbICE5 at each time interval between the cold-tolerant and cold-sensitive rubber tree clones (Figure 7).
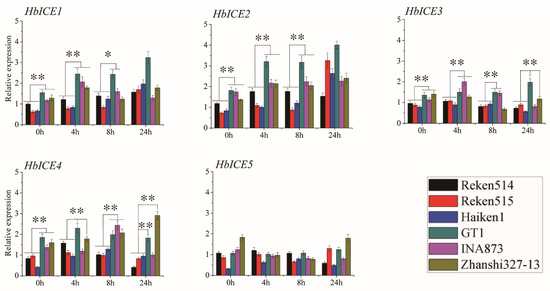
Figure 7.
Expression profiles of five HbICEs genes in three cold-sensitive rubber tree clones (Haiken1, Reken514, and Reken515) and three cold-tolerant rubber tree clones (GT1, Zhanshi327-13, and INA873). The asterisks (**) and (*) respectively represented very significant difference (p < 0.01) and significant difference (p < 0.05).
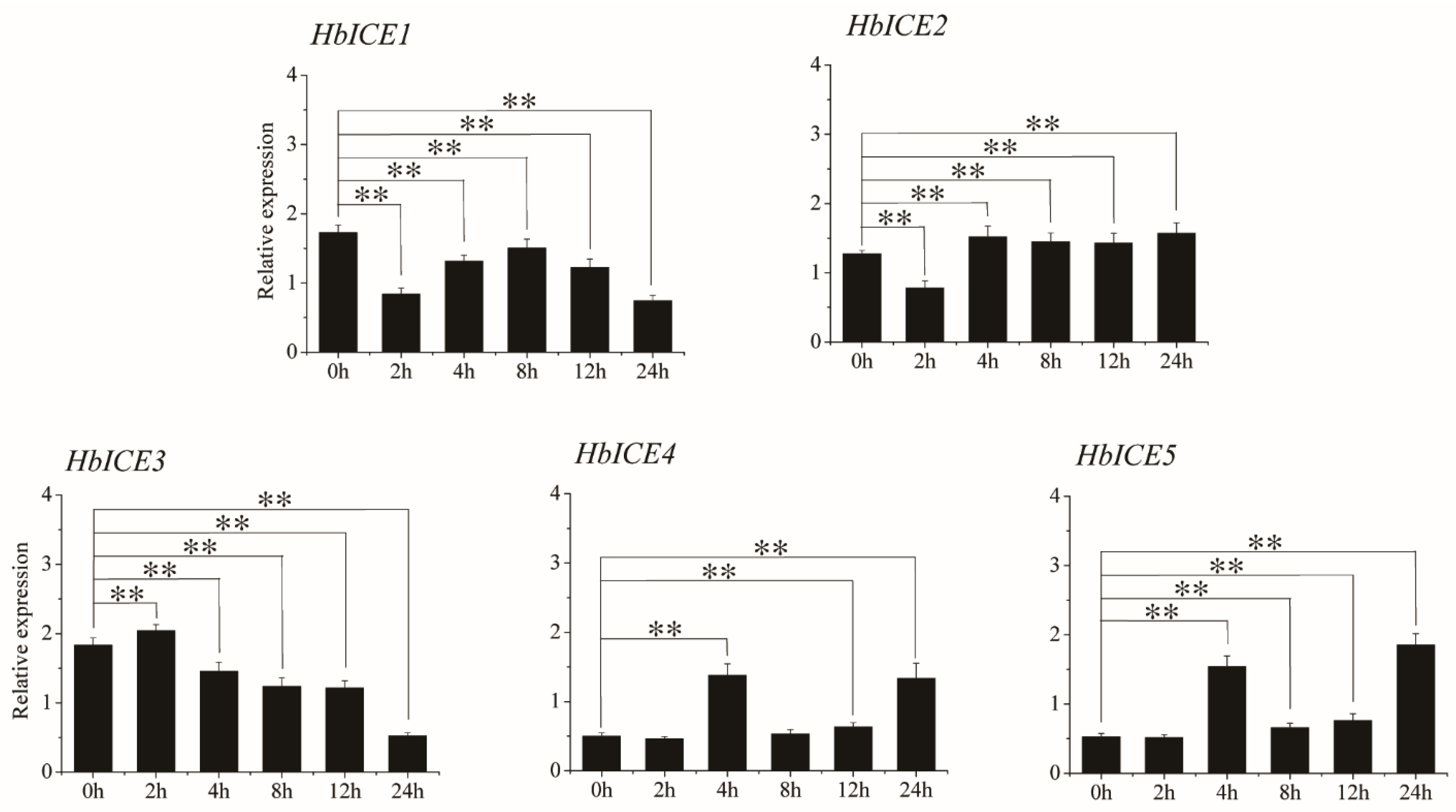
HbICE2, HbICE3, HbICE4, and HbICE5, which could significantly enhance the drought survival of transgenic Arabidopsis, were selected to further analyze their express patterns in 93-114 under drought stress by qPCR. The results showed that HbICE2 was significantly down-regulated at 2 h during drought treatment in contrast to the background. After 2 h of drought treatment, the expression of HbICE2 was continuously up-regulated by drought stress in 93-114. On the contrary, HbICE3 was significantly up-regulated at 2 h of drought treatment in contrast to the background. After 2 h of drought treatment, the HbICE3 expression was continuously down-regulated by drought stress in 93-114. The expression pattern of HbICE4 was similar to HbICE5 in 93-114 under cold stress. There was no significant difference in the expressions of HbICE4 and HbICE5 at 2 h of drought stress in contrast to the background, and the expressions of HbICE4 and HbICE5 were significantly increased at 4 h, down-regulated at 8 h, and then significantly up-regulated at 12 h and 24 h of drought stress. However, the expression level of HbICE1 was significantly down-regulated upon drought stress in comparison with the background (Figure 8).
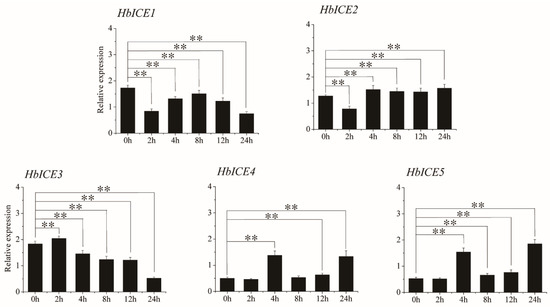
Figure 8.
Expression profiles of five HbICE genes in rubber tree clone 93-114 under drought stress. The asterisks (**) represented very significant difference (p < 0.01).
4. Discussion
ICE is a positive regulator of plant response to cold stress, and plays a major role in cold signaling pathways in various plants such as Arabidopsis [6,13], wheat [7], rice [10], banana [30,31,32], Vitis amurensis [12], tomato [11], Zoysia japonica [9], trifoliate orange [8], and Pyrus ussuriensis [33]. Therefore, it is reasonable to deduce that the putative ICE homolog, HbICEs, also plays a major role in the cold response in the rubber tree. Thus, characterization and function analysis of the HbICE genes are crucial to understand the cold response pathway in the rubber tree. According to the version of rubber tree Reyan7-33-97 genome [34], five HbICE genes were isolated and verified by a RT-PCR and nucleic acid sequencing technique in the present study. HbICE1 was identical to any HbICE2 that has ever been reported [35], and the other four HbICE genes were noval HbICE homologs.
Multiple sequence alignments showed that HbICEs contained typical features of the ICE protein such as the bHLH domain, ZIP (zipper region), and ACT-like domain and possessed the varied N termini. All HbICEs had the highly conserved sequence KMDRA/TSILGDA/TID/EYLKELL that is specific to the ICEs of plants but not to the MYC proteins as previously described [3]. The potential sumoylation site, which is previously identified in Arabidopsis and Vitis amurensis [12,22], was also present in HbICE1, HbICE2, HbICE3, HbICE4, and HbICE5 of rubber tree. SIZ1-mediated sumoylation is essential to AtICE1 stability [21]. The presence of potential sumoylation site suggested that sumoylation is important for HbICEs stability. The S-rich regions are a putative site of phosphorylation. OST1/SnRK2.6 interacts with transcription factor ICE1 and increases its stability and transcriptional activity by phosphorylation [20]. The presence of S-rich regions in HbICE1, HbICE2, and HbICE3 suggests a similar regulatory mechanism. The ACT-like domain was present in the C termini of HbICE proteins, which participates in the dimerization of bHLH transcription factors [36,37]. HbICE4 and HbICE5 lacked parts of the varied N termini, but possessed the typical ICE protein features like the grape VvICE4 protein [38]. HbICE4 and HbICE5 shared 88.46% similarity at the amino acid level and 71.93% and 70.97% similarity to VvICE4 respectively [38].
In addition to the modification-regulated stability and activity of ICE proteins, transcriptional regulation is also important for enhanced cold tolerance. Overexpression of HbICE1 confers the cold tolerance of Arabidopsis and yeast [35,39]. Overexpression of Pyrus ussuriensis PuICE1 enhances cold tolerance of tomato under cold stress [33]. Overexpression of rice OsICE1 and OsICE2 improves the cold tolerance of Adrabidopsis seedlings [10]. Likewise, overexpression of Adrabidopsis AtICE1 significantly enhances cold tolerance of indica rice under cold treatment [6]. In the present study, functional identification of five HbICE genes was conducted by evaluating the cold tolerance of HbICEs-overexpressed Arabidopsis and tobacco plants and the expression level of HbICEs in cold-tolerant and sensitive rubber tree clones. The cold tolerance of both the HbICE3-overexpressed Arabidopsis and tobacco plants like that of wild-type plants and shared similar physiological parameters and were lower in SPAD value, but higher in relative conductivity and relative water content in comparison with the HbICE1-, HbICE2-, HbICE4-, and HbICE5-overexpressed Arabidopsis and tobacco plants. However, there was a difference to some extent in the cold tolerance of the four HbICEs-overexpressed Arabidopsis and tobacco plants. The overexpression of HbICE2 conferred the strongest cold tolerance, followed by the overexpression of HbICE1 and HbICE4 in Arabidopsis, while overexpression of HbICE1 and HbICE4 conferred the strongest cold tolerance, followed by the overexpression of HbICE2 in tobacco plants. The expression levels of HbICE1, HbICE2, and HbICE4 were significantly higher in cold-tolerant rubber tree clones than that in cold-sensitive rubber tree clones both in normal conditions and upon cold stress, suggesting that the cold tolerance of rubber tree clones was positively correlated with expression level of the three genes. Similarly, the expression level of ICE genes was significantly higher in cold-tolerant Vitis riparia than that in cold-sensitive Vitis vinifera [38]. Although the expression level of HbICE3 was significantly higher in cold-tolerant rubber tree clones than that in cold-sensitive rubber tree clones, overexpression of HbICE3 had no effect on enhancing the cold tolerance of either Arabidopsis or tobacco transgenic plants. By contrast, there was no difference in the expression level of HbICE5 between the cold-tolerant and cold-sensitive rubber tree clones, but the overexpression of HbICE5 conferred stronger cold tolerance than overexpression of HbICE3 in either Arabidopsis or tobacco plants. The difference in the evaluation of cold tolerance by different experimental systems indicates that the transcriptional regulation of ICE genes on cold tolerance is species-specific to some extent.
Previous studies demonstrate that the expression of ICE genes is up-regulated by drought, such as PtrICE1 [8], TaICEs [7], and ZjICE1 [9]. Overexpression of AtICE1 enhances drought tolerance of transgenic indica rice [6]. Similarly, HbICE2, HbICE4, and HbICE5 were up-regulated by drought and overexpression of the three genes obviously enhanced the drought tolerance of transgenic Arabidopsis. It is noted that drought down-regulated the expression of HbICE1, which is different from the other HbICE genes and the ICE genes in other plant species that are up-regulated by drought [9]. The HbICE1 had little effect in enhancing the drought tolerance of transgenic Arabidopsis. Five ICE genes were isolated from rubber tree. They are different in transcript abundance and expression patterns in response to cold and drought stresses and among different rubber tree clones. The cold tolerance of rubber tree clones is, perhaps, positively controlled by the expression level of HbICE1, HbICE2, and HbICE4.
5. Conclusions
To understand the cold tolerance mechanism of rubber tree, five ICE genes were isolated from rubber tree. They are different in transcript abundance and expression patterns in response to cold and drought stresses and among different rubber tree clones. Overexpression of HbICE1, HbICE2, and HbICE4 significantly enhanced the cold tolerance of transgenic Arabidopsis and tobacco, which showed a significant increase in chlorophyll content and decrease in relative water content and conductivity at the early stage of cold stress in comparison with wild-type plants. Furthermore, overexpression of HbICE2 and HbICE4, but also HbICE1 enhanced drought tolerance in transgenic Arabidopsis. The cold tolerance of rubber tree clones is, perhaps, positively controlled by the expression level of HbICE1, HbICE2, and HbICE4.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f13010052/s1, Supplementary Table S1: Primer sequences were used in this paper. Supplementary Table S2: Summary information of HbICE family genes in the rubber tree.
Author Contributions
Y.L. conceived the project; C.Q., S.Y. and Y.L. conducted the experiments; M.S. and S.W. analyzed the data; J.W. cultivated plantlets of rubber-tree clones and provided materials for experiments; Y.L. and W.T. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Key-Area Research and Development Program of Guangdong Province (No: 2020B020217002) and the Earmarked Fund for China Agriculture Research System (CARS-33-YZ1).
Acknowledgments
The authors would like to thank Yang JH for providing experimental materials and reviewers for their careful reading and valuable suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Badawi, M.; Reddy, Y.V.; Agharbaoui, Z.; Tominaga, Y.; Danyluk, J.; Sarhan, F.; Houde, M. Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol. 2008, 49, 1237–1249. [Google Scholar] [CrossRef]
- Chinnusamy, V. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, J.; Takashi Yuasa, T.; Huong, T.T.; Harano, K.; Tanaka, S.; Iwata, T.; Phan, T.; Iwaya, M. Rice homologs of inducer of CBF expression (OsICE) are involved in cold acclimation. Plant Biotechnol. 2011, 28, 303–309. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Hu, Y.R.; Jiang, L.Q.; Wang, F.; Yu, D. Jasmonate regulates the inducer of CBF expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.K.; Kumar, V.V.S.; Yadav, S.K.; Kumar, T.S.; Rao, M.V.; Chinnusamy, V. Overexpression of Arabidopsis ICE1 enhances yield and multiple abiotic stress tolerance in indica rice. Plant Signal. Behav. 2020, 15, 1814547. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ren, Y.; Tang, Z.; Shi, W.; Zhou, M. Characterization and expression profiling of the ICE-CBF-COR genes in wheat. Peer J. 2019, 7, e8190. [Google Scholar] [CrossRef]
- Huang, X.S.; Zhang, Q.H.; Zhu, D.X.; Fu, X.Z.; Wang, M.; Zhang, Q.; Moriguchi, T.; Liu, J.H. ICE1 of Poncirus trifoliata functions in cold tolerance by modulating polyamine levels through interacting with arginine decarboxylase. J. Exp. Bot. 2015, 66, 3259–3274. [Google Scholar] [CrossRef]
- Zuo, Z.F.; Kang, H.G.; Park, M.Y.; Jeong, H.; Sun, H.J.; Song, P.S.; Lee, H.Y. Zoysia japonica MYC type transcription factor ZjICE1 regulates cold tolerance in transgenic Arabidopsis. Plant Sci. 2019, 289, 110254. [Google Scholar] [CrossRef]
- Deng, C.; Ye, H.; Fan, M.; Pu, T.; Yan, J. The rice transcription factors OsICE confer enhanced cold tolerance in transgenic Arabidopsis. Plant Signal. Behav. 2017, 12, e1316442. [Google Scholar] [CrossRef]
- Feng, H.L.; Ma, N.N.; Meng, X.; Zhang, S.; Wang, J.R.; Chai, S.; Meng, Q.W. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol. Biochem. 2013, 73, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Jiao, Y.; Li, R.; Zhang, N.; Xiao, D.; Ding, X.; Wang, Z. Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type bHLH transcription activators that regulate cold tolerance in Arabidopsis. PLoS ONE 2014, 9, e102303. [Google Scholar] [CrossRef]
- Kim, Y.S.; Lee, M.; Lee, J.H.; Lee, H.J.; Park, C.M. The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 2015, 89, 187–201. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Nozawa, R. Overexpression of SIZ1 enhances tolerance to cold and salt stresses and attenuates response to abscisic acid in Arabidopsis thaliana. Plant Biotechnol. 2014, 31, 167–172. [Google Scholar] [CrossRef][Green Version]
- Miura, K.; Jin, J.B.; Lee, J.; Yoo, C.Y.; Stirm, V.; Miura, T.; Ashworth, E.N.; Bressan, R.A.; Yun, D.J.; Hasegawa, P.M. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar] [CrossRef]
- Doherty, C.J.; Van Buskirk, H.A.; Myers, S.J.; Thomashow, M.f. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.H.; Agarwal, M.; Zhang, Y.; Xie, Q.; Zhu, J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 2006, 103, 8281–8286. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Li, Y.; Yu, W.C.; Chen, Y.Y.; Yang, S.G.; Wu, S.H.; Chao, J.Q.; Wang, X.L.; Tian, W.M. Genome-wide identification and characterization of heat-shock transcription factors in rubber tree. Forests 2019, 10, 1157–1169. [Google Scholar] [CrossRef]
- Li, Y.; Yang, S.G.; Shi, M.J.; Zhang, S.X.; Wu, S.H.; Chen, Y.Y.; Li, W.G.; Tian, W.M. HbARF2 and HbARF16.3 function as negative regulators for the radial trunk growth of rubber tree. Ind. Crop Prod. 2020, 158, 112978. [Google Scholar] [CrossRef]
- Sambe, M.A.; He, X.; Tu, Q.; Guo, Z. A cold-induced myo-inositol transporter-like gene confers tolerance to multiple abiotic stresses in transgenic tobacco plants. Physiol Plant. 2015, 153, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Gao, J.; Dou, T.; He, W.; Sheng, O.; Bi, F.; Deng, G.; Gao, H.; Dong, T.; Li, C.; Zhang, S.; et al. MaMAPK3-MaICE1-MaPOD P7 pathway, a positive regulator of cold tolerance in banana. BMC Plant Biol. 2021, 21, 97. [Google Scholar] [CrossRef]
- Peng, H.H.; Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Molecular characterization of cold-responsive basic helix-loop-helix transcription factors MabHLHs that interact with MaICE1 in banana fruit. Planta 2013, 238, 937–953. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.L.; Wang, J.N.; Shan, W.; Fan, J.G.; Kuang, J.F.; Wu, K.Q.; Li, X.P.; Chen, W.X.; He, F.Y.; Chen, J.Y.; et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 2012, 36, 30–51. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, K.; Jin, C.; Zhang, S. ICE1 of Pyrus ussuriensis functions in cold tolerance by enhancing PuDREBa transcriptional levels through interacting with PuHHP1. Sci. Rep. 2015, 5, 17620. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef]
- Chen, W.J.; Wang, X.; Yan, S.; Huang, X.; Yuan, H.M. The ICE-like transcription factor HbICE2 is involved in jasmonate-regulated cold tolerance in the rubber tree (Hevea brasiliensis). Plant Cell Rep. 2019, 38, 699–714. [Google Scholar] [CrossRef]
- Feller, A.; Hernandez, J.M.; Grotewold, E. An ACT-like domain participates in the dimerization of several plant basic-helix-loop-helix transcription factors. J. Biol. Chem. 2006, 281, 28964–28974. [Google Scholar] [CrossRef]
- Lee, Y.S.; Herrera-Tequia, A.; Silwal, J.; Geiger, J.H.; Grotewold, E. A hydrophobic residue stabilizes dimers of regulatory ACT-like domains in plant basic helix-loop-helix transcription factors. J. Biol. Chem. 2021, 296, 100708. [Google Scholar] [CrossRef]
- Rahman, M.A.; Moody, M.A.; Nassuth, A. Grape contains 4 ICE genes whose expression includes alternative polyadenylation, leading to transcripts encoding at least 7 different ICE proteins. Environ. Exp. Bot. 2014, 106, 70–78. [Google Scholar] [CrossRef]
- Deng, X.M.; Wang, J.; Li, Y.; Wang, J.; Tian, W.M. Characterization of a cold responsive HbICE1 gene from rubber trees. Trees 2016, 31, 137–147. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).