Abstract
Clarifying the genetic relationship and divergence among Curcuma L. (Zingiberaceae) species around the world is intractable, especially among the species located in China. In this study, Reduced Representation Sequencing (RRS), as one of the next generation sequences, has been applied to infer large scale genotyping of major Chinese Curcuma species which present little differentiation of morphological characteristics and genetic traits. The 1295 high-quality SNPs (reduced-filtered SNPs) were chosen from 997,988 SNPs of which were detected from the cleaned 437,061 loci by RRS to investigate the phylogeny and divergence among eight major Curcuma species locate in the Hengduan Mountains of the Qinghai–Tibet Plateau (QTP) in China. The results showed that all the population individuals were clustered together within species, and species were obviously separated; the clustering results were recovered in PCA (Principal Component Analysis); the phylogeny was (((((C. Phaeocaulis, C. yunnanensis), C. kwangsiensis), (C. amarissima, C. sichuanensis)), C. longa), (C. wenyujin, C. aromatica)); Curcuma in China originated around ~7.45 Mya (Million years ago) in the Miocene, and interspecific divergence appeared at ca. 4–2 Mya, which might be sped up rapidly along with the third intense uplift of QTP.
1. Introduction
Curcuma, commonly known as turmeric, has been an important flowering plant genus with medicinal, edible, and horticultural utilizations in the Orient from ancient times. Usually Curcuma are distributed in the subtropical and tropical area of Asia [1,2,3], and there are about 10 species in China (C. longa, C. sichuanensis, C. amarissima, C. yunnanensis, C. phaeocaulis, C. kwangsiensis, C. aromatica, C. wenyujin, C. flaviflora, C. exigua, and C. viridiflora) [4]. Among them, C. viridiflora and C. exigua may be extinct [5], C. yunnanensis and C. flaviflora are scarce and difficult to collect [4]. The worldwide Curcuma represent a paraphyletic group involving Hitchenia, Stahlianthus, and Smithatris in Zingibereae according to matK and ITS [6], and it has been classified into four lineages (Hitcheniopsis, Pierreana, Curcuma, and Ecomata) by using ITS and three chloroplast regions (trnL-trnF, psbA-trnH, matK) [2]. Recently, partial lineages in Curcuma have been clarified, though the phylogeny and divergence among some species are still intractable, especially species in China due to the intense uplift of the Qinghai–Tibet Plateau (QTP) [7,8]. Chinese Curcuma species are distributed to increase ranging from Tibet (only herbarium specimens represent this species now) to South China, most growing in the Hengduan Mountains Areas of QTP, which play an important role on species distribution, evolution, and divergence in eastern Asia [9,10,11,12,13].
The internal micro-anatomy of the rhizomes and leaves of Chinese Curcuma show few differences, and different anatomy traits can support different classification. The Chinese Curcuma species were divided into three groups according to the traits of oil cells, vascular bundles, and the number and diameter of xylem vessels (Group I: C. longa and C. sichuanensis, Group II: C. kwangsiensis and C. exigua, Group III: C. wenyujin, C. aromatica, C. phaeocaulis C. zedoaria, and C. yunnanensis) [14,15]. The hair distribution, stoma density and size, epidermic cell shape and size in leaf, were also suggested to be unique features among Chinese Curcuma species (Group I: C. longa, C. wenyujin, and C. sichuanensis; Group II: C. kwangsiensis and C. exigua; Group III: C. aromatica, C. chuanyujin, C. zedoaria, C. phaeocaulis, and C. yunnanensis). The study on pollen morphology showed that Chinese Curcuma species can be divided into two groups, Group I (pollen < 3 μm) including C. amarrisima, C. flaviflora, C. phaecaulis, C. sichuanensis, and C. wenyujin, and Group II (pollen > 3 μm) including C. aromatica, C. yunnanensis, and C. longa [16]. Therefore, the phylogeny of Chinese Curcuma are always controversial and ambiguous for poor diagnostic characters of morphology (aerial part, underground part, floral morphology, etc.) [17]. Cytological studies supported that some species in this genus were polyploid [18,19,20,21]. Chromosome number in Chinese Curcuma were studied to find out that C. kwangsiensis is 2n = 4x = 84 and C. flaviflora is 2n = 2x = 42, and the left of C. longa, C. aromatica, C. sichuanensis, C. elata, C. wenyujin, and C. phaeocaulis are triploid (2n = 3x = 63) [19]. Polyploidy is commonly distributed in Curcuma, e.g., C. longa 2n = 2x/3x/4x = 32/63/64 [19,22,23,24], C. kwangsiensis 2n = 4x = 64/84 [19,23], and C. aromatica 2n = 2x/3x = 42/86 [25,26], making it hard to distinguish them hardly based on ploidy and chromosome number [18,25,26].
In the past decades, molecular phylogeny of Curcuma around the world has been improved, while the phylogeny and divergence of Chinese Curcuma are still unclear [1,2,7]. Two groups, (Group I: C. chuanhuangjiang, C. aromatica, C. yunnanensis, C. kwangsiensis, and C. phaeocaulis; Group II: C. longa, C. sichuanensis, C. amarissima, and C. wenyujin), were divided by using six mtDNA genes [1]. The matK, rbcL, trnH-psbA, trnL-F, and ITS2 were applied to test barcodes within Curcuma collected from Myanmar and China, and without enough evidence to identify each species (no barcoding gaps found in four chloroplast regions and little gaps in ITS2) [7]. Based on chloroplast genome, a recent study showed that eight Curcuma species in China were divided into two groups (Group I including C. wenyujin, C. phaeocaulis, and C. aromatica, Group II including C. longa, C. yunnanensis, C. amarissima, and C. sichuanensis), and C. flaviflora is far away from the aforementioned seven species, clustered together with Zingiber spectablie (a kind of ginger) and Z. officinale [27]. In all, most DNA regions lack enough information on Chinese Curcuma species identification, particularly among related species, such as C. longa and C. sichuanensis, C. aromatica, and C. wenyujin. Therefore, new technologies need to be introduced to infer the phylogeny and divergence of Chinese Curcuma [3,28,29]. High-throughput and next-generation sequencing (NGS) can provide large amounts of genomic data and make great progress on phylogeny, divergence, and biogeography [30,31,32,33]. Reduced Representation Sequencing (RRS), one of the NGS approaches, can produce numerous sequence tags, and is successfully used to polymorphism detection in multi-species coalescent-based tree and to deal tricky plant’s phylogeny and biogeography [34,35,36,37,38,39,40].
The phylogeny and divergence of Chinese Curcuma are still under debate due to similar morphological characteristics, unique geographical and climate conditions of the QTP [5,28,41,42,43]. The rising and climatic fluctuation of Hengduan Mountain Range (presently affected by monsoons from both the Indian and Pacific oceans) and Yunnan–Guizhou Plateau (affected mainly by Pacific monsoons) were induced with rapid uplift of the Himalayas and the QTP [44,45]. In the Pliocene, the third intense uplift of QTP changed the geographical environment, climate, and species distribution/divergence in China and eastern Asia [9,10,46,47]. Curcuma in China are distributed in the QTP generally, such as Sichuan, Yunnan, Zhejiang, Guangdong, and Guangxi, even in Tibet without living individuals collected in recent years (herbarium presented: 02010668, 02010669, 00074714, 00074715, 01376872, 01376873, 01376874, and 01376875 in Herbarium, Institute of Botany, Chinese Academy of Sciences). The uplift of QTP might play a key role in Curcuma evolution and divergence in China.
In this study, RRS was utilized (1) to evaluate the phylogeny of Curcuma species in China and (2) to explore the impact on the evolution and divergence of Chinese Curcuma because of uplift of QTP.
2. Materials and Methods
2.1. Material and Sequencing
A total 60 specimens of eight Curcuma species were used in this study, and Hedychium coronarium (commonly known as white ginger) was used as outgroup (specimens information in Table 1). All specimens used in this study were identified by Prof. Ruiwu Yang with the Sichuan Agricultural University, and were cultivated at the Sichuan Agricultural University Farmland.

Table 1.
Sample of species, with source details and total loci after processing with ipyRAD, of the 61 individuals used in phylogenetic analyses. Each sample has a sample name, species name, collecting information, and loci information.
The genomic DNA isolation was carried out on fresh leaves by the CTAB method [48]. The RRS libraries for each sample were prepared using the protocol outline as previously described [49]. We used the restriction enzyme PstI (CTGCAG) to digest the extracted genomic DNA from each individual, and then ligated the resulting fragments to a barcode adaptor and a common adaptor with the correct sticky ends. Then, a Qiagen MinElute 96-well PCR purification kit was used in the clean-up step to clean up the products. After PCR, the PicoGreen and a qPCR machine were used to examine the quality of the PCR products. All individuals were pooled into a single RRS library. Sample sequencing was done on Illumina Hiseq PE150 sequencer in Genepioneer Biotechnologies Co. Ltd., Nanjing, China. Raw Reads of the founder lines were deposited in the National Center for Biotechnology Information (NCBI) BioProject ID: PRJNA557061.
2.2. Clustering
The software of pipeline ipyRAD 0.7.29 [50] was used to process the raw data from the Illumina FASTQ files. The pipeline is focusing on preparing RADseq type data for population level analyses [51]. Following seven sequential steps, the ipyRAD pipeline can obtain species or higher variation across clades in clustering and alignment method based on specific parameters in the ipyRAD documentation (https://github.com/dereneaton/ipyrad (accessed on 21 January 2019)). The ipyRAD standard parameter settings were as follows: Nucleotides with Phred scores of <20 were coded as unknown bases (N), and sequences with >5% N’s were thrown out. Sequences were clustered within individuals by 90% similarity via the uclust function in USEARCH [52]. Clusters of less than 10 sequences were discarded and the minimum number of individuals per cluster was set to 5. Heterozygous loci among more than two individuals were discarded. The remaining clusters were treated as loci and assembled into a phylogenetic matrix.
2.3. Concatenation-Based Species Tree Inference
The reduced-filtered SNPs were acquired from the filtered SNPs (QC) via quality control by using PLINK 1.9 with standard parameter settings. The standard parameter settings were as follows: Missingness per marker was set to 0.05. Minor allele frequency was set to 0.05. The model of substitution for data was run in MrModeltest [53] by the Akaike Information Criterion (AIC) and obtained the best model of GTR. The maximum likelihood (ML) was implemented under the GTR nucleotide substitution model in RAxML8.2.8 [54]. The maximum parsimony (MP) tree branch with booststrap support was done in the software PAUP * 4.0a134. Optimal MP trees were searched by a heuristic strategy with 1000 random sequence additions and TBR branch swapping. Bootstrap values were calculated using 1000 replicates, 10 random additions per replicate, and TBR branch swapping. Bayesian inference (BI) tree was carried out using MrBayes 3.1.2 [55]. Markov chain Monte Carlo (MCMC) searches were started from a random tree and run for 3,000,000 generations, where the topologies were sampled every 100 generations. Furthermore, 25% of our individuals (which the first 2500 trees) were discarded as burnin. The Bayesian posterior probabilities of the nodal supports were inferred and the 50% majority-rule consensus tree was constructed based on the rest of trees.
2.4. Population Structure and Divergence Time Inference
Principal component analysis (PCA) is a purely mathematical method that reflects the clustering between groups and is based on the degree of SNPs in different individuals by EIGENSOFT version 7.2.1 [56]. Population structure of 60 Curcuma individuals was obtained by ADMIXTURE 1.3.0 [57]. We predefined the ancestral proportions (K) from 3 to 12, and ran the cross-validation error (CV) procedure. The default settings and methods were used for other parameters.
On the basis of the high-quality SNPs, the BEAST version 2.5.0 was used to estimate Chinese Curcuma species divergence time, and the Bayesian tree was dated by setting divergence time between Hedychium coronarium and Curcuma as 42.4 Mya (37.4–47.4 Mya) [58]. The GTR model for nucleotide substitution and the “Bayesian skyline” tree prior model was confirmed with a standard normal prior. Substitution model and site heterogeneity model were used the optimal model based on the Bayesian analysis to select the model “relaxed clock” and MCMC runs 200 million generations, and every 1000 steps in the individuals were to ensure effective sample size (ESS) in each parameter greater than 100. The output file assessed convergence in Tracer 1.5. The phylogenetic tree used TreeAnnotator 1.5.3 to discard 25% as burnin. Finally, the divergence time was analyzed and obtained in FigTree 1.1.2 [59].
3. Results
3.1. Sequences Discovery and Characterization
A total of 53.15 Gb raw data were produced by RRS (Raw Reads in NCBI BioProject ID: PRJNA557061). Then, 437,061 unique RRS loci across all the individuals were revealed by using the denove clustering method in ipyRAD. The reduced-filtered SNPs (1295 bp) were obtained under quality control (QC) in filtered SNPs by using Plink 1.9 to analyze the phylogeny, evolution, and divergence.
3.2. Phylogenetic Analyses
The MP, ML, and BI trees were obtained by reduced-filtered SNPs with strong support values (Figure 1), and the same clustering results were recovered in PCA (Figure 2). The phylogeny shown respectively in MP, ML, and BI tree were consistent in this study.
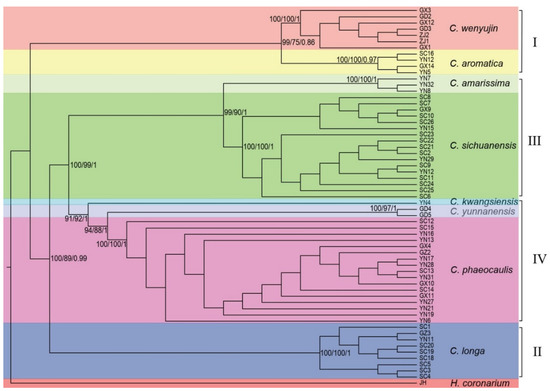
Figure 1.
The ML, MP, and BI trees of Chinese Curcuma. The values (maximum likelihood bootstrap/maximum parsimony bootstrap/Bayesian support value) were on the species clade. The Group I consisted of C. wenyujin and C. aromatica; Group II consisted of C. longa; The Group III consisted of C. amrissima and C. sichuanensis; The Group IV consisted of C. kwangsiensis, C. yunnanensis, and C. phaeocaulis.
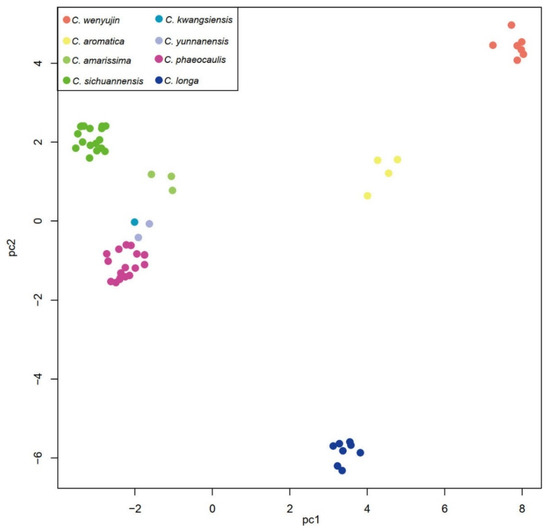
Figure 2.
The principle components analysis of Chinese Curcuma. Principal component analysis (PCA) of the 60 individuals and different species with different colors.
In the three phylogenetic trees, all population specimens from the same species were clustered together firstly and named as a clade in this study, and the eight Curcuma species were divided into four groups (including eight clades) with robust support values: Group I (including C. wenyujin clade and C. aromatica clade) with MP BS = 75, ML BS = 99, and BI PP = 86; Group II (C. longa clade) with MP BS = 100, ML BS = 100, and BI PP = 100; Group III (including C. amarissima clade and C. sichuanensis clade) with MP BS = 90, ML BS = 99, and BI PP = 100; and Group IV (including C. kwangsiensis clade, C. yunnanensis clade, and C. phaeocaulis clade) with MP BS = 92, ML BS = 91, and BI PP = 100. C. longa formed a monophyletic branch with MP BS = 89, ML BS = 100, and BI PP = 99; C. wenyujin clade (MP BS = 100, ML BS = 100, and BI PP = 100) was sister to C. aromatica clade (MP BS = 100, ML BS = 100, and BI PP = 97); C. amarissima clade (MP BS = 100, ML BS = 100, and BI PP = 100) clustered with C. sichuanensis clade (MP BS = 100, ML BS = 100, and BI PP = 100); C. kwangsiensis clade, the first branch in Group IV, was sister to C. yunnanensis clade (MP BS = 97, ML BS = 100, and BI PP = 100) and C. phaeocaulis clade (MP BS = 100, ML BS = 100, and BI PP = 100).
To test the evolution of Chinese Curcuma, a Bayesian clustering algorithm with admixed models was used to estimate the ancestral proportions (K) for each specimen (Figure 3). Based on CV error, the K = 12 represented the best model for these 60 samples. When K = 3, the eight species belong to three gene pools (blue gene pools, green gene pools, and red gene pools). C. longa had independent gene pools (blue). The species of C. amarissima, C. sichuanensis, C. kwangsiensis, C. yunnanensis, and C. phaeocaulis shared green gene pools. The red gene pools were shared by C. wenyujin and C. aromatica; When K = 4 to 12, C. longa could be distinguished by constitution of gene pool, and Group I (C. wenyujin and C. aromatica) is close to Group IV (C. kwangsiensis, C. yunnanensis, and C. phaeocaulis).
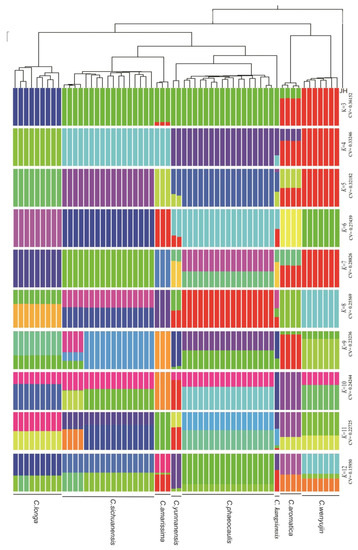
Figure 3.
Population genetic structure of Chinese Curcuma. When the kinship (K) = 3–12, each vertical bar represented a Curcuma sample and different color represented different putative ancestral background in putative.
3.3. Divergence Time Inference
Curcuma occurred in the Miocene (~7.45 Mya) in China (Figure 4). C. longa appeared around 6.43 Mya, the intraspecific of C. longa diversified at ~2.45 Mya from the late Pliocence to Quaternary. Group I (C. amarissima and C. sichuanensis) and II (C. kwangsiensis, C. yunnanensis and C. phaeocaulis) separated at ~4.83 Mya. The divergence in Group I, occurred at ~3.57 Mya in the Pliocence, and the intraspecific diversification of C. sichuanensis and C. amarissima were at ~1.89 Mya and ~0.67 Mya, respectively. In Group II, C. kwangsiensis occurred at ~3.74 Mya, C. yunnanensis and C. phaeocaulis emerged and separated ca 2.79 Mya, and the diversification within species of C. yunnanensis and C. phaeocaulis were at ~0.81 Mya and ~1.45 Mya. C. wenyujin and C. aromatica originated at ~6.08 Mya during the Miocene and Pliocence. The intraspecific diversification within C. wenyujin and C. aromatica were at ~1.17 Mya and ~0.56 Mya, respectively.
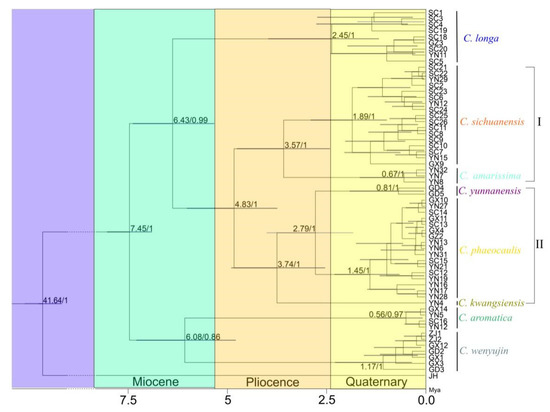
Figure 4.
Phylogeny tree resulting from the Bayesian inference and divergency time of Chinese Curcuma in BEAST (Divergence time/Bayesian support). Group I consisted of C. amarissima, C. sichuanensis, C. kwangsiensis, C. yunnanensis, and C. phaeocaulis. Group II was made up of C. kwangsiensis, C. yunnanensis, and C. phaeocaulis.
4. Discussion
4.1. Application of “Next-Generation” Sequencing in Estimating the Phylogeny and Biological Implication of Curcuma
The recent rapid speciation of species might lead to a mistake in inference by using a single phylogenetic tree, and this might be presented a poor fit in the multi-species coalescent supermatrix data and model [60,61]. A large amount of sequence data is ideal for establishing and reconstructing phylogenetic trees [5,28,41,62,63,64]. In this study, RRS was firstly used to produce big data to analyze the phylogeny and divergence of Chinese Curcuma. More loci and variable sites might improve the phylogeny reconstruction of Chinese Curcuma [5,41]. Compared to limited individuals and data size introduced in some previous studies, more individuals were involved to produce big data for mining more accurate information to recover a well-resolved phylogeny in this study [31].
4.2. Phylogeny Inference
The phylogeny of Curcuma species is difficult to resolve by depending on traditional approaches for their complicated origins (hybridization, introgression, and common appearance in species) [2,8,17]. The genetic relationship among Chinese Curcuma species is very close in this study, which is consistent with several previous studies [1,3,7].
Studies based on various morphological evidences reveal different results and lead to unreliable classifications of Curcuma: Based on pollen grains, Chen and Xia believed C. aromatica, C. yunnanensis, and C. longa had a close relationship, and the relationships among C. amarrisima, C. elata, C. flaviflora, C. phaecaulis, C. sichuanensis, and C. wenyujin were very close and hard to distinguish [16]. These species in Xiao et al. were delineated as Group I: C. longa, C. xanthorrhiza, and C. sichuanensis, Group II: C. kwangsiensis and C. exigua, Group III: C. wenyujin, C. aromatica, C. phaeocaulis C. zedoaria, and C. yunnanensis by using oil cells and vascular bundles [14]; and the morphology of leaves produced different results (Group I: C. longa, C. xanthorrhiza, C. wenyujin, and C. sichuanensis; Group II: C. kwangsiensis and C. exigua; and Group III: C. aromatica, C. chuanyujin, C. zedoaria, C. phaeocaulis, and C. yunnanensis) [15].
On the basis of molecular marker data (isoenzyme and DNA barcode), Deng et al. (2011a and 2011b) and Li et al. believed the relationship between C. longa and C. sichuanensis was very close, and supported that C. sichuanensis originated from the cultivated mutation of C. longa [1,3,65]. The genetic relationships among C. kwangsiensis, C. yunnanensis, C. aromatica, and C. phaeocaulis were poorly supported, and C. wenyujin clustered with C. longa, C. sichuanensis, and C. amarissima [5]. Chloroplast genomes study showed C. aromatica, C. wenyujin, and C. phaeocaulis have a close relationship and supported C. yunnanensis, C. amarissima, C. sichuanensis, and C. longa as a group [27]. In this study, the phylogeny of Chinese Curcuma is (((((C. Phaeocaulis, C. yunnanensis) C. kwangsiensis), (C. amarissima, C. sichuanensis)), C. longa), (C. wenyujin, C. aromatica)). C. longa is a single clade (MP BS = 100, ML BS = 100, and BI PP = 100), separated from C. sichuanensis clade, was sister to C. amarissima clade with strong support (MP BS = 90, ML BS = 99, and BI PP = 100), was inconsistent with the study of Deng et al. (C. sichuanensis clustered together with C. longa) [3]. The clade of C. wenyujin was close to C. aromatica clade [5] in Group I with MP BS = 75, ML BS = 99, and BI PP = 86. Based on RAMP and ISSR markers, C. phaeocaulis and C. yunnanensis had a close relationship [66,67]. In this study, C. kwangsiensis, C. yunnanensis, and C. phaeocaulis clustered together (Group IV) with robust support values (MP BS = 92, ML BS = 91, and BI PP = 100).
4.3. Divergence Time
The estimates of divergence indicated that the earliest appearance of Curcuma in China was in the Miocene. In the Miocene, large-scale orogenesis and geological events frequently emerged and influenced the speciation of plants living in QTP area during that times [68]. During the time, the drought climate (the Asian Monsoon) was related to intense uplift of QTP [69,70]. Curcuma lies dormant in winter, rhizomes fleshy with tuber-bearing roots, and blooms in the rainy season, which is similar to the drought-resistant plants to satisfy such drought climate [17]. The interspecific divergence of Chinese Curcuma occurred in ca. 4–2 Mya, which was coincided with the drought-resistant and deciduous plant sprang up and expanded rapidly in Hengduan Mountains ca. 4–2 Mya [71]. Chinese Curcuma expanded rapidly during this period, and the third intense uplift of QTP sped up their interspecific divergence.
In addition, hybridization and polyploidy play an important role on speciation and diversification [72,73]. According to previous studies on Curcuma, the distribution range overlapped, and the introgressive hybridization existed extensively [74]. Several Chinese Curcuma species were most likely to be of hybrid origin (C. aromatica and C. kwangsiensis) [8]. The cytomixis and chromosome doubling could promote the production of a large number of new phenotypes in a short time, and polyploids tend to be more adaptive than their parents [75,76,77]. Hybridization and polyploidization in plants are commonly distributed in the QTP regions, where the species overlap generally and the environment vary frequently, to satisfy the harsh environment of the QTP regions [78,79]. Chinese Curcuma species are confirmed to be polyploidy, which might help to adapt the harsh living environment at that time [27,74].
4.4. Future Directions
The phylogeny of major Chinese Curcuma species was improved in this study. Chinese Curcuma species are only a subset of Curcuma around the world. More species with population samples, as well as species from the genera of Hitchenia, Stahlianthus, and Smithatris, and more data should be involved to analyze the phylogeny, evolution, and diversification of Curcuma [6].
In addition, Curcuma has a complex evolutionary history; hence, more new methods such as ddRRS, ddRad, and transcriptome should be used to ensure the reliability of data for the phylogeny, evolution, and diversification reconstruction on such recent radiated genera [80].
5. Conclusions
The RRS was firstly involved to improve the phylogeny and divergence of Chinese Curcuma. The third intense uplift of QTP might speed up the interspecific divergence of Curcuma in China. Overall, this study provides valuable information on the origin of Chinese Curcuma.
Author Contributions
H.L. and R.Y. designed the experiments and analyzed the data. H.L. wrote the original manuscript. G.G. and J.D. collected the plant materials. K.X. and R.Y. identified the materials. L.Z., C.D., and H.W. assisted with manuscript preparation. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National modern agricultural industry technology system Sichuan innovation team (SCCXTD-2020-19), the Science and Technology Foundation of Guizhou Province (Qiankehejichu [2017]1138), and the Education Department of Guizhou Province for youth growth on science and technology (No. Qianjiaohe KY[2016](221)).
Data Availability Statement
These plant materials are required for the collection of plant individuals. The plant materials are maintained in accordance with the institutional guidelines of the College of Life Sciences, Sichuan Agricultural University, China. The Raw Reads are available in NCBI BioProject ID: PRJNA557061.
Acknowledgments
This manuscript has been released as a pre-print at https://www.researchsquare.com/article/rs-5880/v1 (accessed on 27 September 2019) [81]. We would like to thank Khawaja Shafique Ahamad (Department of Botany, University of Poonch Rawalakot, Azad Jammu, and Kashmir Pakistan) for improving the quality of the manuscript. We are grateful to Chuanbei Jiang (Genepioneer Biotechnologies Co. Ltd., Nanjing) for assistance in RRS library preparation. We also thank Zhimemg Wang (College of Life Science, Peking University, Beijing) for helping us run pyRAD and kindly providing additional help in assembly programs. My sincere gratitude also goes to Keyan Zhang (Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Yunnan) for valuable comments for this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Deng, J.B.; Ding, C.B.; Zhang, L.; Zhou, Y.H.; Yang, R.W. Relationships among six herbal species (Curcuma) assessed by four isozymes. Phyton 2011, 80, 181–188. [Google Scholar]
- Zaveska, E.; Fer, T.; Sida, O.; Krak, K.; Marhold, K.; Leong-Skornickova, J. Phylogeny of Curcuma (Zingiberaceae) based on plastid and nuclear sequences: Proposal of the new subgenus Ecomata. Taxon 2012, 61, 747–763. [Google Scholar] [CrossRef]
- Deng, J.B.; Ding, C.; Zhang, L.; Yang, R.; Zhou, Y. Authentication of three related herbal species (Curcuma) by DNA barcoding. J. Med. Plants Res. 2011, 5, 6401–6406. [Google Scholar] [CrossRef]
- Nian, L.; Telin, W. Notes on Curcuma in China. J. Trop. Subtrop. Bot. 1999, 7, 146–150. [Google Scholar] [CrossRef]
- Deng, J.B.; Gao, G.; Ahmad, K.; Luo, X.; Zhang, F.; Li, S.; Yang, R. Evaluation on genetic relationships among China’s endemic Curcuma L. herbs by mtDNA. Phyton Int. J. Exp. Bot. 2018, 87, 156–161. [Google Scholar]
- Kress, W.J.; Prince, L.M.; Williams, K.J. The phylogeny and a new classification of the gingers (Zingiberaceae): Evidence from molecular data. Am. J. Bot. 2002, 89, 1682–1696. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, J.T.; Xia, N.H. Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. Mol. Ecol. Resour. 2014, 2, 337–348. [Google Scholar]
- Zaveska, E.; Fer, T.; Sida, O.; Marhold, K.; Leong-Skornickova, J. Hybridization among distantly related species: Examples from the polyploid genus Curcuma (Zingiberaceae). Mol. Phylogenet. Evol. 2016, 100, 303–321. [Google Scholar] [CrossRef] [PubMed]
- Li, J. The Qinghai-Tibet Plateau Uplifting and Environmental Evolution in Asia: Article Collection of Academician; Science Press: Beijing, China, 2006. [Google Scholar]
- Liu, X.; Dong, B. Influence of the Tibetan Plateau uplift on the Asian monsoon-arid environment evolution. Chin. Sci. Bull. 2013, 58, 4277–4291. [Google Scholar] [CrossRef]
- Hoorn, C.; Mosbrugger, V.; Mulch, A.; Antonelli, A. Biodiversity from mountain building. Nat. Geosci. 2013, 6, 154. [Google Scholar] [CrossRef]
- Wen, J.; Zhang, J.Q.; Nie, Z.L.; Zhong, Y.; Sun, H. Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 2014, 5, 4. [Google Scholar] [CrossRef]
- Favre, A.; Päckert, M.; Pauls, S.U.; Jähnig, S.C.; Uhl, D.; Michalak, I.; Muellner-Riehl, A.N. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. Camb. Philos. Soc. 2015, 90, 236–253. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.H.; Shu, G.M.; Li, L.Y.; Fang, Q.M.; Su, Z.W. Histological and morphological studies on the rhizomes of Curcuma. China J. Chin. Mater. Med. 2004, 20, 84–87. [Google Scholar]
- Xiao, X.H.; Zhao, Y.L.; Cheng, J.; Shu, G.M.; Shu, Z.W. Histological and morphological studies on leaves of Curcuma in China. China J. Chin. Mater. Med. 2004, 29, 203–207. [Google Scholar]
- Chen, J.; Xia, N.H. Pollen morphology of Chinese Curcuma L. and Boesenbergia Kuntz (Zingiberaceae): Taxonomic implications. Flora 2011, 206, 458–467. [Google Scholar] [CrossRef]
- Leong-Skornickova, J.; Šída, O.; Záveská, E.; Marhold, K. History of infrageneric classification, typification of supraspecific names and outstanding transfers in Curcuma (Zingiberaceae). Taxon 2015, 64, 362–373. [Google Scholar] [CrossRef]
- Leong-Škorničková, J.; Šída, O.; Jarolímová, V.; Sabu, M.; Fér, T.; Trávníček, P.; Suda, J. Chromosome Numbers and Genome Size Variation in Indian Species of Curcuma (Zingiberaceae). Ann. Bot. 2007, 100, 505–526. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xia, N.; Zhao, J.; Chen, J.; Henny, R.J. Chromosome Numbers and Ploidy Levels of Chinese Curcuma Species. Hortscience 2013, 48, 525–530. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Huang, X.X.; Huang, S.F. A report on chromosome numbers on Chinese Zingiberaceae (5). Guihaia 1988, 8, 143–147. [Google Scholar]
- Chen, Z.Y.; Chen, Z.S.; Huang, F.S. A report on chromosome numbers on Chinese Zingiberaceae (2). Guihaia 1984, 4, 13–18. [Google Scholar]
- Sato, D. The karyotype analysis in Zingiberales with special reference to the protokaryotype and stable karyotype. Sci. Papers Coll. Gen. Educ. Univ. Tokyo 1960, 10, 225–243. [Google Scholar]
- Nian, L. The Taxonomic Study of Curcuma L. from China; South China Botanical Garden: Guangzhou, China, 1985. [Google Scholar]
- Sugiura, T. Studies on the chromosome numbers in higher plants. Cytologia 1936, 7, 544–595. [Google Scholar] [CrossRef]
- Raghavan, T.; Venkatasubban, K. Cytological studies in the family Zingiberace with special reference to chromosome number and Cyto-Taxonomy. In Proceedings of the Indian Academy of Sciences—Section B; Springer: New Delhi, India, 1943; Volume 17, pp. 118–132. [Google Scholar] [CrossRef]
- Ramachandran, K. Chromosome numbers in the genus Curcuma Linn. Curr. Sci. 1961, 30, 194–196. [Google Scholar]
- Liang, H.; Zhang, Y.; Deng, J.; Gao, G.; Ding, C.; Zhang, L.; Yang, R. The complete chloroplast genome sequences of 14 Curcuma species: Insights into genome evolution and phylogenetic relationships within Zingiberales. Front. Genet. 2020, 11, 802. [Google Scholar] [CrossRef]
- Cao, H.; Sasaki, Y.; Fushimi, H.; Komatsu, K. Molecular analysis of medicinally-used Chinese and Japanese Curcuma based on 18S rRNA gene and trnK gene sequences. Biol. Pharm. Bull. 2001, 24, 1389–1394. [Google Scholar] [CrossRef]
- Newmaster, S.G.; Subramanyam, R. Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Mol. Ecol. Resour. 2009, 9, 172–180. [Google Scholar]
- Lemmon, E.M.; Lemmon, A.R. High-throughput identification of informative nuclear loci for shallow-scale phylogenetics and phylogeography. Syst. Biol. 2012, 61, 745–761. [Google Scholar] [CrossRef]
- Harvey, M.G.; Smith, B.T.; Glenn, T.C.; Faircloth, B.C.; Brumfield, R.T. Sequence capture versus restriction site associated DNA sequencing for shallow systematics. Syst. Biol. 2016, 65, 910–924. [Google Scholar] [CrossRef]
- Godden, G.T.; Jordon-Thaden, I.E.; Chamala, S.; Crowl, A.A.; Garcia, N.; Germain-Aubrey, C.C.; Heaney, J.M.; Latvis, M.; Qi, X.; Gitzendanner, M.A. Making next-generation sequencing work for you: Approaches and practical considerations for marker development and phylogenetics. Plant Ecol. Divers. 2012, 5, 427–450. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.-L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef]
- Nemati, Z.; Blattner, F.R.; Kerndorff, H.; Erol, O.; Harpke, D. Phylogeny of the saffron-crocus species group, Crocus series Crocus (Iridaceae). Mol. Phylogenet. Evol. 2018, 127, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E.; Orban, L. A robust, simple Genotyping-by-Sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Kumar, S.; Filipski, A.J.; Battistuzzi, F.U.; Pond, S.L.K.; Tamura, K. Statistics and Truth in Phylogenomics. Mol. Biol. Evol. 2012, 29, 457–472. [Google Scholar] [CrossRef]
- Ward, J.A.; Bhangoo, J.; Fernández-Fernández, F.; Moore, P.; Swanson, J.D.; Viola, R.; Velasco, R.; Bassil, N.; Weber, C.A.; Sargent, D.J. Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genom. 2013, 14, 1471–2164. [Google Scholar] [CrossRef]
- Bernhardt, N.; Brassac, J.; Dong, X.; Willing, E.M.; Poskar, C.H.; Kilian, B.; Blattner, F.R. Genome-wide sequence information reveals recurrent hybridization among diploid wheat wild relatives. Plant J. 2020, 102, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Perez-Escobar, O.A.; Bogarin, D.; Schley, R.; Bateman, R.M.; Gerlach, G.; Harpke, D.; Brassac, J.; Fernandez-Mazuecos, M.; Dodsworth, S.; Hagsater, E.; et al. Resolving relationships in an exceedingly young Neotropical orchid lineage using Genotyping-by-sequencing data. Mol. Phylogenet. Evol. 2020, 144, 12. [Google Scholar] [CrossRef]
- Deng, J.B.; Liu, J.; Ahmad, K.; Ding, C.; Zhang, L.; Zhou, Y.; Yang, R. Relationships evaluation on six herbal species (Curcuma) by dna barcoding. Pak. J. Bot. 2015, 47, 1103–1109. [Google Scholar]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.-X.; Fu, C.-X.; Comes, H. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 2011, 59, 225–244. [Google Scholar] [CrossRef]
- Fan, D.M.; Yue, J.P.; Nie, Z.L.; Li, Z.M.; Comes, H.P.; Sun, H. Phylogeography of Sophora davidii (Leguminosae) across the ‘Tanaka-Kaiyong Line’, an important phytogeographic boundary in Southwest China. Mol. Ecol. 2013, 22, 4270–4288. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Ree, R.H. Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. USA 2017, 114, 3444–3451. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fang, X. Uplift of the Tibetan Plateau and environmental changes. Chin. Sci. Bull. 1999, 44, 2117–2124. [Google Scholar] [CrossRef]
- Yu, S.-H.; Wen, Q.-Z. Geochemical evolution and environmental changes of Qinghai—Xizang Plateau Since Late Cenozoic. Acta Geochim. 1998, 17, 258–264. [Google Scholar]
- Doyle, J. DNA Protocols for Plants-CTAB Total DNA Isolation; Springer: Berlin, Germany, 1991; Volume 283–293. [Google Scholar]
- Grabowski, P.P.; Morris, G.P.; Casler, M.D.; Borevitz, J.O. Population genomic variation reveals roles of history, adaptation and ploidy in switchgrass. Mol. Ecol. 2014, 23, 4059–4073. [Google Scholar] [CrossRef] [PubMed]
- Eaton, D.A.; Overcast, I. ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 2020, 36, 2592–2594. [Google Scholar] [CrossRef]
- Catchen, J.M.; Amores, A.; Hohenlohe, P.; Cresko, W.; Postlethwait, J.H.; De Koning, D.J. Stacks: Building and genotyping loci de novo from short-read sequences. G3 Genes Genomes Genet. 2011, 1, 171–182. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest v2.(Program Distributed by the Author) Evolutionary Biology Centre; Uppsala University: Uppsala, Sweden, 2004; Available online: http://www.ebc.uu.se/systzoo/staff/nylander.html (accessed on 11 September 2018).
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Drummond, A.J. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 2010, 27, 570–580. [Google Scholar]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Durand, E.Y.; Patterson, N.; Reich, D.; Slatkin, M. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 2011, 28, 2239–2252. [Google Scholar] [CrossRef]
- Zheng, M.L. A Phylogenetic Study on the Tribe Zingibereae (Zingiberaceae). Ph.D. Thesis, Xishuangbanna Tropical Botanical Garden Chinese Academy of Sciences, Xishuangbanna, China, 2010. [Google Scholar]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Joseph, L.; Edwards, S.V. A species tree for the australo-papuan fairy-wrens and allies (Aves: Maluridae). Syst. Biol. 2012, 61, 253–271. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, J.; Knowles, L.L. Multispecies coalescent delimits structure, not species. Proc. Natl. Acad. Sci. USA 2017, 114, 1607–1612. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.H.; Cutler, J.S.; Friel, J.P.; Dening Touokong, C.; Coop, G.; Wainwright, P.C. Complex histories of repeated gene flow in Cameroon crater lake cichlids cast doubt on one of the clearest examples of sympatric speciation. Evolution 2015, 69, 1406–1422. [Google Scholar] [CrossRef]
- Meier, J.I.; Marques, D.A.; Mwaiko, S.; Wagner, C.E.; Excoffier, L.; Seehausen, O. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Moore, M.J.; Soltis, P.S.; Bell, C.D.; Burleigh, J.G.; Soltis, D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 2010, 107, 4623–4628. [Google Scholar] [CrossRef]
- Li, Y.Y.; Lei, Y.X.; Gao, G.; Zhang, Y.; Deng, J.B.; Tong, S.S.; Yang, R.W. Genetic Diversity of Chinese Curcuma based on RAMP Makers. Genom. Appl. Biol. 2015, 34, 1784–1790. [Google Scholar]
- Xiao, X.H.; Liu, F.Q.; Shi, C.H.; Li, L.Y.; Qiao, C.Z.; Su, Z.W. RAPD polymorphism and Authentication of Medicinal Plants from Turmeri (Curcuma L. ) in China. Chin. Tradit. Herb. Drugs 2000, 31, 209–212. [Google Scholar]
- Li, Y.Y.; Lei, Y.X.; Gao, G.; Zhang, Y.; Deng, J.B.; Tong, S.S.; Yang, R.W. Genetic diversity analysis of Chinese Curcuma based on ISSR makers. Mol. Plant Breed. 2016, 14, 1189–1194. [Google Scholar]
- Zhou, Z.; Huang, J.; Ding, W. The impact of major geological events on Chinese flora. Biodivers. Sci. 2017, 25, 17–23. [Google Scholar] [CrossRef]
- Li, J.J.; Fang, X.M.; Pan, B.T.; Zhao, Z.J.; Song, Y.G. Late cenozoic intensive uplift of Qinghai-Xizang plateau and its impacts on environments in surrounding area. Quat. Sci. 2001, 21, 381–391. [Google Scholar]
- Yi, G.Z.; Ji, J.; Balsam, W.; Liu, L.; Chen, J. Mid-pliocene asian monsoon intensification and the onset of northern hemisphere glaciation. Geology 2009, 37, 599–602. [Google Scholar]
- Biasatti, D.; Yang, W.; Feng, G.; Xu, Y.; Flynn, L. Paleoecologies and paleoclimates of late cenozoic mammals from Southwest China: Evidence from stable carbon and oxygen isotopes. J. Asian Earth Sci. 2012, 44, 48–61. [Google Scholar] [CrossRef]
- Mallet, J. Hybrid speciation. Nature 2007, 446, 279–283. [Google Scholar] [CrossRef]
- Abbott, R.J.; Albach, D.; Ansell, S.; Arntzen, J.W.; Zinner, D. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef]
- Gui, L.; Jiang, S.; Xie, D.; Yu, L.; Huang, Y.; Zhang, Z.; Liu, Y. Analysis of complete chloroplast genomes of Curcuma and the contribution to phylogeny and adaptive evolution. Gene 2020, 732, 144355. [Google Scholar] [CrossRef]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef]
- Chao, D.-Y.; Dilkes, B.; Luo, H.; Douglas, A.; Yakubova, E.; Lahner, B.; Salt, D.E. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 2013, 341, 658–659. [Google Scholar] [CrossRef] [PubMed]
- Selmecki, A.M.; Maruvka, Y.E.; Richmond, P.A.; Guillet, M.; Shoresh, N.; Sorenson, A.L.; De, S.; Kishony, R.; Michor, F.; Dowell, R. Polyploidy can drive rapid adaptation in yeast. Nature 2015, 519, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Yang, Z. Testing hybridization hypotheses based on incongruent gene trees. Syst. Biol. 2000, 48, 422–434. [Google Scholar]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef] [PubMed]
- Escallon, E.V.; Richardson, J.E.; Kidner, C.A.; Madriñán, S.; Stone, G.S. Transcriptome mining for phylogenetic markers in a recently radiated genus of tropical plants ( Renealmia L.f., Zingiberaceae). Mol. Phylogenet. Evol. 2017, 119, 13–24. [Google Scholar]
- Liang, H.; Zhang, Y.; Deng, J.; Gao, G.; Ding, C.; Zhang, L.; Yu, X.; Zhou, Y.; Yang, R. Application of genotyping-by-sequencing data on inferring the phylogeny of Curcuma (Zingiberaceae) from China. Res. Sq. 2019. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).