Recovery of Soil-Denitrifying Community along a Chronosequence of Sand-Fixation Forest in a Semi-Arid Desertified Grassland
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Location and Site Description
2.2. Experimental Design and Soil Sampling
2.3. Soil Property Analysis
2.4. Clone Library Construction
2.5. Quantifications of nosZ, nirK, and nirS Genes
2.6. Data Analysis
3. Results
3.1. Soil Properties
3.2. Diversities of nirK, nirS, and nosZ Gene Libraries
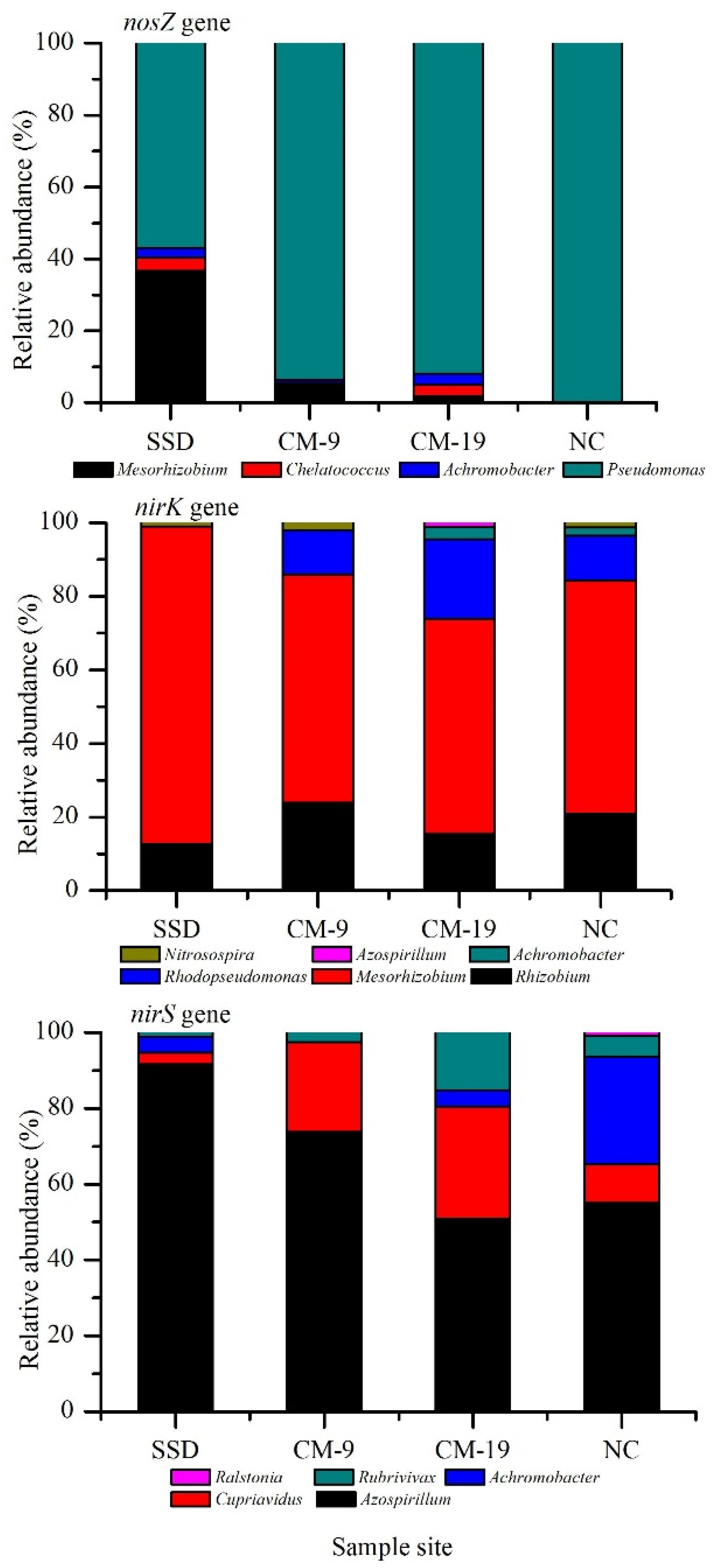
3.3. Abundance of the nirK, nirS, and nosZ Genes
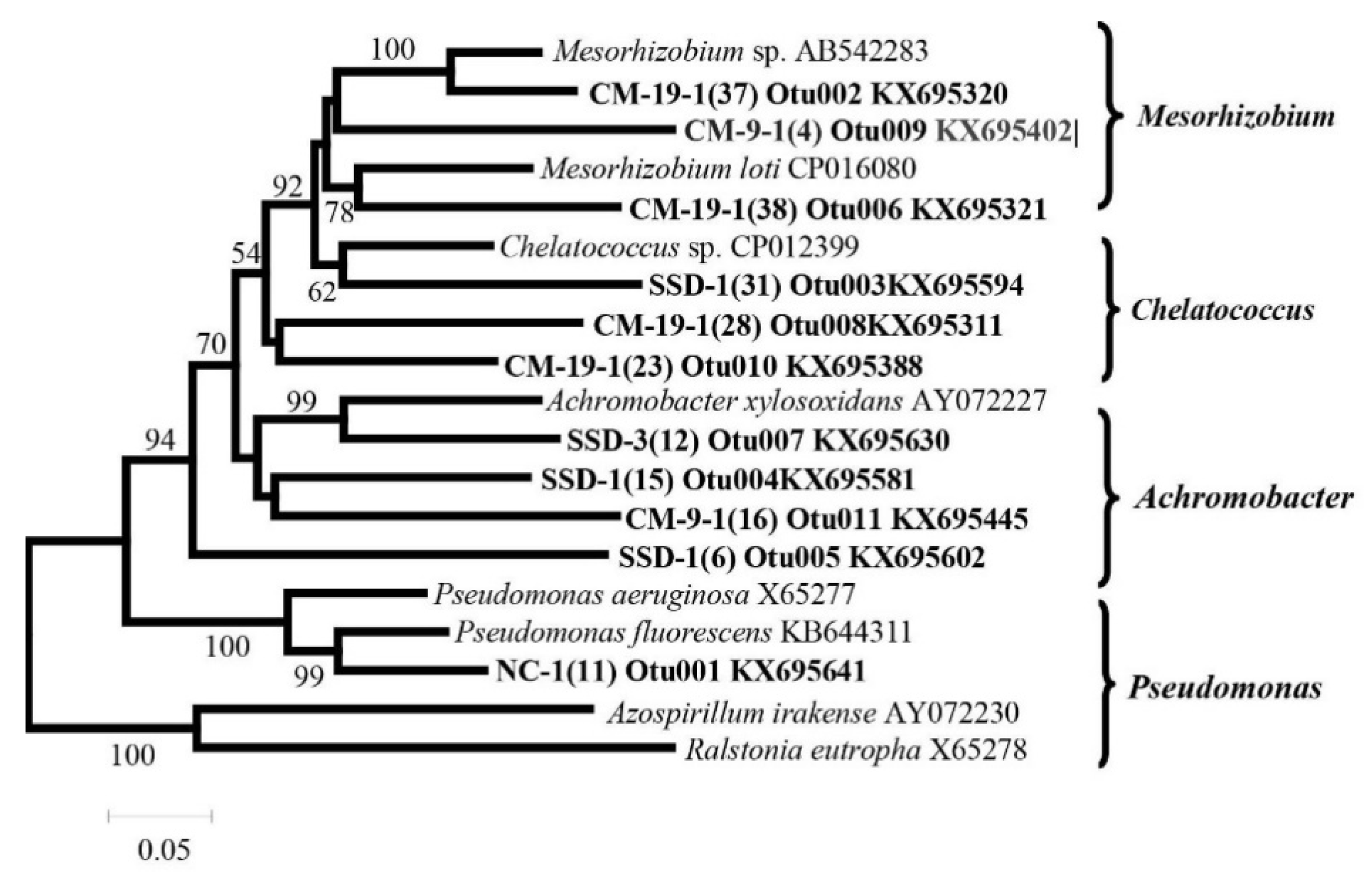
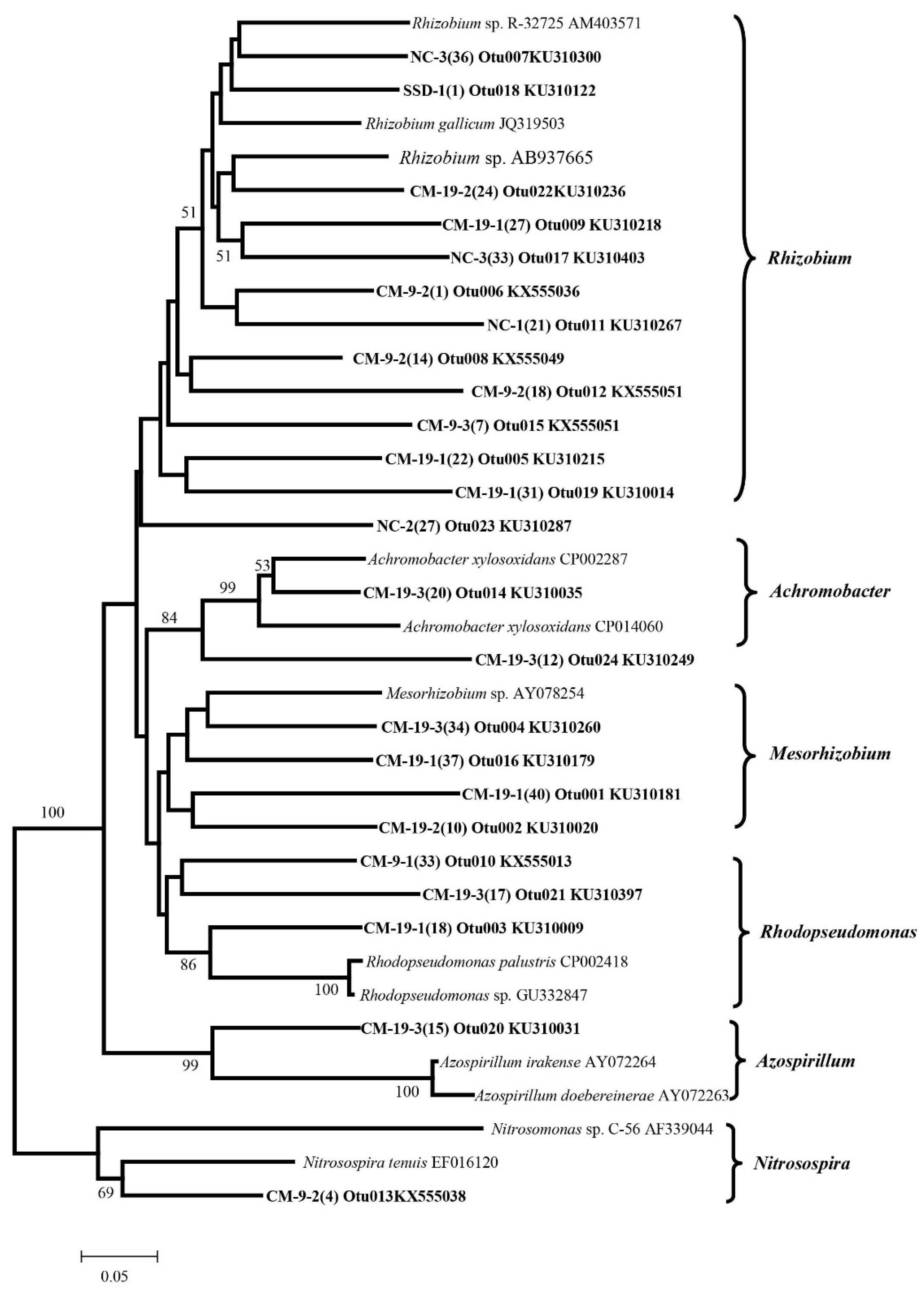
3.4. Detection of Denitrifying Bacterial Taxa
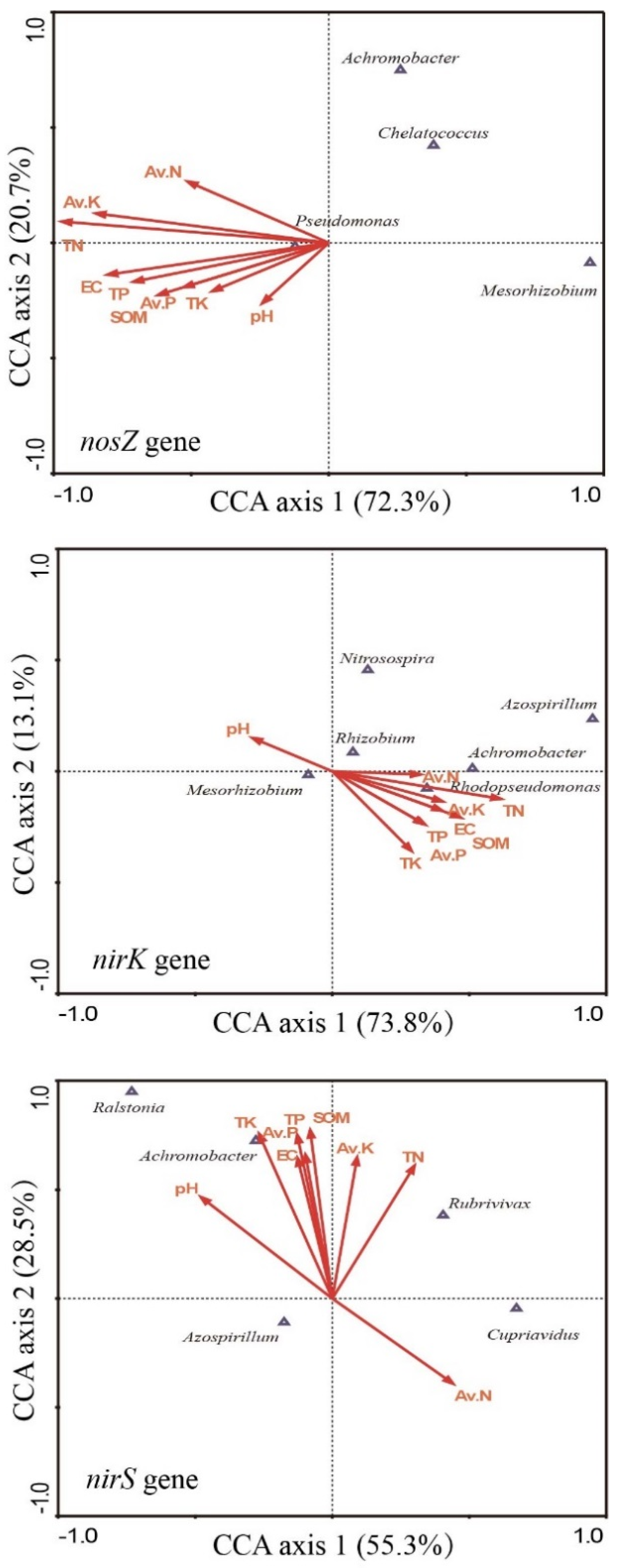
3.5. Relationship between the Composition of the Denitrifying Community and Soil Properties
4. Discussion
4.1. Improvement of Sand-Fixation Forest on Soil Nutrients
4.2. Effect of Revegetation on Soil-Denitrifying Communities
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, C.Y.; Jiang, D.M.; Teng, X.H.; Jiang, Y.; Liang, W.J.; Cui, Z.B. Soil chemical and microbiological properties along a chronosequence of Caragana microphylla Lam. plantations in the Horqin Sandy Land of Northeast China. Appl. Soil Ecol. 2008, 40, 78–85. [Google Scholar] [CrossRef]
- Liu, X.M.; Zhao, H.L. Comprehensive Strategy for Eco-Environmental Control in Horqin Sand Land; Gansu Science and Technology Publishing House: Lanzhou, China, 1993. [Google Scholar]
- Jiang, F.Q.; Cao, C.Y.; Zeng, D.H. Degradation and Restoration of Ecosystems on Horqin Sand Land; Chinese Forestry Publishing House: Beijing, China, 2002. [Google Scholar]
- Cao, C.Y.; Jiang, S.Y.; Zhang, Y.; Zhang, F.X.; Han, X.S. Spatial variability of soil nutrients and microbiological properties after the establishment of leguminous shrub Caragana microphylla Lam. plantation on sand dune in the Horqin Sandy Land of Northeast China. Ecol. Eng. 2011, 37, 1467–1475. [Google Scholar] [CrossRef]
- Cao, C.Y.; Jiang, D.M.; Alamsa; Luo, Y.M.; Kou, Z.W. Ecological processes of vegetation restoration of Caragana mirophylla in the sand fixing area. Chin. J. Appl. Ecol. 2000, 11, 349–354. [Google Scholar]
- Guan, P.T.; Zhang, X.K.; Yu, J.; Ma, N.N.; Liang, W.J. Variation of soil nematode community composition with increasing sand-fixation year of Caragana microphylla: Bioindication for desertification restoration. Ecol. Eng. 2018, 81, 93–101. [Google Scholar] [CrossRef]
- Su, Y.Z.; Zhao, H.F. Soil properties and plant species in an age sequence of Caragana microphylla plantations in the Horqin Sandy Land, north China. Ecol. Eng. 2003, 20, 223–235. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, G.L.; Sun, Y.J.; Yang, Y.; Zhou, H.J. Effects of Thinning on microbial community structure in the organic horizon of Chinese pine plantations in Badaling, Beijing, China. Forests 2019, 10, 828. [Google Scholar] [CrossRef]
- Cao, C.Y.; Zhang, Y.; Cui, Z.B.; Feng, S.W.; Wang, T.T.; Ren, Q. Soil bacterial community responses to revegetation of moving sand dune in semi-arid grassland. Appl. Microbiol. Biotechnol. 2017, 101, 6217–6228. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.M.; Cao, C.Y.; Zhang, Y.; Cui, Z.B.; Han, X.S. Plantations of native shrub species restore soil microbial diversity in the Horqin Sandy Land, northeastern China. J. Arid Land 2014, 6, 445–453. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, C.Y.; Peng, M.; Xu, X.J.; Zhang, P.; Yu, Q.J.; Sun, T. Diversity of nitrogen-fixing, ammonia-oxidizing, and denitrifying bacteria in biological soil crusts of a revegetation area in Horqin Sandy Land, Northeast China. Ecol. Eng. 2014, 41, 71–79. [Google Scholar] [CrossRef]
- Liu, L.C.; Liu, Y.B.; Hui, R.; Xie, M. Recovery of microbial community structure of biological soil crusts in successional stages of Shapotou desert revegetation, northwest China. Soil Biol. Biochem. 2017, 107, 125–128. [Google Scholar] [CrossRef]
- Zhelezova, A.; Chernov, T.; Tkhakakhova, A.; Xenofontova, N.; Semenov, M.; Kutovaya, O. Prokaryotic community shifts during soil formation on sands in the tundra zone. PLoS ONE 2019, 14, e0206777. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.A.; Zhao, H.L.; Zhao, X.Y.; Zhang, T.H.; Guo, Y.R.; Wang, S.K.; Drake, S. Spatial pattern and heterogeneity of soil properties in sand dunes under grazing and restoration in Horqin Sandy Land, Northern China. Soil Tillage Res. 2008, 99, 202–212. [Google Scholar] [CrossRef]
- Che, R.; Qin, J.; Tahmasbian, I.; Wang, F.; Zhou, S.; Xu, Z.; Cui, X.Y. Litter amendment rather than phosphorus can dramatically change inorganic nitrogen pools in a degraded grassland soil by affecting nitrogen-cycling microbes. Soil Biol. Biochem. 2018, 120, 145–152. [Google Scholar] [CrossRef]
- Zumft, W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Hojberg, O. Dissimilatory nitrate reductases in bacteria. Biochim. Et Biophys. Acta-Gene Struct. Expressopn 1999, 1446, 1–23. [Google Scholar] [CrossRef]
- Bru, D.; Sarr, A.; Philippot, L. Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 2007, 73, 5971–5974. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.; Baudoin, E.; Lopez-Gutierrez, J.C.; Martin-Laurent, F.; Brauman, A.; Philippot, L. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 2004, 59, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Horn, M.A.; Drake, H.L.; Schramm, A. Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar. Appl. Environ. Microbiol. 2006, 72, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.; Tortosa, G.; Granados, A.; Delgado, A.; Bedmar, E.J.; Delgado, M.J. Involvement of Bradyrhizobium japonicum denitrification in symbiotic nitrogen fixation by soybean plants subjected to flooding. Soil Biol. Biochem. 2011, 43, 212–217. [Google Scholar] [CrossRef]
- Li, H.L.; Zhang, Y.; Wang, T.T.; Feng, S.W.; Ren, Q.; Cui, Z.B.; Cao, C.Y. Responses of soil denitrifying bacterial communities carrying nirS, nirK, and nosZ genes to revegetation of moving sand dunes. Ecol. Indictors 2019, 107, 105541. [Google Scholar] [CrossRef]
- FAO. FAO/ IUSS Working Group WRB, World reference base for soil resources. In World Soil Resources Reports103; FAO: Rome, Italy, 2006. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon and Organic Matter, Part 2. In Methods of Soil Analysis, 2nd ed.; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy: Madison, WI, USA, 1982; Volume 9, pp. 539–577. [Google Scholar]
- Institute of Soil Science, Chinese Academy of Sciences (ISSCAS). Physical and Chemical Analysis Methods of Soils; Shanghai Science Technology Press: Shanghai, China, 1978; pp. 7–59. [Google Scholar]
- Rich, J.J.; Heichen, R.S.; Bottomley, P.J.; Cromack, K.; Myrold, D.D. Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl. Environ. Microbiol. 2003, 69, 5974–5982. [Google Scholar] [CrossRef]
- Throbäck, I.N.; Enwall, K.; Jarvis, A.; Hallin, S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 2004, 49, 401–417. [Google Scholar] [CrossRef]
- Tippmann, H.F. Analysis for free: Comparing programs for sequence analysis. Brief. Bioinform. 2004, 5, 82–87. [Google Scholar] [CrossRef]
- Han, M.Q.; Li, Z.Y.; Zhang, F.L. The ammonia oxidizing and denitrifying prokaryotes associated with sponges from different sea areas. Microb. Ecol. 2013, 66, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Nei, M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004, 5, 150–163. [Google Scholar] [CrossRef]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006, 72, 5181–5189. [Google Scholar] [CrossRef]
- Kandeler, E.; Deiglmayr, K.; Tscherko, D.; Bru, D.; Philippot, L. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 2006, 72, 5957–5962. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Li, S.G.; Harazono, Y.; Zhao, H.L.; He, Z.Y.; Chang, X.L.; Zhao, X.Y.; Zhang, T.H.; Oikawa, T. Micrometeorological changes following establishment of artificially established Artemisia vegetation on desertified sandy land in the Horqin sandy land, China and their implication on regional environmental change. J. Arid Environ. 2002, 52, 101–119. [Google Scholar] [CrossRef]
- Wang, H.L.; Deng, N.; Wu, D.Y.; Hu, S.; Kou, M. Long-term net transformation and quantitative molecular mechanisms of soil nitrogen during natural vegetation recovery of abandoned farmland on the Loess Plateau of China. Sci. Total Environ. 2017, 607, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Ligi, T.; Truu, M.; Truu, J.; Nolvak, H.; Kaasik, A.; Mitsch, W.J.; Mander, U. Effects of soil chemical characteristics and water regime on denitrification genes (nirS, nirK, and nosZ) abundances in a created riverine wetland complex. Ecol. Eng. 2014, 42, 47–55. [Google Scholar] [CrossRef]
- Kara, O.; Bolat, I.; Cakiroglu, K.; Ozturk, M. Plant canopy effects on litter accumulation and soil microbial biomass in two temperate forests. Biol. Fertil. Soils 2008, 45, 193–198. [Google Scholar] [CrossRef]
- Jangid, K.; Williams, M.A.; Franzluebbers, A.J.; Schmidt, T.M.; Coleman, D.C.; Whitman, W.B. Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties. Soil Biol. Biochem. 2011, 43, 2184–2193. [Google Scholar] [CrossRef]
- Prescott, C.E.; Grayston, S.J. Tree species influence on microbial communities in litter and soil: Current knowledge and research needs. For. Ecol. Manag. 2013, 309, 19–27. [Google Scholar] [CrossRef]
- Liu, J.L.; Dang, P.; Gao, Y.; Zhu, H.L.; Zhu, H.N.; Zhao, F.; Zhao, Z. Effects of tree species and soil properties on the composition and diversity of the soil bacterial community following afforestation. For. Ecol. Manag. 2018, 427, 342–349. [Google Scholar] [CrossRef]
- Jurksiene, G.; Janusauskaite, D.; Baliuckas, V. Microbial community analysis of Nnative Pinus sylvestris L. and alien Pinus mugo L. on dune sands as determined by Ecoplates. Forests 2020, 11, 1202. [Google Scholar] [CrossRef]
- Yoshida, M.; Ishii, S.; Otsuka, S.; Senoo, K. Temporal shifts in diversity and quantity of nirS and nirK in a rice paddy field soil. Soil Biol. Biochem. 2009, 41, 2044–2051. [Google Scholar] [CrossRef]
- Katsuyama, C.; Kondo, N.; Suwa, Y.; Yamagishi, T.; Itoh, M.; Ohte, N.; Kimura, H.; Nagaosa, K.; Kato, K. Denitrification activity and relevant bacteria revealed by nitrite reductase gene fragments in soil of temperate mixed forest. Microbes Environ. 2008, 23, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Priemé, A.; Braker, G.; Tiedje, J.M. Diversity of nitrite reductase (nirK and nirS) gene fragments in forested upland and wetland soils. Appl. Environ. Microbiol. 2002, 68, 1893–1900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cao, C.Y.; Han, X.S.; Jiang, S.Y. Soil nutrient and microbiological property recoveries via native shrub and semi-shrub plantations on moving sand dunes in Northeast China. Ecol. Eng. 2013, 53, 1–5. [Google Scholar] [CrossRef]
- Molina, L.; Ramos, C.; Duque, E.; Ronchel, M.C.; Garcia, J.M.; Wyke, L.; Ramos, J.L. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 2000, 32, 315–321. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, C.Y.; Cui, Z.B.; Qian, W.; Liang, C.P.; Wang, C.M. Soil bacterial community restoration along a chronosequence of sand-fixing plantations on moving sand dunes in the Horqin sandy land in northeast China. J. Arid Environ. 2019, 165, 81–87. [Google Scholar] [CrossRef]
- Sun, Y.F.; Shen, J.P.; Zhang, C.J.; Zhang, L.M.; Bai, W.M.; Fang, Y.; He, J.Z. Responses of soil microbial community to nitrogen fertilizer and precipitation regimes in a semi-arid steppe. J. Soils Sediments 2018, 18, 762–774. [Google Scholar] [CrossRef]
- Urbanova, M.; Snajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Graham, E.B.; Wieder, W.R.; Leff, J.W.; Weintraub, S.R.; Townsend, A.R.; Cleveland, C.C.; Philippot, L.; Nemergut, D.R. Do we need to understand microbial communities to predict ecosystem function? A comparison of statistical models of nitrogen cycling processes. Soil Biol. Biochem. 2014, 68, 279–282. [Google Scholar] [CrossRef]


| Items | SSD | CM-9 | CM-19 | NC | ANOVA in Response to Age | |
|---|---|---|---|---|---|---|
| Fregression | p | |||||
| Vegetation coverage (%) | <5 | 55 | 70 | 75 | – | – |
| Crown diameter (cm × cm) | – | 70 × 75 | 80 × 85 | 140 × 155 | – | – |
| Mean height (cm) | – | 81.21 | 90.52 | 130.21 | – | – |
| Shoot number (N) | – | 18.55 | 19.62 | 40.54 | – | – |
| pH | 6.70 ± 0.05 | 6.75 ± 0.05 | 6.63 ± 0.02 | 6.89 ± 0.07 | 9.196 | 0.013 |
| Electrical conductivity (µs·cm−1) | 23.90 ± 4.36 | 40.78 ± 10.0 | 40.10 ± 3.11 | 58.52 ± 2.06 | 34.247 | <0.001 |
| Organic matter (g·kg−1) | 0.05 ± 0.002 | 0.26 ± 0.015 | 0.29 ± 0.08 | 0.74 ± 0.034 | 24.109 | 0.001 |
| Total N (g·kg−1) | 0.08 ± 0.004 | 0.62 ± 0.004 | 0.71 ± 0.005 | 0.79 ± 0.006 | 12.243 | 0.006 |
| Total P (g·kg−1) | 0.32 ± 0.021 | 0.59 ± 0.020 | 0.64 ± 0.103 | 0.99 ± 0.198 | 29.767 | <0.001 |
| Total K (%) | 1.46 ± 0.05 | 1.49 ± 0.28 | 1.66 ± 0.11 | 2.22 ± 0.47 | 16.614 | 0.002 |
| Available N (mg·kg−1) | 1.41 ± 0.22 | 3.49 ± 0.34 | 2.80 ± 0.39 | 1.86 ± 0.57 | 5.346 | 0.030 |
| Available P (mg·kg−1) | 5.09 ± 0.93 | 6.83 ± 2.12 | 7.30 ± 0.88 | 9.01 ± 1.21 | 12.165 | 0.006 |
| Available K (mg·kg−1) | 425.3 ± 2.98 | 444.5 ± 2.99 | 447.9 ± 5.24 | 459.6 ± 4.50 | 30.817 | <0.001 |
| Site | Sequencing Results | Diversity Estimates | ||||||
|---|---|---|---|---|---|---|---|---|
| Total Sequences | Total OTUs | ACE | Chao | Shannon | Simpson | Coverage | ||
| nosZ | SSD | 109 | 15 | 17.33 | 16.90 | 2.15 | 0.17 | 0.96 |
| CM-9 | 105 | 15 | 48.94 | 32.24 | 0.87 | 0.69 | 0.89 | |
| CM-19 | 108 | 19 | 30.28 | 31.09 | 1.78 | 0.31 | 0.90 | |
| NC | 95 | 8 | 11.32 | 7.61 | 0.65 | 0.68 | 0.97 | |
| nirK | SSD | 102 | 25 | 50.26 | 32.00 | 2.48 | 0.12 | 0.90 |
| CM-9 | 109 | 29 | 48.43 | 44.75 | 3.26 | 0.04 | 0.88 | |
| CM-19 | 116 | 61 | 100.67 | 111.67 | 3.40 | 0.07 | 0.82 | |
| NC | 98 | 61 | 151.99 | 139.09 | 3.52 | 0.06 | 0.86 | |
| nirS | SSD | 102 | 9 | 16.64 | 14.00 | 0.70 | 0.73 | 0.95 |
| CM-9 | 125 | 19 | 35.13 | 32.75 | 1.29 | 0.54 | 0.91 | |
| CM-19 | 94 | 24 | 164.24 | 48.00 | 2.09 | 0.25 | 0.83 | |
| NC | 98 | 13 | 59.93 | 27.00 | 1.42 | 0.36 | 0.92 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, C.; Zhang, Y.; Cui, Z.; Li, H.; Wang, T.; Ren, Q. Recovery of Soil-Denitrifying Community along a Chronosequence of Sand-Fixation Forest in a Semi-Arid Desertified Grassland. Forests 2021, 12, 354. https://doi.org/10.3390/f12030354
Cao C, Zhang Y, Cui Z, Li H, Wang T, Ren Q. Recovery of Soil-Denitrifying Community along a Chronosequence of Sand-Fixation Forest in a Semi-Arid Desertified Grassland. Forests. 2021; 12(3):354. https://doi.org/10.3390/f12030354
Chicago/Turabian StyleCao, Chengyou, Ying Zhang, Zhenbo Cui, Hailong Li, Tingting Wang, and Qing Ren. 2021. "Recovery of Soil-Denitrifying Community along a Chronosequence of Sand-Fixation Forest in a Semi-Arid Desertified Grassland" Forests 12, no. 3: 354. https://doi.org/10.3390/f12030354
APA StyleCao, C., Zhang, Y., Cui, Z., Li, H., Wang, T., & Ren, Q. (2021). Recovery of Soil-Denitrifying Community along a Chronosequence of Sand-Fixation Forest in a Semi-Arid Desertified Grassland. Forests, 12(3), 354. https://doi.org/10.3390/f12030354







