Abstract
We aimed to extract DNA and amplify PCR fragments at the mitochondrial DNA Nad7.1 locus and 11 nuclear microsatellite loci in nine circa 11,000-year-old individuals of Scots pine found at the bottom of the Baltic sea and test the genetic associations with the present-day gene pool of Scots pine in Lithuania. We followed a strict anticontamination protocol in the lab and, simultaneously with the aDNA specimens, tested DNA-free controls. The DNA was extracted by an ATMAB protocol from the ancient wood specimens sampled underwater from Scots pine stumps located circa 20–30 m deep and circa 12 km ashore in western Lithuania. As the references, we used 30 present-day Lithuanian populations of Scots pine with 25–50 individuals each. The aDNA yield was 11–41 ng/μL. The PCR amplification for the mtDNA Nad7.1 locus and the nDNA loci yielded reliable aDNA fragments for three and seven out of nine ancient pines, respectively. The electrophoresis profiles of all the PCR tested DNA-free controls contained the sizing standard only, indicating low likelihood for contamination. At the mtDNA Nad7.1 locus, all three ancient Scots pine individuals had the type A (300 bp) allele, indicating postglacial migration from the refugia in Balkan peninsula. The GENECLASS Bayesian assignment tests revealed relatively stringer and consistent genetic associations between the ancient Scots pine trees and the present-day southern Lithuanian populations (assignment probability 0.37–0.55) and several wetlands in Lithuania. Our study shows that salty sea water efficiently preserves ancient DNA in wood at the quality levels suitable for genetic testing of trees dated back as far as 11,000 years before present.
1. Introduction
Understanding the temporal and spatial distribution of the genetic variation of such a widespread coniferous species as Scots pine is of fundamental interest in revealing the evolution and present-day genetic structure of this species [1,2]. Especially interesting in this respect are the first waves of postglacial migrants, such as the pines in the study dated back to 11,000 before present (BP) [3,4].
During the geological period of Pleistocene, that begun circa 2 million years ago, the Earth experienced repeated ice age periods of circa 100,000 years interspaced by circa 10,000-years-long interglacial warmings. The last Pleistocene glaciation in Europe started and ended circa 110,000 to 12,500 years BP. During the last glacial maximum (LGM) circa 20,000 years BP, the southernmost edge of the polar ice sheet reached northern Poland, Germany, and the Baltic states [5]. The tundra zone, with permafrost suppressing any significant assemblies of woody vegetation, stretched to southern France, northern Italy and the Caspian and Black seas [5]. Studies of the fossil pollen abundance in soil sedimental horizons and DNA polymorphism of maternally inherited plastid genomes show that the surviving wooden vegetation spread northward from three main southern glacial refugia in Iberian and Italian peninsulas and the Balkans, as well as in a smaller eastern refugium in the present-day eastern Ukraine [6,7,8]. The temperatures started and continue to increase since circa 14,000 years BP, giving the start of the Holocene period [9]. Radiocarbon dating of the fossil pollen in Europe suggests that, during the period from 14,000 to 12,000 years BP, the trees expanded northward relatively fast [9]. Scots pine, especially, as a pioneer, spread northward with a rate of circa 150 km per century, mainly forming pine–birch forests [6,10]. Scots pine was already present in the eastern Baltic seacoast circa 13,000 years BP [4]. Scots pine was most widespread in Europe circa 9500–7000 years BP, followed by a decline in southern Europe and expansion further north, reaching northernmost Fennoscandia by circa 8500 BP [10,11]. Since 4500 BP, likely due to a large-scale climatic drift toward wetter conditions, Scots pine experienced a marked reduction in range, especially in northwestern Europe but to lesser extent in Finland [12,13] and Lithuania [4].
Presently, Scots pine is one of the most widespread pioneer forest tree species tolerating a wide range of ecological conditions in Eurasia, one of a few well-growing species on nutrient-poor, sandy soils. In Scots pine, the mitochondrial DNA (mtDNA) genome is maternally inherited [14,15]. Therefore, it is likely to represent distinct genetic lineages corresponding to separate glacial refugia [16]. Based on the polymorphism of the mitochondrial DNA marker, coded here as Nad7-1 [8], two distinct glacial refugia for Scots pine in the temperate and boreal zones of Eurasia were revealed: (a) multiple refugia south of the ice sheet cover longitudinally, stretching over the whole of Europe [17,18], referred here as universal type A mitochondrial DNA haplotype; (b) northern refugium, much smaller in geographical area, likely located in the present-day eastern Ukraine and found mainly in the Baltic, Karelian (northwestern part of Russia), and Finnish populations, referred here as the northern type B haplotype. Our earlier study on geographical variation of the mitochondrial DNA Nad7.1 locus among 54 populations of Scots pine from the eastern European range revealed the type A with the 300 bp allele and type B with 295 bp allele mtDNA variants [19]. Based on [8], the 295 bp type B variant was identified as representing the northerly glacial refugium. Geographical distribution of the Nad7-1 northern B variant was not random, indicating its glacial refugium centered at about 300 km southeastward of Moscow at the interception of latitude 50–55° N and longitude 35–45° E. The major postglacial migration route from this northern refugium is located northwestward toward Karelia and northern Finland [19]. It is likely that the species migration occurred in waves and from distinct refugia, leading to a higher genetic diversity of the central over the to the peripheral populations [5].
Several questions arise here—how well are the pioneer Scots pine gene pools preserved during the subsequent migration waves? Are there any ecological niches, such as wetlands, where ancient gene pools may have had favorable conditions to persist until present? To answer these questions, we benefitted from the genetic material of Scots pine that, over the past 11,000 years, has been well-conserved at the bottom of the salty Baltic sea [3,20,21]. The objectives of our study were to (a) identify the most likely postglacial refugium of the Scots pine trees found at the bottom of the Baltic sea dated back to 11,000 BC by studying the maternally inherited mitochondrial DNA haplotypes and (b) assess the genetic associations among the ancient underwater Scots pine trees with the present-day gene pools of Scots pine in Lithuania by using a set of nuclear microsatellite DNA markers. We also tested the temporal genetic associations with Scots pine populations on wetland sites at three locations in Lithuania.
2. Material and Methods
2.1. The Material
A dozen stumps of large pine trees were found at the bottom of the Baltic sea circa 25-m depth and circa 12–14 km ashore from the nearest coast of Neringa spit near Juodkrante resort (abbreviated as JUOD) in the western Lithuania (described as sites RF-I and RF-III in detail by [21]). Wood from nine of these stumps was sampled by diving and detaching small sections of the stump wood. The sampled stumps are located within a circa 2 km radius. After detaching while underwater, the detached samples were placed into plastic containers, which were tightly closed. The containers containing the sea water and the wood samples were stored in darkness at 5 °C for a few days until transporting to laboratory. Upon transportation to the lab, the wood samples were replaced into sterile sealed plastic bags and stored in −20 °C until DNA extraction. Microscope analysis of the wood samples indicated that these are coniferous species, likely pines. We confirmed Scots pine as the species of these underwater trees by successful PCR amplification of the 11 nuclear microsatellite loci used by us for studying over 1000 samples of Scots pine in earlier studies [22]. As a part of another study, the wood samples were 14C radiocarbon dated [3].
As the references for the association between the ancient and the present-day Scots pine gene pools, we genotyped 25 to 60 mature individuals (age 80 to 250 years) in each of 27 autochthonous populations of Scots pine on normally irrigated common for Scots pine sites in major Scots pine-dominated large forest tracts in Lithuania (Table 1, Figure 1 and Figure S1). In addition, we compared three wetland Scots pine populations located in different parts of Lithuania (Figure 1 and Figure S1). The wetland sites are raised bogs with natural regeneration of Scots pine of area 2435 ha in KAMA, 3645 ha in BARG and 6847 ha in CIAP (the latter is one of the largest raised bogs in Lithuania). We sampled the dry site populations right at the edge of each wetland within a range of circa 10 to 300 m. from the wetland. All the reference populations were genotyped with the same set of nuclear microsatellites, identical PCR protocol and on the same DNA sequencer as the ancient Scots pine individuals (see below).

Table 1.
Location and sample size of the reference populations as abbreviated in Figure 1 and the GENECLASS assignment probabilities of the seven ancient Scots pine individuals to these reference populations, based on nuclear microsatellite markers. Nobs is the number of trees. The assignment probabilities vary from 0 to 1, where the values closer to 1 indicate strong genetic associations. Mean4 is the mean assignment probability of the four sea pine samples (B_1, C_2, T_3, P_2) that gave marked, above-zero associations with the inland populations (high values highlighted in yellow). Note that ancient pine samples B_2, E_1, and E_2 are circa 300 years older than the remaining ancient pines (Table 2).
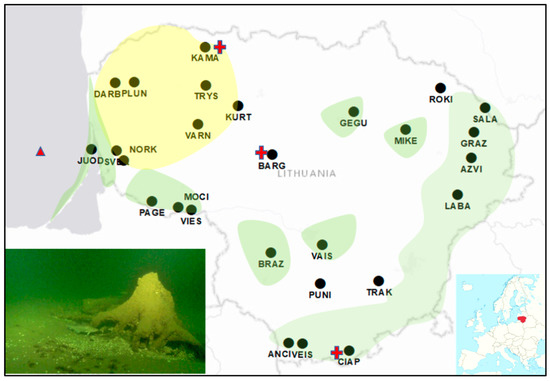
Figure 1.
The location of the reference populations of Scots pine (filled circles) and the ancient underwater stumps of Scots pine found circa 12 km ashore in the Baltic sea (triangle). The three wetland Scots pine populations are marked by red crosses, located at the vicinity of the populations on normally irrigated soils of KAMA (north), BARG (near center) and CIAP populations (south). The lower right map shows the position of Lithuania in Europe. An underwater tree stump is shown in the lower left photo. The greenish darkened areas indicate the main Scots pine-dominated continuous forest tracts. These greenish areas also well coincide with the location of the poor, sandy soils. The yellowish shades outline the areas where Scots pine is common in patchy, small Norway spruce-dominated forest tracts. The deciduous forests prevail on rich, moist soils of the midland lowland.
Note that the present-day Juodkrante population (JUOD) of age circa 200–250 years is in the Neringa spit, circa 1 km from the seashore, right at the spot where, 12 km seaward, the ancient pines were found.
2.2. DNA Extraction
The DNA was extracted from circa 100 mg of wood saw dust by a modified ATMAB protocol [23]. All the DNA extraction procedures were carried out in a separate room from the lab, where no DNA manipulation is carried out. Before the extraction procedure, all tools and labware were disinfected under UV chamber for 30 min. The wood saw dust was obtained by boring 1–2 cm deep into the wood samples with an electric drill (bore diameter 3 mm). The sawdust was further ground with three metal beads placed into 2 mL “Eppendorf” tubes for 4 min. in “Retsch MM400” mill; circa 80 mg of powdered wood sampled the DNA extraction. We removed the metal beads and added 1 mL DNA extraction buffer (warm, 55 °C temperature.), 50 µL DTT (1M) and 2 µL RNAse and mixed the solution and stored the tubes in a water bath for 12 h at 55 °C. After, we let them cool for 10 min. and added 400 µL of dichlormethan, vortexed and opened the lid to remove the air from the tubes (the work is done under the fume hood). The tubes were centrifuged for 10 min, and the upper layer was pipetted into another 1.5 mL “Eppendorf” tubes (amount ~600–800 µL); if the upper phase was not clear, the “dichlormethan precipitation” step was repeated (for some samples 3–5 times). After, 400 µL cold isopropanol was added to each cup and shaken softly until DNA was precipitated (and visible). The 1.5 mL “Eppendorf” tubes were kept at −20 °C overnight (12 h at least). The tubes were centrifuged for 10 min. Then all the liquid was discharged, and we let the remaining DNA-pellet dry for 5 min. After drying, 1 mL cold ethanol (76%) was added, and the DNA pellet (e.g., by vortexing) was released. Cups were centrifuged for 10 min (13,000 rpm, at 4 °C) again, and then all the liquid was removed, and the remaining DNA pellet dried for an hour. After drying, 50 µL 1 × TE buffer was added to force the DNA pellet to float; then the DNA pellet was resuspended overnight (in the fridge or at room temperature). The DNA was stored in a freezer at −20 °C. The DNA concentration was measured with a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.3. Mitochondrial DNA Markers
To detect the polymorphism at the mtDNA Nad7-1 locus, we optimized the procedure to avoid the restriction step in the laboratory analyses described by [8]. We used the GeneBank deposited sequences of the [8] mtDNA Nad7-1 haplotypes (DQ665913 to DQ665915) to design the primers flanking the 5 bp deletion, so that the PCR products would result in the following two segments [8]: 300 bp for the universal A haplotype and 295 bp for the northern B haplotype, to be resolved on a DNA sequencer. The primer sequences for the Nad7-1 locus designed with the BLAST primer designing tool were as follows: Nad7.1 forward 5′-ATACCGTCTGGCGAAAACGCCG-3′ and Nad7.1 reverse 5′-GGCCTCTCCATTTCCAATGACCCG-3′.
The PCR was run in a separate room, where no DNA manipulation is carried out. The PCR procedure was run as described in [19]. To test against contamination during the PCR, along with the PCR wells with the PCR mix, buffer, the primers, the size standard and the DNA of ancient pine samples, we included a DNA-free control with the same chemical compounds as other DNA-containing wells (6 samples with DNA and 3 DNA-free samples). The controls were placed in the PCR plate immediately after each sample with the aDNA. The plate with the post-PCR products was wrapped into folium and kept for a 10–20 min. in an empty freezer at −20 °C until the electrophoresis with the ABI PRISM® 310 Genetic Analyze (Applied Biosystems, Waltham, MA, USA) by using the GeneScan 500 LIZ ladder standard. We scored the alleles with the GeneMapper® software (Version 4.0, Applied Biosystems, Waltham, MA, USA).
2.4. Nuclear DNA Markers
We used 11 nuclear microsatellite loci with 2 to 3 nucleotide repeats that were used earlier by us on a set of Lithuanian Scots pine populations: Psyl57, Psyl57, Psyl2, Psyl18, Psyl42, Psyl25, Psyl16 (EST-SSRs, [24]), Spac7.14, Spac12.5, Spac11.4 (genomic SSRs developed for Pinus sylvestris, [25]) and PtTX4011, PtTX4001 (genomic SSRs developed for Pinus taeda, [26]). We combined the loci into three multiplexes for the PCR and the two multiplexes for the capillary electrophoresis. We carried out the PCR with the same cycling settings for all three multiplexes in a total volume of 10 µL PCR mix, containing 5 µL Platinum Multiplex PCR Master Mix (ThermoFisher Scientific), 4 µL RNAse free water, 0.5 µL primer F, 0.5 µL primer R and 1 µL of sample DNA. Thermal cycler GeneAmp PCR System9700 was used for PCR reaction, with the following settings: initial denaturation at 95 °C for 15 min, followed by 30 cycles of 94 °C for 30 s, 57 °C for 1.30 min, 72 °C for 30 s and a final elongation step at 60 °C for 30 min. We run the capillary electrophoresis on the ABI PRISM® 310 Genetic Analyze (Applied Biosystems, Waltham, MA, USA) by using the GeneScan 500 LIZ ladder standard and scored the alleles with the GeneMapper® software (Version 4.0, Applied Biosystems, Waltham, MA, USA).
2.5. Statistical Analyses
The reference population data set was checked with MICRO-CHECKER software [27] for null alleles and misprints. Null allele frequencies were low, below 0.05 for all the loci.
To test the genetic associations between the of the ancient underwater Scots pine individuals with present-day Scots pine gene pools in Lithuania, we used the Bayesian assignment approach implemented in the GENECLASS2 software [28]. We chose the [29] assignment method of individuals by calculating the individual assignment probabilities for the multilocus genotype of each ancient pine individual into the reference populations, based on Monte Carlo resampling of 10,000 simulated individuals [30]. The individual assignment probabilities were averaged for each present-day population.
3. Results
3.1. The Mitochondrial DNA Analysis
The DNA concentration of all nine ancient underwater Scots pine (abbreviated as sea pine) samples was sufficient for PCR amplification and ranged between 20–40 ng/μL (Table 2).

Table 2.
Characteristics and summary of the DNA testing success of the ancient underwater individuals of Scots pine (details on the PCR quality at the microsatellite loci are given in the Table S1). “PCR attempts” are the number of independent PCR runs (sampled from the “maternal” extracted DNA sample). “Fine” means PCR fragments above 50 relative florescence units (RFU) found at least in two replications (RFU rages from 0 to 6000, where all DNA fragments below 10 RFU is set as noise). NA means no amplification. “Not used” means not used in the analysis. RF codes refer to the sea sampling site according to [21]. Original lab tree ID is given in the brackets for reference with other studies.
The PCR in all four DNA-free control samples amplified no DNA fragments above 10 relative fluorescence units (RFU), whereas the size standard in the DNA-free control samples was of high quality, above 2500 RFU. This result indicates a low likelihood of contamination of the DNA samples from the underwater Scots pine stumps.
All six replications of the three ancient underwater individuals of Scots pine contained the 300 bp allele at the mitochondrial DNA Nad7-1 locus (Table 3). The PCR amplification quality was high, well above 3000 relative fluorescence units (RFU), near the upper margin for fragment detection by the ABI310 Genetic Analyzer (Table 2). Based on [8,19], the 300 bp Nad-7 allele belongs to the Scots pine from the southern glacial refugium (Figure 2).

Table 3.
PCR amplification of the alleles at the Nad7.1 locus for the three ancient Scots pine trees found in the Baltic sea. Controls are the DNA-free samples containing the same constituents and included in the PCR and capillary electrophoresis runs together with the ancient pine. NA—no amplification of DNA of any kind above 10 RFU. Sample ID refers to two independent samples from a single wood specimen (individual tree).
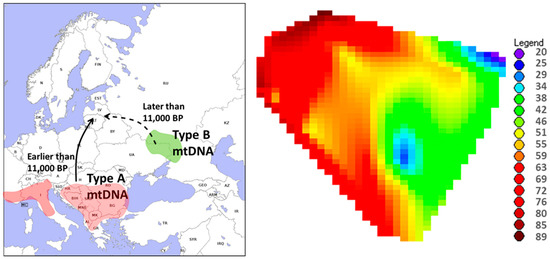
Figure 2.
(Left). The likely sources of postglacial migration of Scots pine into Baltic region marked by darker areas (modified from [8,9]). In our study, we found no mitochondrial DNA haplotype B (mtDNA type B) in the DNA samples from the ancient underwater stumps of Scots pine, indicating that the Scots pine from the southern European refugia (represented by the mtDNA type A) reached the Baltic region earlier than Scots pine from the northern refugium (mtDNA type B). (Right). A contour plot of the present-day Scots pine population percentage of mtDNA type A plotted on a simplified map of Lithuania (the legend shows the percentage of the mtDNA type A in each of the 27 populations, modified from [33]). As there were only two mtDNA haplotypes found (type A and type B), the percentages below 50 (green to yellow on the map to the right) show areas with the prevalence of the mtDNA type B. Clearly, the mtDNA type A dominates the southwestern part of the country.
3.2. The Nuclear DNA Analysis
Over the two replicated runs, we successfully amplified acceptable quality alleles at the 11 nuclear microsatellite loci in seven out of nine ancient pine individuals (Table 2). In the remaining two individuals, repeated testing gave acceptable amplification in 6 out of 11 loci, only. Therefore, in this paper, we analyzed only the seven individuals, which had quality scores at all 11 loci (Table 2). The quality of most of the amplified PCR fragments in these seven sea Scots pine samples varied between 100 and 2000 RFU (Table S1).
The PCR on the DNA-free controls that were run with the same constituents as the rest except of the sample DNA amplified quality fragments of the standard with no other DNA fragments above 10 relative fluorescence units (RFU, where 10 RFU is considered as the lowest acceptable PCR fragment quality bound). This indicates low likelihood for contamination in the ancient DNA containing samples.
Comparison of the locus wise allele frequency distribution among the present-day and the 12,000 years old Scots pine trees revealed a trend for the low size alleles in the ancient pines in the most variable loci of the Spac series (Figure 3). For the Psyl and Pttx loci, no such tendency was observed (Figure 3).
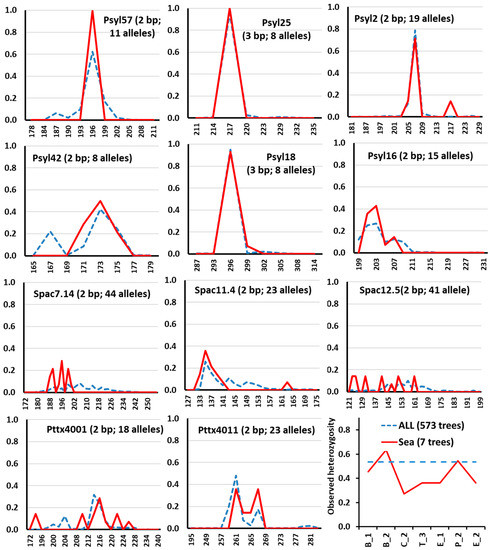
Figure 3.
The observed heterozygosity (of all 11 loci, lower right) and locus allele frequencies of the 12,000-year-old sea pines (red solid line, seven trees, abbreviated as Sea) and the present-day Scots pine trees in Lithuania (dashed line, n = 573 trees, ALL). The microsatellite motive (in bp) and the number of different alleles per locus are given in the brackets at the locus name at the top of each plot. The Psyl loci are the expressed sequence SSR loci. The Spac loci are the genomic SSR loci. The most variable genomic SSR loci of Spac series display a consistent tendency for low allele sizes in the ancient Scots pine trees.
The GENECLASS Bayesian assignment test showed that the three ancient Scots pine individuals (B_2, E_1 and E_2) had close to zero assignment probabilities to all the present-day reference populations in Lithuania (Table 1). For the remaining four ancient Scots pine individuals (P_2, C_2, B_1 and T_3), the assignment probabilities ranged between 0 and 0.81, indicating a highly variable dataset for the association assignment (Table 1). The highest mean assignment probability of the above mentioned four ancient Scots pine individuals was obtained to the present-day southern and, to a lesser extent, western Lithuanian Scots pine populations (ranging between 0.36 and 5.43, Figure 4, Table 1). When examining the individual assignment properties of the four ancient Scots pine individuals separately, a consistent similarity to the present-day southern Lithuanian population was obvious at the assignment probability, reaching as high values as 0.5–0.7, especially with the southern ANCI, BRAZ and CIAP dry site populations (Table 1). In addition to the southern populations, ancient Scots pine individuals P_2 and B_1 showed similarity with northeastern, while T_3 and B_1 did so with the western, Lithuanian populations, especially PLUN (Table 1). The two ancient Scots pine individuals of B_1 and T_3 had high assignment probabilities to the seaside population of JUOD, which is geographically the closest present-day inland population to the ancient pines (0.42–0.58, Table 1). However, the individuals C_2 and P_2 were less similar to JUOD population, causing a moderate mean association probability with JUOD of 0.33 (Table 1).
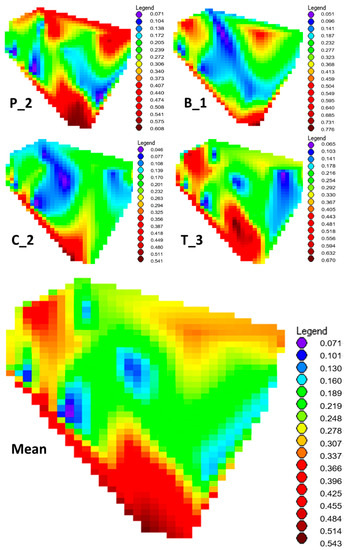
Figure 4.
The interpolated contour plots of the GENECLASS assignment probabilities of the four ancient sea pine tree samples (P_2, B_1, C_2, C_3) to each of the 27 autochthonous present-day reference populations of Scots spine plotted on a simplified map of Lithuania. The mean assignment probability of these four sea pine trees is given in the lowermost contour map.
At two of the three wetland sites, the four ancient Scots pine samples (P_2, B_1, C_2 and T_3) were markedly more strongly associated with the bog than with the adjacent dry site populations of Scots pine (Figure 5, Table 1). The strongest association differential was found for the BARG wetland located in central Lithuania: the mean association probability (p) with the BARG bog population was 0.49, while that with the adjacent dryland population dropped to 0.09 (see BARG in Table 1, Figure 5). However, in the southern CIAP wetland, the result was the opposite: the ancient pines were more strongly associated with the dry site (p = 0.42) than with the bog populations (p = 0.16, Table 3).
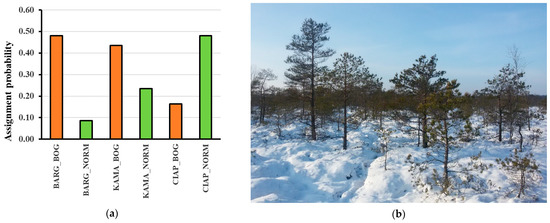
Figure 5.
(a) The mean GENECLASS assignment probabilities of the four ancient Scots pine individuals (P_2, B_1, C_2 and T_3) to the pairs of the adjacent wetland (BOG) and dryland (NORM) populations of Scots pine at three locations in Lithuania (KAMA, north; BARG, center; and CIAP, south of the country, Figure 1). (b) The reference population Scots pine in in the wetland of Bargailiai (during sampling in the raised bog, age of Scots pine trees circa 100 years, estimated by sampling the increment cores). The adjacent dryland reference population of BARG is seen at the horizon.
4. Discussion
4.1. DNA Contamination Avoidance
Being aware of possible contamination with contemporary DNA, e.g., from pollen [34], we did the DNA testing during December–January when the least pollen is present in the air [1]. We did not bleach nor cleaned with laser irradiation the ancient wood samples. Our strategy was to rely on the DNA-free controls and high yields of comparably less degraded DNA while preserved in the salty water than wood laying below ground, such as in [34]. We did not expect such contaminants as pollen to outperform the amplification of the template DNA. Most of the PCR fragments in our study were above 100 RFU, which is well above the expected amplification strength of DNA contaminants, such as PCR amplicons or pollen [35]. Pollen grain outer walls are extremely resistant to chemical detergents reducing already-low pollen DNA concentrations. Another argument against the pollen contamination is that pollen contamination is expected to produce multiple PCR fragments, because pollen grains contain different genotypes [1]. This, however, was not the case on our PCR profiles, where we observed no more than two fragments in all 11 nDNA loci.
We used an ATMAB DNA extraction protocol, which is economically and laboratory-handy and easily available. The high aDNA yields we have obtained from the circa 11,000-year-old underwater pine wood indicates that the salty sea water is a very efficient preservative of ancient wood remains. Therefore, geneticists may strongly cooperate with underwater archeologists and pay attention to secure prompt and proper handling of wood samples after underwater sampling at this and other sites [36]. Nuclear DNA is used to be better preserved than the organelle DNA in ancient plant materials, and its yield is markedly higher [37], which may explain a higher amplification success of nDNA versus mtDNA in our study (Table 2).
4.2. Evolutionary Origin of Scots Pine
The regression of the historical Yoldia sea, the predecessor of the modern Baltic sea, was at its maximum during 11,000 to 10,700 years BP, allowing the forest vegetation 33–55 m below the present-day sea level, which includes the locations of ancient pines investigated in our study [38,39]. Thus, our samples aged circa 11,000 BP are very likely to represent the first postglacial migration waves of Scots pine into the eastern Baltic region. Our positive results on the type A mtDNA haplotype in the ancient pine samples indicate that the postglacial migration of Scots pine from the southern European refugia may have occurred earlier than from the northeastern refugium located in southern part of European Russia (Figure 2). This finding supports Scots pine postglacial migration theories reporting a relatively later contribution of the northerly type B local refugium [8,40]. It is likely that the cold receded later in the northeastern refugium, allowing for a subsequent migration from these areas northward. In the study of present-day Lithuanian Scots pine gene pool, ref. [33] reported gradual decrease of the type B mtDNA haplotype westward (Figure 2), which well corresponds with our findings. However, note that our assumptions are based on three individuals, which provides indicative rather than concussive results.
4.3. Genetic Associations with the Present-Day Gene Pools of Scots Pine
Considering the time span of circa 11,000 years between the present-day and ancient Scots pine gene pools in our study, it is interesting to examine the debatable mutation models of the DNA base pair repeat motives at the microsatellite loci. Our results indicate that, in contrast to the conservative EST-SSRs, the highly variable, presumably neutral SSR loci tend to mutate in a stepwise manner by increasing the allele sizes with new insertions over the past 11,000 years (Figure 3). The relatively lower observed heterozygosity of the ancient sea pines may be explained by diversity-enriching migration events from distinct gene pools expected to increase the heterozygosity of the modern generations [5].
Scots pine is a widespread wind pollinated conifer distinguishing from the other forest tree species by the strongest geneflow among the populations [41]. This strong geneflow results in low differentiation among populations and high genetic diversity within populations of Scots pine [42]. The neutral DNA markers mainly reflect zones of shared gene pools of Scots pine populations via phenology synchrony [42]. These phenology-based structures are indirectly associated with the adaptive variation, because phenology is a strong adaptive trait for Scots pine [43]. Our reference populations cover the main Scots pine forests in the country, and the sample sizes of 20 to 50 individuals are large enough to represent the local gene pools by using the microsatellite DNA markers [22]. Therefore, we believe we used a reliable data set for the association analysis. Nevertheless, in our study, three of the seven ancient pines had no associations with all the present-day Scots pine populations in Lithuania (Table 1). This may be because of (a) high within-population diversity of the ancient Scots pine gene pool, or (b) the ancient Scots pine samples represent separate generations of forests originating from different postglacial migration sources. Based on radiocarbon dating of fossil pollen, P. sylvestris reached the northernmost part of Scandinavia circa 7800 BP [6,44]. The fossil pollen records near the Lithuanian coast of Baltic sea indicate the dominance of birch forests during the early Preboreal period circa 11,500 years BP, followed by a drastic increase of Scots pine forests since the late Preboreal circa 10,500 years BC [4,45,46]. Our genetic material of the 10,400–11,200 years BP may include multiple migration waves during of the postglacial Scots pine expansion.
What could be the reasons for the relatively stronger long-term temporal associations namely with the present-day southern Lithuanian Scots pine populations? These could be random associations, because the environmental stochasticity over many generations could markedly change the allele frequencies. However, in particular cases, neutral genomic variants may be locally conserved in large populations over long time [47]. Such a specific case could be Scots pine with the neutral part of the genome several times exceeding that of the other tree species. Neutral allele frequencies in a widespread, wind-pollinated species with large population sizes likely to be free of strong defects of mutations, drift and natural selection and temporally stay under the Hardy-Weinberg equilibrium over many generations [47]; why not over circa 100 nonoverlapping generations, given 100 years of natural aging in Scots pine growing at its edaphic optimum? We found that the temporal associations were consistently strong with all present-day southern Lithuanian populations genotyped in our study. In case of random associations, we would have observed strong temporal associations scattered evenly over Lithuania, but this was not the case in our study. Another interesting observation is that the geographical patters of the long-term temporal genetic associations (Figure 5), well-correspond with the Scots pine pollen abundance maps from 13,300 to 500 BP, based on fossil pollen dating records in Lithuania (Fig. 3.6.14 and 3.6.15 in [46]). The [4] study revealed that Scots pine population in southern Lithuania remained stable from 13,300 BP up to the present day, while, in the other parts of the country, the Scots pine presence was variable over time. In southern Lithuania, the ancient Scots pine gene pool may have been better preserved, because of their dominance of the poorest sandy soils better suited for Scots pine than for any other forest tree species (lichen-rich Cladiniosum ground vegetation types).
We expected a closer genetic association between the ancient pines and the old growth Scots pine trees located closest to the sea sampling site in JUOD forest, circa 1 km from the seacoast. Two of the four ancient pine individuals were associated with JUOD population with comparably high probability between 0.42 to 0.58 (Table 1). Thus, there is a similarity; however, sampling in more populations, especially those closest to the seashore, would help in concluding on the remnants of the ancient gene pools in Neringa. There were several large-scale deforestation events in Neringa during middle centuries, followed by sandstorms and wind erosion and human afforestation initiatives. Thus, we may have multiple founder effects manifesting in Scots pine gene pool of Neringa.
Another consistent finding is the relatively stronger associations between the ancient pines and the wetland populations of Scots in central (BARG) and northern (KAMA) Lithuanian wetlands. Peat layers with remains of Cyperaceae, Menyanthes, Euquisetum, Polypodiaceae and Sphagnum plant species were found at the bottom of the Baltic sea within the sampling area of the ancient underwater Scots pine trees studied here [3,20]. Within the sampling site RF-I, the radiocarbon dating of the peat specimens sampled 29 m below the present sea level at the bottom of the underwater peat layer returned ages of circa 11,770 years BP. This indicates cotemporary presence of Scots pine and wetlands that formed in the zone of regression/transgression of the pre-Baltic sea water pools [3]. The present-day Scots pine-dominated wetland sites are raised bogs of circa 5000 ha that are sufficient for maintaining distinct gene pools, due to phenology differences between the pines on wet and dry landscapes [48]. During hot and dry summer periods, the upper layer of desiccated moss and peat reaches 1 to 2 m in depth in these raised bogs. This is a strong selective force at the juvenile age for Scots pines tolerant to drought. In southern Lithuania, CIAP wetland population did not provide such associations. A possible reason for that could be already strong associations with the southern Lithuanian Scots pine gene pool, itself.
In conclusion, our study showed that salty sea water efficiently preserves ancient DNA in wood at the quality suitable for genetic testing of trees dated back as far as 11,000 years before present. The ancient Scots pine individuals found the bottom of the Baltic sea contain the type A (300 bp) allele at the mitochondrial DNA Nad7.1 locus, indicating their origin from the southern glacial refugia in southern Europe. The Bayesian assignment tests based on nuclear DNA microsatellite markers revealed relatively stronger genetic associations between the ancient Scots pine trees found underwater in the Baltic sea and the present-day Scots pine populations of southern Lithuania and several wetlands in Lithuania. A possibility of nonrandom long-term temporal genetic associations and presence of populations better conserving the founder gene pools in Scots pine was discussed.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/12/3/317/s1, Figure S1: The present-day reference Scots pine populations sampled in Lithuania. Upper left, 250-year-old Scots pine trees in Punia (PUNI) in southern Lithuania. Upper right, a 90-year-old Scots pine in the wetland of Bargailai (BARG) central Lithuania. Lower left, circa 200-year-old trees in Marcinkonys (MARC) in southern Lithuania and circa 250-year-old tree in Juodkrante (JUOD) in the seaside spit of Neringa, the closest to the underwater trees, Table S1: The DNA concentration (DNAC in ng/mL) and the size of the amplified PCR fragments for the two microsatellite alleles per locus given in the relative fluorescence units (RFU) from the output of the GeneMapper ver. 4.0 software. The RFU values between 100 and 2000 RFU are considered as optimal (manual of GeneMapper ver. 4.0 soft).
Author Contributions
D.D. conceptualization, data analysis, manuscript writing, review and editing; J.B. laboratory analyses, V.Ž. conceptualization, funding acquisition, project administration, underwater sample collection; L.D. conceptualization, data curation, A.G. underwater sample collection, review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Social Fund through the Research Council of Lithuania according to the activity “Improvement of researchers’ qualification by implementing world-class R&D projects” of measure no. 09.3.3-LMT-K-712 (grant application code: 09.3.3-LMT-K-712-01-0171, agreement no. DOTSUT-251).
Data Availability Statement
Data is contained within the article or Supplementary Materials Table S1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gugerli, F.; Parducci, L.; Petit, R.J. Ancient plant DNA: Review and prospects. New Phytol. 2005, 166, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Gutaker, R.M.; Burbano, H.A. Reinforcing plant evolutionary genomics using ancient DNA. Curr. Opin. Plant Biol. 2017, 36, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Žulkus, V.; Girininkas, A. Drowned Early Mesolithic Landscapes on the Baltic Sea bed in the Lithuanian Waters. J. Environ. Sci. Eng. 2014, 3, 274–289. [Google Scholar] [CrossRef]
- Balakauskas, L. Vėlyvojo Ledynmečio ir Holoceno Miškų Augalijos Raida Lietuvoje LRA (Kraštovaizdžio Atkūrimo Algoritmo) Modeliavimo Duomenimis; Forest Vegetation during Late Glaciation Holocene in Lithuania Mased on LRA Modeling PhD Dissertation; Vilnius University Press: Vilnius, Lithuania, 2013; p. 150, (In Lithuanian with English Summary). [Google Scholar]
- Hewitt, G.M. Postglacial recolonization of European biota. Biol. J. Linn. Soc. 1999, 68, 87–122. [Google Scholar] [CrossRef]
- Huntley, B.; Birks, H.J.B. An Atlas of Past and Present Pollen Maps for Europe 0–13,000 Years Ago; Cambridge University Press: Cambridge, UK, 1983; p. 668. ISBN 0521237351. [Google Scholar]
- Tollefsrud, M.M.; Sønstebø, J.H.; Brochmann, C.; Johnsen, Ø.; Skrøppa, T.; Vendramin, G.G. Combined analysis of nuclear and mitochondrial markers provide new insight into the genetic structure of North European Picea abies. Heredity 2009, 102, 549–562. [Google Scholar] [CrossRef]
- Naydenov, K.; Senneville, S.; Beaulieu, J.; Tremblay, F.; Bousquet, J. Glacial variance in Eurasia: Mitochondrial DNA evidence from Scots pine for a complex heritage involving genetically distinct refugia at mid-northern latitudes and in Asia Minor. BMC Evol. Biol. 2007, 7, 233. [Google Scholar] [CrossRef]
- Kvist, L. Phylogeny and Phylogeography of European Parids; Department of Biology, Faculty of Science, University of Oulu: Oulu, Finland, 2000; p. 44. ISBN 951-42-5536-4. Available online: http://urn.fi/urn:isbn:9514255364 (accessed on 5 February 2021).
- Huntley, B. European post-glacial forests: Compositional changes in response to climatic change. J. Veg. Sci. 1990, 1, 507–518. [Google Scholar] [CrossRef]
- Birks, H.J.B. Holocene isochrone maps and patterns of tree-spreading in the British Isles. J. Biogeogr. 1989, 16, 503–540. [Google Scholar] [CrossRef]
- Eronen, M.; Huttunen, P. Radiocarbon-dated sub-fossil pines from Finnish Lapland. Geogr. Ann. Ser. A Phys. Geogr. 1987, 69, 297–304. [Google Scholar] [CrossRef]
- Gear, A.J.; Huntley, B. Rapid changes in the range limits of Scots pine 4000 years ago. Science 1991, 251, 544–547. [Google Scholar] [CrossRef]
- Neatle, D.B.; Sederoff, R.R. Paternal inheritance of chloroplast DNA and maternal inheritance of mitochondrial DNA in loblolly pine. Theor. Appl. Genet. 1989, 77, 212–216. [Google Scholar] [CrossRef]
- Jaramillo-Correa, J.P.; Bousquet, J.; Beaulieu, J.; Isabel, N.; Perron, M.; Bouillé, M. Cross-species amplification of mitochondrial DNA sequence-tagged-site markers in conifers: The nature of polymorphism and variation within and among species in Picea. Theor. Appl. Genet. 2003, 106, 1353–1367. [Google Scholar] [CrossRef] [PubMed]
- Sperisen, C.; Büchler, U.; Gugerli, F.; Mátyás, G.; Geburek, T.; Vendramin, G.G. Tandem repeats in plant mitochondrial genomes: Application to the analysis of population differentiation in the conifer Norway spruce. Mol. Ecol. 2001, 10, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Chernova, G.M.; Mikhailov, N.N.; Denisenko, V.P.; Kozyreva, M.G. Some questions of paleogeography of Holocene of South Eastern Altai. Izv. All-Union Geogr. Soc. 1991, 2, 140–146. [Google Scholar]
- Cheddadi, R.; Vendramin, G.G.; Litt, T.; François, L.; Kageyama, M.; Lorentz, S.; Laurent, J.M.; de Beaulieu, J.L.; Sadori, L.; Jost, A.; et al. Imprints of glacial refugia in the modern genetic diversity of Pinus sylvestris. Glob. Ecol. Biogeogr. 2006, 15, 271–282. [Google Scholar] [CrossRef]
- Buchovska, J.; Danusevicius, D.; Stanys, V.; Šikšnianienė, J.B.; Kavaliauskas, D. The location of the northern glacial refugium of Scots pine based on mitochondrial DNA markers. Balt. For. 2013, 19, 2–12. [Google Scholar]
- Žulkus, V.; Girininkas, A. The Coasts of the Baltic Sea 10,000 Years Ago; Klaipėda University Press: Klaipėda, Lithuania, 2012; p. 55. [Google Scholar]
- Žulkus, V.; Girininkas, A. The eastern shores of the Baltic Sea in the Early Holocene according to natural and cultural relict data. Geo Geogr. Environ. 2020, 7, 1–16. [Google Scholar] [CrossRef]
- Danusevičius, D.; Kavaliauskas, D.; Fussi, B. Optimum Sample Size for SSR-based Estimation of Representative Allele Frequencies and Genetic Diversity in Scots Pine Populations. Balt. For. 2016, 22, 194–202. [Google Scholar]
- Dumolin, S.; Demesure, B.; Petit, R.J. Inheritance of chloroplast and mitochondrial genomes in pedunculated oak investigated with anefficient PCR method. Theor. Appl. Genet. 1995, 91, 1253–1256. [Google Scholar] [CrossRef]
- Sebastiani, F.; Pinzauti, F.; Kujala, S.T.; González-Martínez, S.C.; Vendramin, G.G. Novel polymorphic nuclear microsatellite markers for Pinus sylvestris L. Conserv. Genet. Resour. 2012, 4, 231–234. [Google Scholar] [CrossRef]
- Soranzo, N.; Provan, J.; Powell, W. Characterization of microsatellite loci in Pinus sylvestris L. Mol. Ecol. 1998, 7, 1260–1261. [Google Scholar] [CrossRef]
- Elsik, G.C.; Minihan, V.T.; Hall, S.E.; Scarpa, A.M.; Williams, C.G. Low-copy microsatellite markers for Pinus taeda L. Genome 2000, 43, 112. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Weetman, D.; Hutchinson, W.F. Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol. Ecol. Notes 2006, 6, 255–256. [Google Scholar] [CrossRef]
- Piry, S.; Alapetite, A.; Cornuet, J.M.; Peatkou, D.; Baudouin, L.; Estoup, A. GENECLASS2: A Software for Genetic Assignment and First-Generation Migrant Detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef]
- Rannala, B.; Mountain, J.L. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA 1997, 94, 9197–9221. [Google Scholar] [CrossRef]
- Paetkau, D.; Slade, R.; Burden, M.; Estoup, A. Genetic assignment methods for the direct, real-time estimation of migration rate using assignment methods: A simulation-based exploration of accuracy and power. Mol. Ecol. 2004, 13, 55–65. [Google Scholar] [CrossRef]
- Ramsey, C.B. Bayesian analysis of radiocarbon dates. Radiocarbon 2009, 51, 337–360. [Google Scholar] [CrossRef]
- Reim Reimer, P.J.; Bard, E.; Bayliss, A.; Beck, J.W.; Blackwell, P.G.; Bronk Ramsey, C.; Grootes, P.M.; Guilderson, T.P.; Haflidason, H.; Hajdas, I.; et al. IntCal13 and Marine13 radiocarbon age calibration curves 0–50,000 years cal BP. Radiocarbon 2013, 55, 1869–1887. [Google Scholar] [CrossRef]
- Kavaliauskas, D. Genetic Structure and Diversity of Scots Pine (Pinus sylvestris L.) Populations in Lithuania. Ph.D. Thesis, Aleksandras Stulginskis University Press, Kaunas, Lithuania, 2016; p. 177. [Google Scholar]
- Lendvay, B.; Hartmann, M.; Brodbeck, S.; Nievergelt, D.; Reinig, F.; Zoller, S.; Parducci, L.; Gugerli, F.; Buntgen, U.; Sperisen, C. Improved recovery of ancient DNA from subfossil wood—application to the world’s oldest Late Glacial pine forest. New Phytol. 2018, 217, 1737–1748. [Google Scholar] [CrossRef]
- Ping-Hua, C.; Yong-Bao, P.; Ru-Kai, C. High-throughput procedure for single pollen grain collection and polymerase chain reaction in plants. J. Integr. Plant Biol. 2008, 50, 375–383. [Google Scholar] [CrossRef]
- Hansson, A. Submerged landscapes in the Hanö Bay: Early Holocene shoreline displacement and human environments in the southern Baltic Basin. Ph.D. Thesis, Lund University, Lund, Sweden, 2018; p. 57. [Google Scholar]
- Banerjee, M.; Brown, T.A. Preservation of nuclear but not chloroplast DNA in archaeological assemblages of charred wheat grains. Anc. Biomol. 2002, 4, 59–63. [Google Scholar] [CrossRef]
- Rosentau, A.; Muru, M.; Gauk, M.; Oja, T.; Liibusk, A.; Kall, T.; Karro, E.; Roose, A.; Sepp, M.; Tammepuu, A.; et al. Sea-Level Change and Flood Risks at Estonian Coastal Zone. Coastline Chang. Balt. Sea South East 2017, 19, 363–388. [Google Scholar] [CrossRef]
- Gelumbauskaitė, L.Ž.; Šečkus, J. Late quaternary shore formations of the Baltic basins in the Lithuanian sector. Geologija 2005, 52, 34–45. [Google Scholar]
- Sinclair, W.T.; Morman, J.D.; Ennos, R.A. The postglacial history of Scots pine (Pinus sylvestris L.) in western Europe: Evidence from mitochondrial DNA variation. Mol. Ecol. 1999, 8, 83–88. [Google Scholar] [CrossRef]
- Lindgren, D.; Paule, L.; Shen, X.; Yazdani, R.; Segerstrom, U.; Vallin, J.-E.; Lejdebro, M.L. Can viable pollen carry Scots pine genes over long distances? Grana 1995, 34, 64–69. [Google Scholar] [CrossRef]
- Tóth, E.G.; Köbölkuti, Z.A.; Pedryc, A.; Höhn, M. Evolutionary history and phylogeography of Scots pine (Pinus sylvestris L.) in Europe based on molecular markers. J. For. Res. 2017, 28, 637–651. [Google Scholar] [CrossRef]
- Eriksson, G. Pinus Sylvestris Recent Genetic Research; Swedish University of Agricultural Sciences: Uppsala, Sweden, 2008; p. 111. ISBN 978-91-85911-90-5. [Google Scholar]
- Willis, K.J.; Bennett, K.D.; Birks, J.B. The late Quaternary dynamics of pines in Europe. In Ecology and Biogeography of Pinus; Richardson, D.M., Ed.; Cambridge University Press: Cambridge, UK, 1998; pp. 107–121. [Google Scholar]
- Kabailienė, M. Lietuvos Holocenas Holocene in Lithuania; Mokslas Press: Vilnius, Lithuania, 1990; p. 176. (In Lithuanian) [Google Scholar]
- Kabailienė, M. Gamtinės Aplinkos Raida Lietuvoje per 14,000 Metų Development of the Natural Environment in Lithuania over 14,000 Years; Vilnius University Press: Vilnius, Lithuania, 2006; p. 471. [Google Scholar]
- Flowers, J.M.; Purugganan, M.D. The evolution of plant genomes: Scaling up from a population perspective. Curr. Opin. Genet. Dev. 2008, 18, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Sirgėdienė, M. Genetic Differentiation of Scots Pine Mitochondrial DNA Haplotypes and Bog Populations Based on Morphology and DNA Markers. Ph.D. Thesis, Vytautas Magnus University, Kaunas, Lithuania, 2020. (In English with Lithuanian Summary). [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).