Genome-Wide Identification and Functional Analysis of the Basic Helix-Loop-Helix (bHLH) Transcription Family Reveals Candidate PtFBH Genes Involved in the Flowering Process of Populus trichocarpa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Phylogenetic Analyses of the bHLH in Poplar
2.2. Chromosome Location and Gene Duplication Analysis
2.3. Protein Properties and Structure Analysis
2.4. Cis-Acting Element Analysis and Multiple Sequence Alignment of the PtbHLHs
2.5. Subcellular Location and Protein–Protein Interaction Network Prediction
2.6. Expression Patterns of PtbHLH Genes in Different Tissues
3. Results
3.1. Identification, Characterization and Subcellular Localization of the bHLHs in Poplar
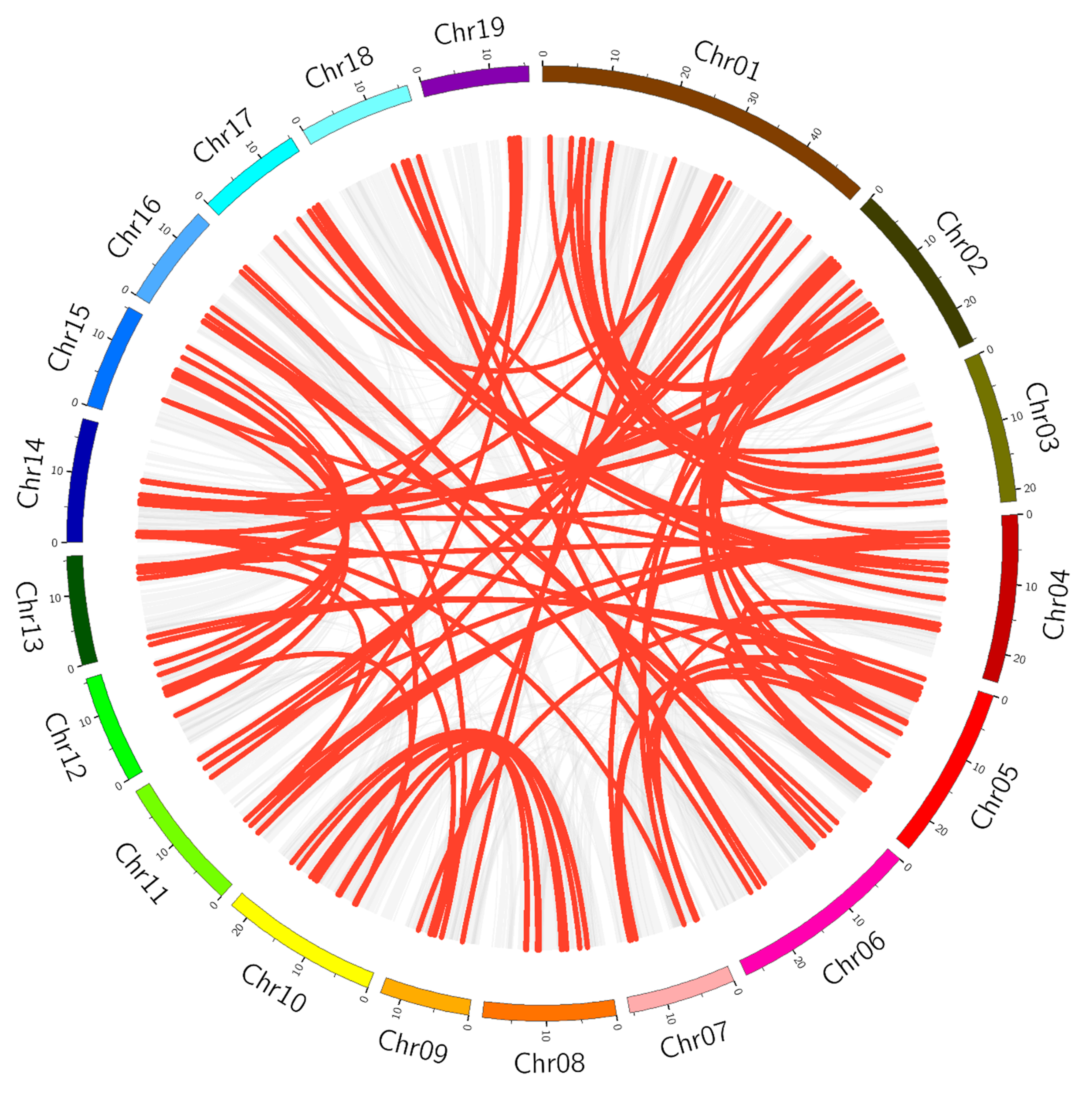
3.2. Chromosomal Distribution and Synteny Analysis of PtbHLH Genes
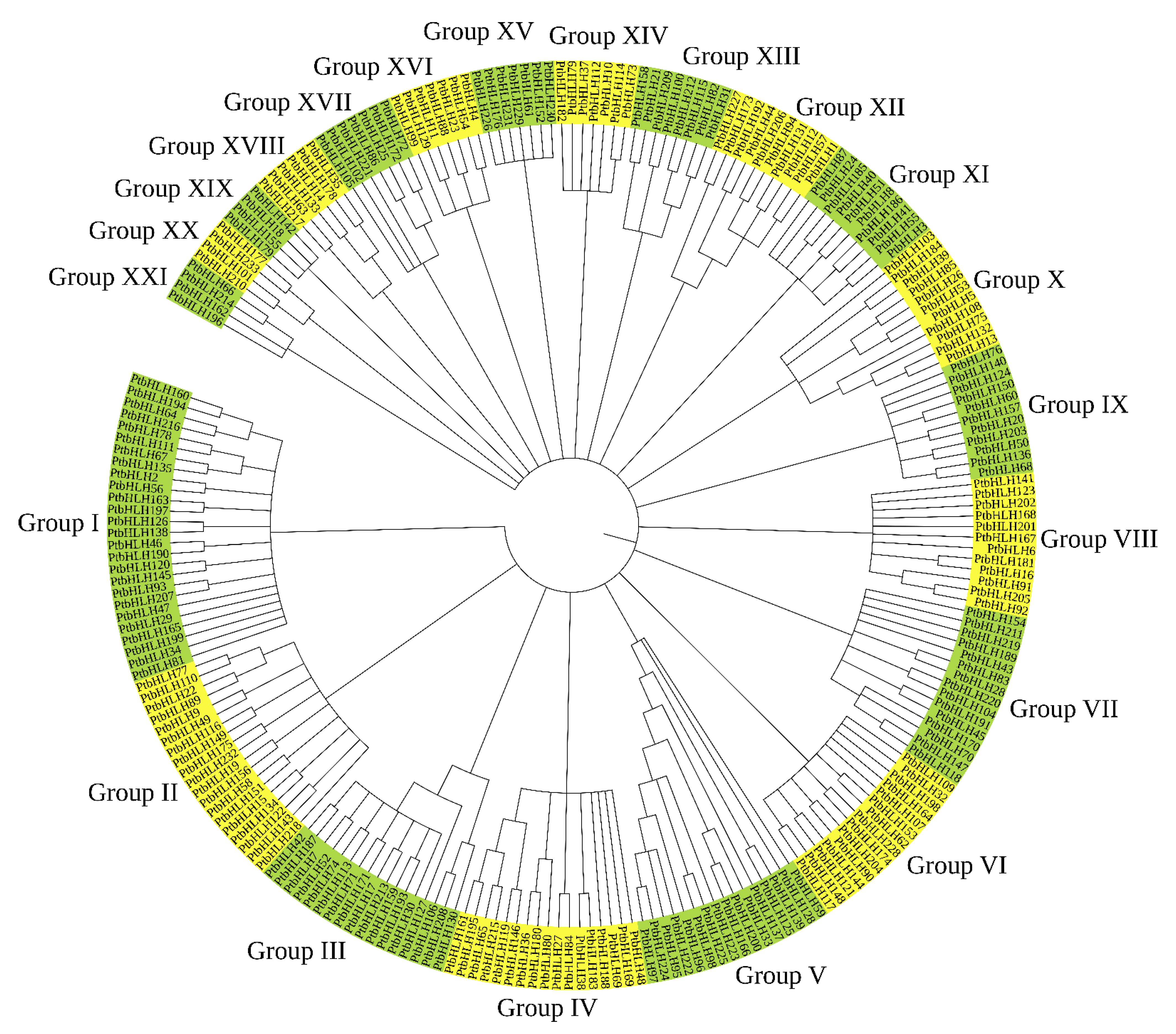
3.3. Phylogenetic Analyses of the PtbHLH in Poplar
3.4. Conserved Motif and Gene Structure Analysis of PtbHLHs
3.5. Cis-Elements Analysis in Promoters of the PtbHLH Genes
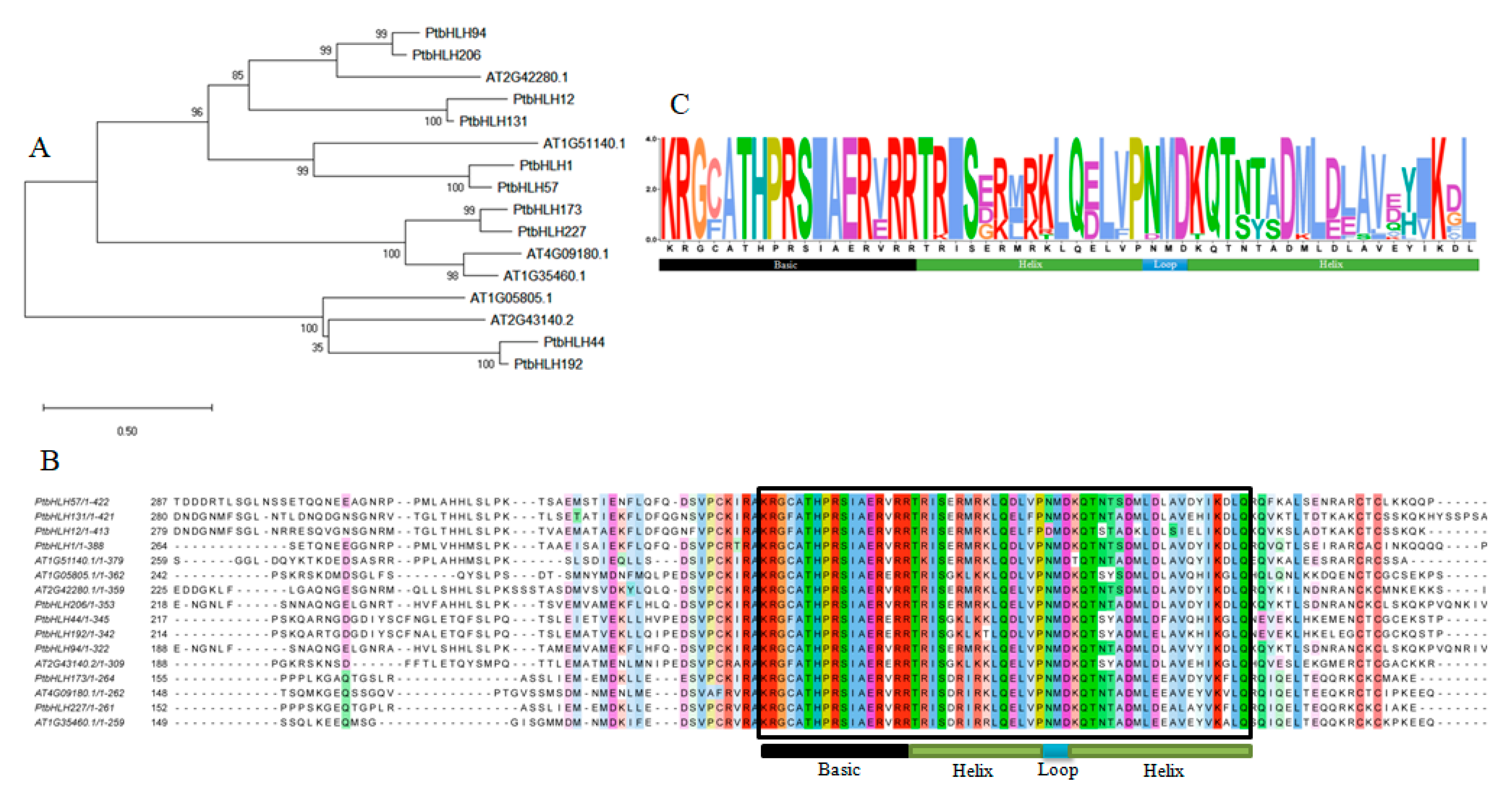
3.6. Multiple Alignment Analysis of the FBH Subfamily
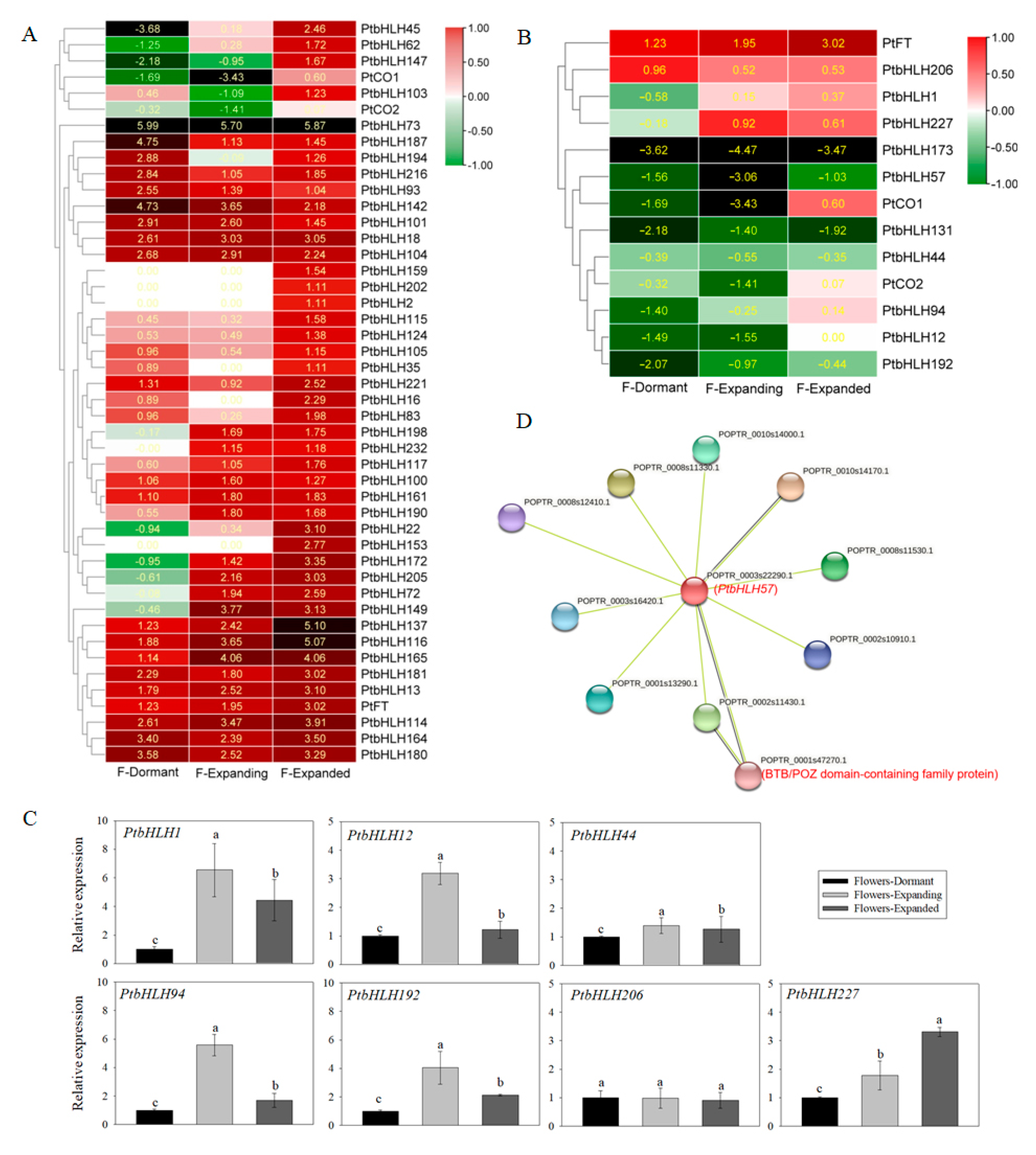
3.7. Expression Profile Analysis of PtbHLH Genes in Different Tissues
3.8. The PtbHLH Protein–Protein Interaction Network Prediction and Some May Be Involved in the Flowering Process
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdullah, M.; Cheng, X.; Cao, Y.; Su, X.; Manzoor, M.A.; Gao, J.; Cai, Y.; Lin, Y. Zinc Finger-Homeodomain Transcriptional Factors (ZHDs) in Upland Cotton (Gossypium hirsutum): Genome-Wide Identification and Expression Analysis in Fiber De-velopment. Front. Genet. 2018, 9, 357. [Google Scholar] [CrossRef]
- Aravind, L.; Anantharaman, V.; Balaji, S.; Babu, M.M.; Iyer, L.M. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev. 2005, 29, 231–262. [Google Scholar] [CrossRef]
- An, J.-P.; Qu, F.-J.; Yao, J.-F.; Wang, X.-F.; You, C.-X.; Hao, Y.-J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Zhao, P.; Kong, N.; Lu, R.; Pei, Y.; Huang, C.; Ma, H.; Chen, Q. Genome-Wide identification and characterization of the potato bhlh transcription factor family. Genes 2018, 9, 54. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Hu, Z.; Zhao, T.; Yang, Y.; Chen, T.; Yang, M.; Yu, W.; Zhang, B. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The Basic Helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004, 5, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in Arabidopsis. PLoS Genet. 2013, 9, e1003861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-García, J.F.; Huq, E.; Quail, P.H. direct targeting of light signals to a promoter element-bound transcription factor. Science 2000, 288, 859–863. [Google Scholar] [CrossRef] [Green Version]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. Flowering bhlh transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, D.H.; Pruneda-Paz, J.L.; Kay, S.A. FBH1 affects warm temperature responses in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2014, 111, 14595–14600. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Chang, X.; Kasuga, T.; Bui, M.; Reid, M.S.; Jiang, C.-Z. A basic helix-loop-helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in petunia. Hortic. Res. 2015, 2, 15059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, F.-Y.; Lai, Y.-C.; Wang, J.; Feng, Y.-R.; Shih, Y.-H.; Chen, J.-H.; Sun, H.-C.; Yang, S.; Li, C.; Wu, K. The Arabidopsis histone demethylase JMJ28 regulates CONSTANS by interacting with FBH transcription factors. Plant Cell 2021, 33, 1196–1211. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Kherawat, B.; Dey, P.; Saha, D.; Singh, A.; Bhatia, S.; Ghodake, G.; Kadam, A.; Kim, H.-U.; Chung, S.M.; et al. Genome-wide identification and characterization of Pin-Formed (PIN) gene family reveals role in developmental and various stress conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021, 22, 7396. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Kabi, M.; Debnath, D.; Saha, D.; Khandual, A.; Rout, S.; Chung, S.M.; et al. Genome-Wide Identification and Characterization of the Brassinazole-resistant (BZR) gene family and its expression in the various developmental stage and stress conditions in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 8743. [Google Scholar] [CrossRef]
- Tong, T.; Fang, Y.X.; Zhang, Z.; Zheng, J.; Zhang, X.; Li, J.; Niu, C.; Xue, D.; Zhang, X. Genome-wide identification and ex-pression pattern analysis of the KCS gene family in barley. Plant Growth Regul. 2021, 93, 1–15. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, H.; Liu, J.; Du, Q.; Lu, S.; Liu, C. Genome-wide identification and functional characterization of natural an-tisense transcripts in Salvia miltiorrhiza. Sci. Rep. 2021, 11, 1–14. [Google Scholar]
- Baruah, P.M.; Krishnatreya, D.B.; Bordoloi, K.S.; Gill, S.S.; Agarwala, N. Genome wide identification and characterization of abiotic stress responsive lncRNAs in Capsicum annuum. Plant Physiol. Biochem. 2021, 162, 221–236. [Google Scholar] [CrossRef]
- Wang, M.; Chen, B.; Zhou, W.; Xie, L.; Wang, L.; Zhang, Y.; Zhang, Q. Genome-wide identification and expression analysis of the AT-hook Motif Nuclear Localized gene family in soybean. BMC Genom. 2021, 22, 1–26. [Google Scholar] [CrossRef]
- Zhao, K.; Li, S.; Yao, W.; Zhou, B.; Li, R.; Jiang, T. Characterization of the basic helix-loop-helix gene family and its tis-sue-differential expression in response to salt stress in poplar. PeerJ 2018, 6, e4502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; López, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Steenwyk, J.L.; Buida, T.J., 3rd; Li, Y.; Shen, X.X.; Rokas, A. ClipKIT: A multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol. 2020, 18, e3001007. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools—An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Clamp, M.; Cuff, J.; Searle, S.M.; Barton, G. The Jalview Java alignment editor. Bioinformatics 2004, 20, 426–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundell, D.; Mannapperuma, C.; Netotea, S.; Delhomme, N.; Lin, Y.C.; Sjodin, A.; Van de Peer, Y.; Jansson, S.; Hvidsten, T.R.; Street, N.R. The Plant Genome Integrative Explorer Resource: PlantGenIE.org. New Phytol. 2015, 208, 1149–1156. [Google Scholar] [CrossRef] [Green Version]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the Basic Helix-Loop-Helix Transcription Factor Gene Family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497–2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Lv, W.; Zhang, H.; Ma, L.; Li, P.; Ge, L.; Li, G. Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) tran-scription factor family in maize. BMC Plant Biol. 2018, 18, 235. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Feng, R.; Ma, R.; Shen, Z.; Cai, Z.; Song, Z.; Peng, B.; Yu, M. Genome-wide analysis of basic helix-loop-helix su-perfamily members in peach. PLoS ONE 2018, 13, e0195974. [Google Scholar]
- Mao, K.; Dong, Q.; Li, C.; Liu, C.; Ma, F. Genome Wide Identification and Characterization of Apple bHLH Transcription Factors and Expression Analysis in Response to Drought and Salt Stress. Front. Plant Sci. 2017, 8, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.F.; Su, L.; Gao, H.H.; Jiang, X.L.; Wu, X.Y.; Li, Y.; Zhang, Q.Q.; Wang, Y.M.; Ren, F.S. Genome-wide characteri-zation of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis. Front. Plant Sci. 2018, 9, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, X.; Li, M.; Li, L.; Yin, H.; Wu, J.; Zhang, S. Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol. 2015, 15, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant tran-scription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Sanagi, M.; Aoyama, S.; Kubo, A.; Lu, Y.; Sato, Y.; Ito, S.; Abe, M.; Mitsuda, N.; Ohme-Takagi, M.; Kiba, T.; et al. Low nitrogen conditions accelerate flowering by modulating the phosphorylation state of flowering BHLH 4 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2022942118. [Google Scholar] [CrossRef]
- Li, H.; Gao, W.; Xue, C.; Zhang, Y.; Liu, Z.; Zhang, Y.; Meng, X.; Liu, M.; Zhao, J. Genome-wide analysis of the bHLH gene family in Chinese jujube (Ziziphus jujuba Mill.) and wild jujube. BMC Genom. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Xu, C.; Park, S.J.; Van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Y.; Xin, H.; Gu, X.; Ma, J.; Li, L. Genome-Wide Identification and Functional Analysis of the Basic Helix-Loop-Helix (bHLH) Transcription Family Reveals Candidate PtFBH Genes Involved in the Flowering Process of Populus trichocarpa. Forests 2021, 12, 1439. https://doi.org/10.3390/f12111439
Ye Y, Xin H, Gu X, Ma J, Li L. Genome-Wide Identification and Functional Analysis of the Basic Helix-Loop-Helix (bHLH) Transcription Family Reveals Candidate PtFBH Genes Involved in the Flowering Process of Populus trichocarpa. Forests. 2021; 12(11):1439. https://doi.org/10.3390/f12111439
Chicago/Turabian StyleYe, Yang, Haodong Xin, Xiting Gu, Jianwen Ma, and Lingli Li. 2021. "Genome-Wide Identification and Functional Analysis of the Basic Helix-Loop-Helix (bHLH) Transcription Family Reveals Candidate PtFBH Genes Involved in the Flowering Process of Populus trichocarpa" Forests 12, no. 11: 1439. https://doi.org/10.3390/f12111439
APA StyleYe, Y., Xin, H., Gu, X., Ma, J., & Li, L. (2021). Genome-Wide Identification and Functional Analysis of the Basic Helix-Loop-Helix (bHLH) Transcription Family Reveals Candidate PtFBH Genes Involved in the Flowering Process of Populus trichocarpa. Forests, 12(11), 1439. https://doi.org/10.3390/f12111439




