Abstract
Derris trifoliata Lour. is an indigenous and associated liana species of mangroves in China; however, its rapid dispersal is threatening mangrove survival. To explore and evaluate their persistence in past disturbances and their potential resistance to future climate and environmental changes, 120 D. trifoliata samples were collected from three sites in Guangdong Province, China, and they were used to develop single nucleotide polymorphic markers using specific-locus amplified fragment sequencing technology. A total of 351.59 Mb reads and 97,998 polymorphic specific-locus amplified fragment sequencing tags were identified, including 360,672 single nucleotide polymorphisms. The principal component analysis, phylogenetic tree, and genetic structure all clustered the samples according to their geographic positions. The three populations showed medium genetic diversity levels and high clonal diversity, indicating that sexual propagation played vital roles in the populations’ succession, although clonal growth was intense within the populations. An association analysis revealed that 9 out of 16 markers were correlated with nitrogen, which indicated the positive roles of nitrogen in population formation and maintenance. This study provides an ecological and molecular basis for understanding the outbreaks of D. trifoliata in mangroves. To control the further expansion of D. trifoliata in mangroves, preventive and control measures should be taken against clonal growth and sexual propagation, respectively; obstructing the clonal growth, especially that of the stolon, should be mainly considered at the junctions of D. trifoliata and mangroves.
1. Introduction
Derris trifoliata Lour. (Leguminosae) is the liana species which usually appears in the shrubs growing along coastal banks and the seaside, and it is also the associated species of mangroves in China. Because of the vital status of mangroves in carbon accumulation [1,2,3] and storm protection [4,5,6], the government invests great amounts of human, material, and financial resources to protect them from external disturbances and destruction caused by domestic and industrial wastewater [7], breeding industries [8], pests [9], and alien species [10]. However, the associated species, as an inside factor of mangroves, were missed. This changed when mangrove managers reported the damage caused by D. trifoliata. The mangroves entangled by D. trifoliata soon died because of their disadvantageous position when competing for light, nutrients, and space. The increases in the biomass and abundance of D. trifoliata, as a tropical liana species, are in accordance with those of other lianas in the world [11,12], which indicated the lianas have increased over the last several years and that this trend will continue for some years.
The D. trifoliata population can proliferate over long distances through seed dispersal, and the populations can also expand in their current habitats by producing rhizomes and stolons. Thus, D. trifoliata reproduce in two ways [13,14,15]. The probability of flowering increases during the first stage and then decreases as the stem diameter increases [16]. Asexual propagation is also called clonality, which is the ability to produce new individuals spontaneously. The generated offspring have the same genetic structure as the parental generation, and the clonality of plants is shown through clonal growth [17]. A genet is a clone or a genetic individual, which is the sum of all the physiological individuals or ramets descended from the same zygote [17]. Clonal architecture refers to the spatial configuration of the ramets from the same genet, which can be divided into two extreme types: guerilla (loose clustering of ramets) and phalanx (distinct compact clumps of ramets) growth forms. There is also a series of intermediate types between the two extreme forms [18]. The clonal architecture not only depends on genetic factors but also the distribution of resources in the habitat. In general, the guerilla growth form tends to acquire the dispersed resources, while the phalanx growth form tends to acquire the centrally distributed resources.
The clonal diversity and clonal structure of a plant population result from adapting to the environment during evolution. Therefore, studying them in-depth may reveal the mechanisms of population formation, maintenance, and recession. Identifying the genet, ensuring the number of genets, analyzing the spatial patterns of genets, and measuring the genetic variation among genets are the bases for studying the evolution mechanisms of a population. In the early period, it is difficult to identify the clonal genet accurately on the basis of phenotypic characteristics in the wild; therefore, the reliability is also highly controversial. Fortunately, the development of molecular marker technology based on PCR has allowed the identification of clonal genets and clonal plant research [19,20,21] for several years. Compared with random amplified polymorphic DNAs [22], amplified fragment length polymorphisms [23], inter-simple sequence repeats [24], and simple sequence repeats [25], single nucleotide polymorphisms (SNPs) are the third-generation molecular markers, and they represent the most abundant and stable type of DNA variation in plant genomes. Additionally, the high-throughput detection of SNPs is rapid [26,27].
Clonal growth provides a competitive advantage in community development, species diversity, and community composition [28,29]. Extensive clonal growth may be a major reason for the rapid diffusion of the D. trifoliata population in mangroves, and similar results were verified for other liana species [28,30,31]. Sexual propagation generates offspring with different genetic structures, and these offspring have the ability and potential to adapt to environmental changes. However, the offspring generated by clonal growth do not adapt to drastic environmental changes because of their limited genetic variations. Thus, the genetic diversity of a population dominated by clonal growth for a long time might decrease; however, many studies found that this was not the case [21,32,33,34].
In 2011, the genetic diversity of D. trifoliata across southern China at the species level was found to be relatively high through the use of inter-simple sequence repeat markers, and the results also suggested that ocean currents, fruit dispersal, and habitat fragmentation played roles in determining the genetic variation [13]. Field investigations revealed that at different sites the D. trifoliata population size and distribution characteristics were different, and the phenotypic traits of pods and seeds also differed [35]. The fast diffusion and spread of the D. trifoliata population in mangroves over the last 10 years severely threatens the survival of mangroves; however, there is limited information on the biology, physiology, ecology, and molecular biology of D. trifoliata.
In this study, we developed SNP markers using specific-locus amplified fragment sequencing (SLAF-seq) technology to identify the genotypes of samples. These markers may also be used to investigate the genetic information within and among D. trifoliata populations. Here, our objectives were: (a) to analyze and demonstrate the genetic information on the basis of phylogenetic trees, a principal component analysis (PCA), and genetic structures; (b) to clarify the clonal diversity, architecture, structure, and clonal growth capability and to explain the functions of reproductive models in population succession; (c) to describe the spatial distribution of genets and examine whether spatial autocorrelations are significant in natural populations; and (d) to evaluate the evolutionary potential and developmental trend using the results of this study.
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
The D. trifoliata samples were collected from three sites (Figure 1), Nansha District (113°33′ E–22°50′ N), Yangjiang City (112°12′ E–21°49′ N), and Zhanjiang City (109°57′ E–21°26′ N), in Guangdong Province, China, in October 2020.
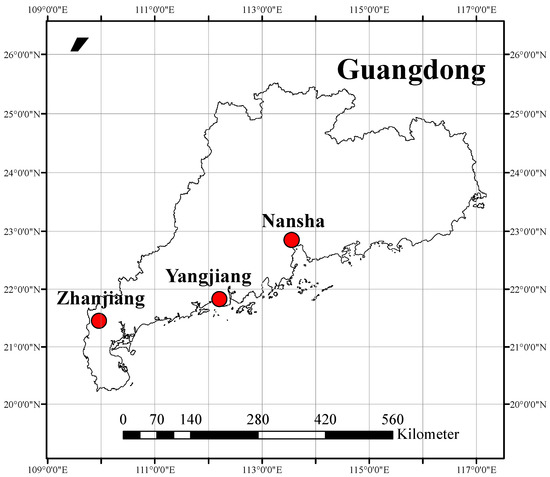
Figure 1.
The location of three sites in Guangdong province, China.
Because the population sizes and distribution characteristics at these three different sites varied, different numbers of samples were collected at the three sites. The population size in the Nansha District is small and sparse relative to the other two sites, and the individual cases occur sporadically along the edge of the mangroves. A total of 12 samples were collected in the Nansha District, and the collected samples represent the population’s genetic information. The biomass and abundance of D. trifoliata in Yangjiang and Zhanjiang cities were great and overdispersed, and the selected populations from these two sites were distributed upstream and downstream along the sea, respectively. A quadrat was set at approximately the middle position along the sea, and 44 samples were collected in this quadrat. An additional 10 samples were collected from upstream to downstream. We also recorded the spatial positions of the 44 samples in the quadrat, and constructed sampling maps of the Yangjiang (Figure 2a) and Zhanjiang (Figure 2b) sites.
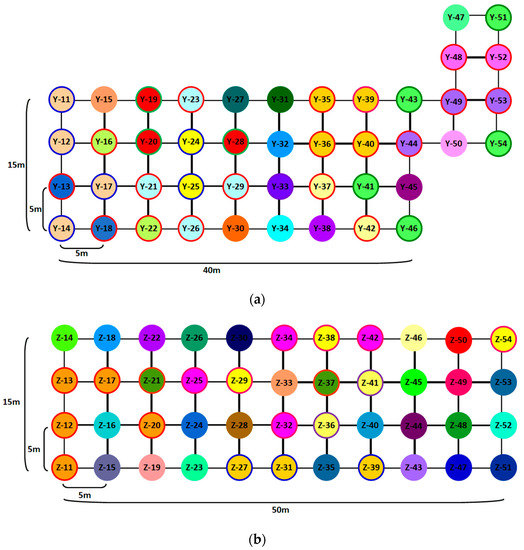
Figure 2.
The sampling maps and clonal structures of Derris trifoliata Lour. populations at the Yangjiang (a) and Zhanjiang (b) sites. Circles of the same color in each population represent individuals from the same genets produced by clonal growth.
The fresh leaf samples from the selected individual plants were preserved in dry ice and taken to the laboratory immediately. The cetyl-trimethyl-ammonium bromide method [36] was used to extract genomic DNA for further sequencing and genotyping.
2.2. SLAF Library Construction and High-Throughput Sequencing
The SLAF library was constructed following the method of Sun et al. with some modifications [26] by the Beijing Biomarker Technologies Corporation (Beijing, China). First, on the basis of the D. trifoliata genome size and GC content, the soybean (Glycine max) genome (978 Mb, with a GC content 34.76%) was chosen as the reference to be used in a predesign experiment to predict suitable restriction enzymes and the sizes of restriction fragments. The criteria for selecting the most suitable enzyme digestion were as follows: (1) an as low as possible proportion of SLAF tags in the repeat sequence; (2) SLAF tags must be evenly distributed throughout the genome; (3) the lengths of the SLAF tags were consistent with the specific experimental system; and (4) the final number of SLAF tags must match that of the expected tags. Second, the restriction enzyme combination, RsaI + HaeIII (NEB, Ipswich, MA, USA), was selected to digest the qualified genomic DNA of D. trifoliata samples. To maintain sequence depth uniformity, 364qua4-bp fragments obtained by the restriction enzyme digestion were excised. Then, an adenine nucleotide (A) overhang was added to the 3uniformity, 364qua4-bp fragments obtained and [37] sequencing connectors were added. Subsequently, the PCR products were amplified, purified, and mixed, and then the target fragments were selected. After a quality inspection of the library, sequencing was performed using the Illumina HiSeqTM 2500 platform (Illumina, Inc, San Diego, CA, USA) at Biomarker Technologies Corporation. During the sequencing process, real-time monitoring was also conducted for each cycle. The effective original reads of all the samples were identified from the raw data using the Dual-Index paired-end sequencing method [37]. After filtering the sequencing read joints, the quality and volume of the sequencing data were evaluated. To evaluate the enzyme digestion efficiency, the mapping efficiency, and the accuracy and validity of the experimental process, Oryza sativa ssp. japonica was selected as the control group, and it was sequenced simultaneously. The O. sativa ssp. japonica sequence was mapped on the genome sequence (http://rapdb.dna.affrc.go.jp/ accessed on 15 march 2021 using SOAP software [38].
2.3. SLAF-seq Data Grouping and SNPs Discovery
The methods and software for SLAF-seq data grouping and genotyping were those described by Sun et al. [26]. Low quality reads (quality scores lower than 30, indicating a 0.1% chance of an error) were discarded, and the high quality SLAF paired-end reads were clustered using sequence similarity [26]. The reads sharing over 90% identity levels were regarded as the same SLAF tags and grouped into the same SLAF locus. The polymorphic SLAF tags can be defined if the SLAF tag sequences among different samples differed, and the polymorphic SLAF tags were mainly caused by SNPs. The deepest sequence of each SLAF tag was treated as the reference sequence, and the paired-end reads were mapped on the reference genome using the Burrows–Wheeler Aligner [39]. The SNPs were discovered using two methods, Genome Analysis Toolkit [40] and SAMtools [41]. Then, the intersection of the two data sets was obtained from the two methods, and the final intersection data set contained the relative reliable SNPs. Finally, the SNPs with minor allele frequency >0.05 were collected and used for subsequent analyses.
2.4. Phylogenetic Analysis
The SNP markers obtained from the three sites and 120 samples were the basis for further genetic analyses. In the present study, a dendrogram was constructed for the 120 samples using MEGA X software [42] with the neighbor-joining algorithm [43] and the Kimura 2-parameter model. The PCA was performed using EIENSOFT software [44]. The genetic structure of the population as indicated by the SNP markers was also analyzed using admixture software [45], and the genetic structure results quantified the numbers of ancestors and predicted the sources of the samples. The genetic structure map of D. trifoliata was constructed by assuming the group number (K value) was between 1 and 10 and producing the corresponding cluster analysis. Then, cross-validation was performed for the cluster results, and finally, on the basis of the valley value of the cross-validation errors, the optimal group number was confirmed. The results of the phylogenetic tree, PCA analysis, and genetic structure were mutually supplemented and verified.
2.5. Genetic Analysis
Genetic diversity indexes, including the average minor allele frequency, expected and observed allele numbers, expected (He) and observed (Ho) heterozygosities, Nei’s gene diversity index (H), the number of polymorphic markers, polymorphic information content (PIC), and Shannon-Wiener index (I) were calculated at the population and species levels using a Perl program.
Clonal diversity was estimated using the following five parameters [46]: (a) The number of genets (G). When the genetic distance between individuals within the population was 0, then we concluded that they were the different ramets from the same genet; (b) mean clonal size (NC):
where N is the total number of samples; (c) the genotype ratio (PD):
(d) Simpson’s diversity index (D):
where is the ramet number of individuals of genotype i [47], and (e) Fager’s index (E) is used to describe the distribution evenness of different genotypes within the population:
Spatial autocorrelation analyses were also performed using GenALEx version 6.5 [48]. Distance classes were selected to have even distance sizes to provide a reliable basis for statistical inference.
2.6. Association Analyses between SNPs and Environmental Factors
The association analyses between SNPs and environmental variables were conducted using BayeScan [49] and Latent Factor Mixed Model (LFMM;[50] programs. For BayeScan, which was conducted using the differences in allele frequencies among different populations, a scatter diagram was constructed on the basis of the relationship between the log10(q-value) of each SNP loci and differentiation index (Fst). The points located on the right of the perpendicular line (false discover rate (FDR) = 0.05) were treated as the selected loci. For the LFMM program, the correlations between SNP loci and the environmental gradient were detected using the MCMC algorithm. The p-values were adjusted using the chi-squared test, and then candidate loci (FDR ≤ 0.01) were selected. A BLAST [51] comparison was conducted using the candidate loci screened by these two methods, and the loci were annotated using multiple databases (including GO [52], KEGG [51], COG [53], NR [54], and Swiss-Prot).
3. Results
3.1. Sequence and Quality Statistics Based on SLAF-seq
In total, 351.59 Mb of paired-end reads were obtained by high-throughput sequencing, and the average Q30 (quality score of 30) and GC (guanine-cytosine) content were 92.56% and 39.33%, respectively (Table 1), which illustrated that the sequenced data were qualified and could be used for further analyses. To evaluate the mapped efficiency, the sequencing reads of O. sativa ssp. japonica was mapped on its reference genome using SOAP software [38], and the paired-end mapped reads efficiency was 90.60%, which illustrated that the mapped results were normal. The enzyme digestion efficiency was determined as being 92.37% in O. sativa ssp. Japonica, which indicated that the restriction enzyme digestion level was normal.

Table 1.
Statistics of SLAF tags and SNP markers for D. trifoliata Lour.
There was no genomic information for D. trifoliata; consequently, we constructed a reference genome by connecting the deepest SLAF tags developed in our research. Then, the sequencing reads were aligned to the constructed reference genome to obtain the SNPs. We finally developed 892,103 SLAF tags, including polymorphic tags, non-polymorphic tags, as well as SLAF tags in repeated sequences throughout the genome. The average sequence depth of each SLAF tag was approximately 12.76×, among which 97,998 polymorphic SLAF tags, accounting for 10.98% of the total, were identified. Then, low integrity (<80%), low minor allele frequency (<5%), and insufficient reads (<90%) were removed, leaving 360,672 SNPs for further analyses.
3.2. Phylogenetic Analysis
The 120 D. trifoliata samples were clustered into three major groups by a neighbor-joining analysis using the software of MEGA X. The samples from the Yangjiang and Zhanjiang sites were further divided into two subgroups, and the samples from the Nansha site were clustered as a single group. In total, there were five SNP marker-related subgroups (Figure 3 and Figure 4). The phylogenetic analysis demonstrated that the samples from each of the geographic locations differed; therefore, they were clustered on the basis of their geographical origins.
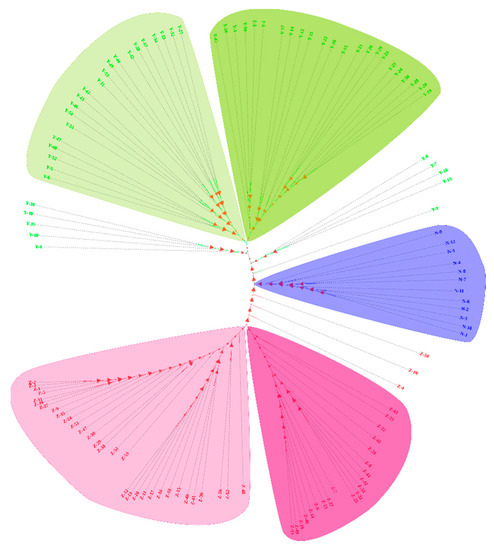
Figure 3.
The phylogenetic tree of 120 D. trifoliata accessions in which five main groups were defined. Each branch represents a sample. The red branch and letter indicate the samples from Zhanjiang, the green branch and letter indicate the samples from Yangjiang, and the blue branch and letter indicate the samples from Nansha. The fuchsia and light fuchsia facets indicate that the samples from Zhanjiang can be divided into two subgroups, the green and light green facets indicate that the samples from Yangjiang can be divided into two subgroups, and the blue facet indicates the samples from Nansha can be grouped separately.
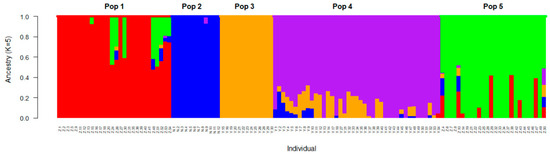
Figure 4.
The population structure analysis of 120 D. trifoliata accessions at a K value of five, with five populations inferred from the admixture analysis. The digits on the x-axis represent the accessions. Pop 1 and pop 5 consist of samples from Zhanjiang, pop 2 consists of samples from Nansha, and pop 3 and pop 4 consist of samples from Yangjiang.
Using 360,672 SNPs from 120 samples, the PCA was performed. The first principal component (PC1) explained 22.1% of the variation in the genotypic data, and PC2 and PC3 explained 29.18% and 34.65% of the observed variances, respectively. A 3D plot (Figure 5) was generated using the first three PCs, and the 120 samples were clearly clustered into three groups on the basis of geographic position, which was consistent with the phylogenetic tree.
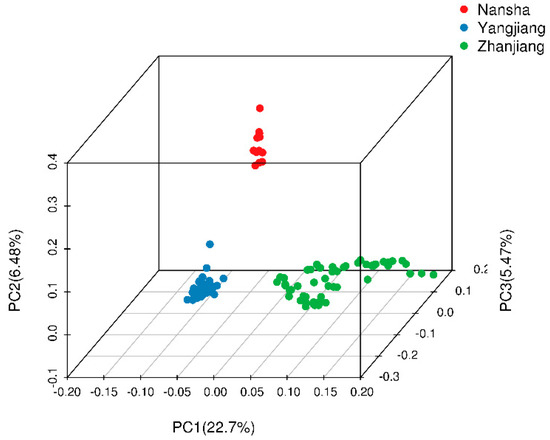
Figure 5.
The principal component analysis of 120 D. trifoliata accessions. The red, blue, and green circles represent Nansha, Yangjiang, and Zhanjiang samples, respectively.
3.3. Genetic Diversity
The populations at the Zhanjiang, Yangjiang, and Nansha sites had the highest, middle, and lowest genetic diversity levels, respectively (Table 2). There was a small range of variation among the three sites in Guangdong Province, China. The value of the observed allele number (Ao) was 2.0 across populations, and the values of expected allele number (Ae) ranged from 1.669 (Nansha) to 1.696 (Zhanjiang), with a mean value of 1.685. The observed heterozygosity (Ho) values were between 0.277 (Zhanjiang) and 0.339 (Nansha), and they were lower than the expected heterozygosity (He) values, which were between 0.379 (Nansha) and 0.391 (Zhanjiang). Nei’s gene diversity index (H) was between 0.393 (Yangjiang) and 0.400 (Nansha), with an average of 0.396. Shannon-Wiener index (I) ranged from 0.558 (Nansha) to 0.572 (Zhanjiang), with an average of 0.566, and the PIC values varied from 0.299 (Nansha) to 0.307 (Zhanjiang), with an average of 0.304. At the species level, the values of He, H, Ho, and I were 0.366, 0.368, 0.207, and 0.543, respectively (Table 2), and the values of He and I in this study were greater than those of Wu [13].

Table 2.
Summary of D. trifoliata genetic diversity parameters assessed using SNP markers.
3.4. Clonal Diversity and Clonal Structure
A total of 120 ramets were assigned to 75 distinct genets. No widespread genet was found across different populations, and all the genets were composed of one to five ramets (Figure 2). The parameters of clonal diversity for the three sites were calculated and are listed in Table 3. The Simpson’s diversity index (D) and genotypic evenness (E) values were 0.91 and 0.97, respectively, for Nansha, 0.98 and 0.84, respectively, for Yangjiang, and 0.93 and 0.85, respectively, for Zhanjiang, which indicated the high level of clonal diversity for D. trifoliata populations.

Table 3.
Clonal diversity parameters of three D. trifoliata populations.
The clonal spatial distribution of D. trifoliata populations in the Yangjiang and Zhanjiang sites are presented in Figure 2a,b, respectively, and the same ramets are usually close to each other, although different genets are frequently found to intermingle locally. We found that the individuals clustered within a short distance and proliferated over a long distance through clonal growth, revealing that the clonal architecture of D. trifoliata had both guerilla and phalanx growth forms and that they were highly adaptable to the environment.
The number of SNPs (360,672) obtained from the three sites was too large to conduct a spatial autocorrelation analysis. Therefore, the SNPs from the three sites were counted and listed separately, and then the intersections of the SNPs among the different three sites were determined. Finally, 31 SNPs were obtained as the polymorphic loci that have the genetic traits associated with the three sites, and the spatial autocorrelations of clonal structures of D. trifoliata populations at the Yangjiang and Zhanjiang sites are shown in Figure 6a,b, respectively.
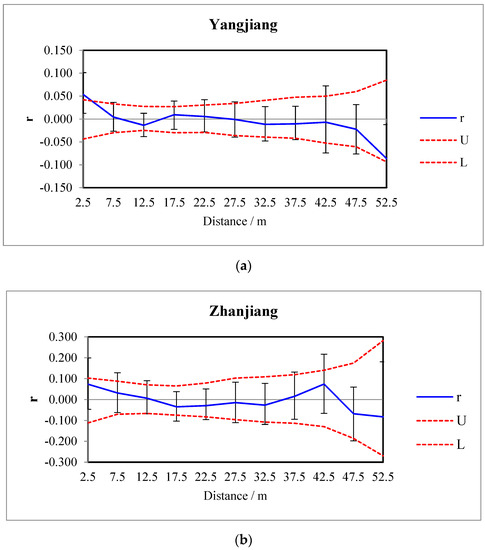
Figure 6.
Correlograms showing the spatial autocorrelation coefficient r of D. trifoliata among individuals at the Yangjiang (a) and Zhanjiang (b) sites. U and L indicate the upper and lower limits, respectively, of the 95% confidence interval.
For the population at the Yangjiang site, when the distance class size was five, there was a significant positive correlation (p < 0.05) among individuals at a spatial distance of less than 5 m. The greatest spatial autocorrelation coefficient was 0.054 in 2.5 m, and there were no significant correlations in other distance classes. There were no significant correlations among individuals in each distance class for the D. trifoliata population at the Zhanjiang site. The intercepts of the x-axis for the D. trifoliata population at the Yangjiang and Zhanjiang sites were 8.643 and 13.267, respectively, which indicated the average minimum lengths of irregular clonal growth.
3.5. Associations between SNP Markers and Environmental Variables
In total, 16 markers were identified using BayeScan, but only Marker 238822 was annotated, which indicated that the marker can be interpreted as surviving long periods of geographical isolation. In total, 86 markers were identified using LFMM, and 15 of them were annotated (Table 4). Two markers (Marker 39224 and Marker 243205) were associated with altitude, whereas Marker 57755 was associated with high temperature. Marker 123428 and Marker 102857 were associated with low temperature, whereas Marker 89512 and Marker 24270 were associated with NH4-N. Additionally, Marker 183381 was associated with organic matter, and seven markers were associated with NH3-N.

Table 4.
Annotation of selected genes associated with SNP markers.
4. Discussion
4.1. Phylogenetic Analysis
Research on D. trifoliata is limited and no previous whole genome has been reported. The present study is the first attempt to use SNPs, as third-generation molecular markers derived from SLAF-seq [55,56], to explore the adaptive mechanisms of D. trifoliata in mangroves. In this study, the minimum cross-validation error occurred at a K-value of 5, which indicated that the samples most likely were derived from five ancestors. Therefore, the optimal number of subpopulations was five. Using the maximum membership probability, 28, 12, 13, 41, and 26 samples were assigned to pops 1–5, respectively. The accessions assigned to pops 1 and 5 were all from Zhanjiang. Pops 3 and 4 were all accessions from Yangjiang, and all 12 accessions from the Nansha District were assigned to pop 2 (Figure 4). According to the determined genetic structures, there were large differences in the accessions from the three different sites, and the accessions from Yangjiang and Zhanjiang were each divided into two subgroups, which was in accordance with the phylogenetic tree. The genetic results were also consistent with the clustering results based on the phenotypic characteristics of pods and seeds [35]. The clustering method applied has been widely used in population analyses [57,58], and this method also has a strong capability to divide the individuals into subpopulations. This provides insights into the evolutionary process.
4.2. Genetic Diversity
The values of Ho and He may reflect the genetic diversity of the population. In this study, the Ho and He values of the three populations ranged from 0.277 to 0.339 and 0.379 to 0.390, respectively. The He values of the three D. trifoliata populations were all higher than those of the three widespread mangrove species Ceriops decandra, Ceriops tagal, and Ceriops australis, which range from 0.023 to 0.053 [59]. They were also higher than the peripheral mangrove species Avicennia marina, which ranges from 0.15 to 0.37 [60]. A relatively higher level of genetic heterozygosity for D. trifoliata in the mangrove habitat might result from the gene transfers between individuals among different populations, as well as anthropogenic activities, such as environmental fragmentation [61,62], which can increase the gene flow between different populations and enhance the genetic diversity level. The PIC value may represent the population’s polymorphism level. The higher the PIC values, the higher the polymorphism level. PIC < 0.25 indicates a low polymorphism level, 0.25 < PIC < 0.50 indicates a median polymorphism level, and PIC > 0.50 indicates a high polymorphism level [55]. In the present study, the PIC values of the three populations were all greater than 0.25 and lower than 0.5, which indicated that they have medium genetic diversity levels in terms of PIC. Overall, the populations of D. trifoliata exhibited higher genetic diversity as assessed by the larger Ae, Ao, He, Ho, PIC, and I values.
4.3. Clonal Diversity and Clonal Structure
The rapid expansion or increase in native plants that exhibit a biological invasion phenomenon may be caused by anthropogenic activities [63], such as environmental fragmentation, water and soil eutrophication [64,65], and deforestation. The PD, E, and D values found in our study for the three sites were all greater than the average values (PD = 0.17, E = 0.68, and D = 0.62) of clonal plants [46], which illustrated the greater clonal diversity of D. trifoliata. Although there was intense clonal growth, the clonal diversity was still maintained at a high level, which indicated that the seedlings were recruited continually [66,67,68], the genets had high survival rates, and there was a cross-pollination breeding system [69,70]. The change in tide levels dispersed the fruit of D. trifoliata, allowing for hybridization events within and among the populations. A study on the forest herb Paris quadrifolia L. indicated that the clonal diversity in moist sites is greater than in dry sites and that clone sizes are smaller in the former than in the latter, which was explained by repeated seedling recruitment and limited seed dispersal within populations [71].
Based on the obtained 31 polymorphic loci of SNPs in the study, the spatial autocorrelation analysis indicated that there was a significant positive correlation (p < 0.05) among individuals at a spatial distance of less than 5 m for the population in the Yangjiang site. Spatial autocorrelation analyses represent an effective method to study the genetic structures of populations on a small scale, and the spatial correlation coefficients (r) were obtained using the clonal genetic distances of all the individuals established on the basis of their geographic distance levels. This method has been widely used in research on clonal plants [34,72,73], but the present study is the first attempt to use the method and SNPs in research on clonal structures.
Natural selection, or the polymorphic loci correlated with environmental factors, has an impact on adaptation [74,75]. Receptor-like protein 12 is a potential candidate involved in adaptation to altitude, and it is involved in the perception of CLV3 and CLV3-like peptides, which act as extracellular signals regulating meristem maintenance [76]. A possible lysine decarboxylase is a potential candidate that correlated with the presence of NH4-N, and the enzyme activity was mainly localized in the roots of the same family of soybean [77]. There were nine out of sixteen markers associated with nitrogen, and they were annotated into the molecular function, biological process, and cellular component categories. Additionally, these nine markers may be associated with proteins that were correlated with the nitrogen fixation of D. trifoliata in mangroves.
The D. trifoliata individuals on the edges of mangroves have supports to climb; therefore, they can allocate more biomass to blooming and fruiting. However, the individuals far away from the mangroves allocate more biomass to stems and branches used for support, and there is little sexual reproduction [16]. Thus, to prevent and control the further expansion of D. trifoliata in mangroves, obstructing the stolon growth that extends into mangroves from the D. trifoliata population, removing the propagative organs of D. trifoliata to reduce seedling recruitment, and cutting the branches of D. trifoliata in the canopy of mangroves should be considered.
5. Conclusions
The present study analyzed the phylogenetic and genetic information of three natural D. trifoliata populations in Guangdong Province, China using SNPs that were developed by SLAF-seq technology. The work was the first attempt to delineate D. trifoliata clone sizes. The phylogenetic analysis illustrated that there were differences among the three populations because of the geographic isolation of the three sites. At the species level, the PIC and I values were 0.209 and 0.543, respectively, which indicated a moderated genetic diversity level in D. trifoliata. The D and E values in the three populations were all greater than the average values of clonal plants, which indicated high clonal diversity levels and that sexual reproduction played vital roles in the populations. There was a significant positive correlation among individuals at a spatial distance of less than 5 m at the Yangjiang site. A total of 16 markers were annotated using a BLAST search, and nine markers correlated with nitrogen. The results in this study formed an ecological and molecular basis for understanding D. trifoliata outbreaks and helped to provide useful measures to control further expansion.
Author Contributions
Conceptualization, Y.Z., B.L. and K.X.; methodology, Y.Z., B.L. and K.X.; software, Y.Z.; validation, Y.Z., B.L. and K.X.; formal analysis, Y.Z., B.L. and K.X.; investigation, Y.Z., B.L., K.X., X.A. and N.S.; resources, Y.Z., B.L., K.X., X.A. and N.S.; data curation, Y.Z.; writing—original draft preparation, Y.Z.; writing—review and editig, Y.Z., B.L. and K.X.; visualization, Y.Z.; supervision, B.L. and K.X.; project administration, K.X. and B.L.; funding acquisition, B.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key-Area Research and Development Program of Guangdong Province (2020B020214001-KT02) and the National Natural Science Foundation of China (41876094).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequence data reported in this study were archived in the Sequence Read Archive (SRA), with the accession number PRJNA734440.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wicaksono, P.; Hartono, D.P.; Nehren, U. Mangrove biomass carbon stock mapping of the Karimunjawa Islands using multispectral remote sensing. Int. J. Remote Sens. 2016, 37, 26–52. [Google Scholar] [CrossRef]
- Collins, D.S.; Avdis, A.; Allison, P.A.; Johnson, H.D.; Hill, J.; Piggott, M.D.; Hassan, M.H.A.; Damit, A.R. Tidal dynamics and mangrove carbon sequestration during the Oligo–Miocene in the South China Sea. Nat. Commun. 2017, 15, 15698. [Google Scholar] [CrossRef]
- Xiong, Y.; Liao, B.; Wang, F. Mangrove vegetation enhances soil carbon storage primarily through in situ inputs rather than increasing allochthonous sediments. Mar. Pollut. Bull. 2018, 131, 378–385. [Google Scholar] [CrossRef]
- Badola, R.; Hussain, S.A. Valuing ecosystem functions: An empirical study on the storm protection function of Bhitarkanika mangrove ecosystem, India. Environ. Conserv. 2005, 32, 85–92. [Google Scholar] [CrossRef]
- Bell, J.; Lovelock, C.E. Insuring Mangrove Forests for Their Role in Mitigating Coastal Erosion and Storm-Surge: An Australian Case Study. Wetlands 2013, 33, 279–289. [Google Scholar] [CrossRef]
- Akber, M.A.; Patwary, M.M.; Islam, M.A.; Rahman, M.R. Storm protection service of the Sundarbans mangrove forest, Bangladesh. Nat. Hazards 2018, 94, 405–418. [Google Scholar] [CrossRef]
- Meziane, T.; Tsuchiya, M. Organic matter in a subtropical mangrove-estuary subjected to wastewater discharge: Origin and utilisation by two macrozoobenthic species. J. Sea Res. 2010, 47, 1–11. [Google Scholar] [CrossRef]
- Hoque, M.M.; Kamal, A.H.M.; Idris, M.H.; Ahmed, O.H.; Saifullah, A.S.M.; Billah, M.M. Status of some fishery resources in a tropical mangrove estuary of Sarawak, Malaysia. Mar. Biol. Res. 2015, 11, 834–846. [Google Scholar] [CrossRef]
- Xin, K.; Xie, Z.; Zhong, C.; Sheng, N.; Gao, C.; Xiao, X. Damage Caused by Sphaeroma to Mangrove Forests in Hainan, Dongzhaigang, China. J. Coast. Res. 2020, 36, 1197–1203. [Google Scholar] [CrossRef]
- Feng, J.; Zhou, J.; Wang, L.; Cui, X.; Ning, C.; Wu, H.; Zhu, X.; Lin, G. Effects of short-term invasion of Spartina alterniflora and the subsequent restoration of native mangroves on the soil organic carbon, nitrogen and phosphorus stock. Chemosphere 2017, 184, 774–783. [Google Scholar] [CrossRef]
- Schnitzer, S.A. A mechanistic explanation for global patterns of liana abundance and distribution. Am. Nat. 2005, 166, 262–276. [Google Scholar] [CrossRef]
- Schnitzer, S.A.; Bongers, F. Increasing liana abundance and biomass in tropical forests: Emerging patterns and putative mechanisms. Ecol. Lett. 2011, 14, 397–406. [Google Scholar] [CrossRef]
- Wu, B.; Geng, S.L.; Shu, B. Genetic variation and the conservation of isolated populations of Derris trifoliata (Leguminosae), a mangrove-associated vine, in southern China. Biochem. Syst. Ecol. 2012, 40, 118–125. [Google Scholar] [CrossRef]
- Raju, A.J.S.; Kumar, R. Pollination ecology of Derris trifoliata (Fabaceae), a mangrove associate in Coringa Mangrove Forest, Andhra Pradesh, India. J. Threat. Taxa 2016, 8, 8788–8796. [Google Scholar] [CrossRef]
- Aluri, J.S.R.; Kumar, R.; Chappidi, P.R. Reproductive biology of mangrove plants clerodendrum inerme, Derris trifoliata, Suaeda maritima, Suaeda monoica, Suaeda nudiflora. Transylv. Rev. Syst. Ecol. Res. 2016, 18, 31–68. [Google Scholar] [CrossRef]
- Zhang, Y.; Liao, B.; Xin, K.; Sheng, N. Allometric equations for liana species Derris trifoliata and the relationship between inflorescence generation and stem diameter. Glob. Ecol. Conserv. 2017, 26, e01511. [Google Scholar] [CrossRef]
- Stuefer, J.F.; Erschbamer, B.; Huber, H.; Suzuki, J. Ecology and Evolutionary Biology of Clonal Plants. Evol. Ecol. 2002, 15, 223–230. [Google Scholar] [CrossRef]
- Callaghan, T.V.; Svensson, B.M.; Bowman, H.; Lindley, D.K.; Carlssom, B.A. Models of clonal plant-growth based on population-dynamics and architecture. Oikos 1990, 57, 257–269. [Google Scholar] [CrossRef]
- Dering, M.; Chybicki, I.J.; Rączka, G. Clonality as a driver of spatial genetic structure in populations of clonal tree species. J. Plant Res. 2015, 128, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Ley, A.C.; Hardy, O.J. Spatially limited clonality and pollen and seed dispersal in a characteristic climber of Central African rain forests: Haumania danckelmaniana (Marantaceae). Biotropica 2016, 48, 618–627. [Google Scholar] [CrossRef]
- Loh, R.; Scarano, F.R.; Alves-erreira, M.; Salgueiro, F. Clonality strongly affects the spatial genetic structure of the nurse species Aechmea nudicaulis (L.) Griseb. (Bromeliaceae). Bot. J. Linn. Soc. 2015, 178, 329–341. [Google Scholar] [CrossRef]
- Ren, M.X.; Zhang, Q.G. Clonal diversity and structure of the invasive aquatic plant Eichhornia crassipes in China. Aquat. Bot. 2007, 87, 242–246. [Google Scholar] [CrossRef]
- de Witte, L.C.; Armbruster, G.F.J.; Gielly, L.; Taberlet, P.; Stöcklin, J. AFLP markers reveal high clonal diversity and extreme longevity in four key arctic-alpine species. Mol. Ecol. 2012, 21, 1081–1097. [Google Scholar] [CrossRef]
- Yang, Y.J.; Ji-Ning, L.I.; Gong, L.; Zhou, J. Clonal Structure and Genetic Diversity of Natural var.Assessed by ISSR. Plant Sci. J. 2013, 31, 85–92. [Google Scholar] [CrossRef]
- Chenault, N.; Arnaud-Haond, S.; Juteau, M.; Valade, R.; Almeida, J.L.; Villar, M.; Bastien, C.; Dowkiw, A. SSR-based analysis of clonality, spatial genetic structure and introgression from the Lombardy poplar into a natural population of Populus nigra L. along the Loire River. Tree Genet. Genomes 2011, 7, 1249–1262. [Google Scholar] [CrossRef]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H. SLAF-seq: An Efficient Method of Large-Scale De Novo SNP Discovery and Genotyping Using High-Throughput Sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef]
- Gong, D.; Huang, L.; Xu, X.; Wang, C.; Ren, M.; Wang, C.; Chen, M. Construction of a high-density SNP genetic map in flue-cured tobacco based on SLAF-seq. Mol. Breed. 2016, 36, 1–12. [Google Scholar] [CrossRef]
- Duchoslavová, J.; Herben, T. Effect of clonal growth form on the relative performance of species in experimental communities over time. Perspect. Plant Ecol. Evol. Syst. 2020, 44, 125532. [Google Scholar] [CrossRef]
- Yu, H.W.; Wang, L.G.; Liu, C.H.; Yu, D.; Qu, J.H. Effects of a spatially heterogeneous nutrient distribution on the growth of clonal wetland plants. BMC Ecol. 2020, 20, 59. [Google Scholar] [CrossRef]
- Yorke, S.R.; Schnitzer, S.A.; Mascaro, J.; Letcher, S.G.; Carson, W.P. Increasing Liana Abundance and Basal Area in a Tropical Forest: The Contribution of Long-distance Clonal Colonization. Biotropica 2013, 45, 317–324. [Google Scholar] [CrossRef]
- Ledo, A.; Schnitzer, S.A. Disturbance and clonal reproduction determine liana distribution and maintain liana diversity in a tropical forest. Ecology 2014, 95, 2169–2178. [Google Scholar] [CrossRef]
- Pluess, A.R.; Stöcklin, J. Population genetic diversity of the clonal plant Geum reptans (Rosaceae) in the Swiss Alps. Am. J. Bot. 2004, 91, 2013–2021. [Google Scholar] [CrossRef]
- Solé, M.; Durka, W.; Eber, S.; Brandl, R. Genotypic and Genetic Diversity of the Common WeedCirsium arvense (Asteraceae). Int. J. Plant Sci. 2004, 165, 437–444. [Google Scholar] [CrossRef][Green Version]
- Namroud, M.C.; Park, A.; Tremblay, F.; Bergeron, Y. Clonal and spatial genetic structures of aspen (Populus tremuloides Michx.). Mol. Ecol. 2005, 14, 2969–2980. [Google Scholar] [CrossRef]
- Zhang, Y.; Xin, K.; Liao, B.W.; Sheng, N.; Ai, X.H. The characteristics of pods and seeds of liana species Derris trifoliata and their relationship with environmental factors in Guangdong, China. Ecol. Indic. 2021, 129, 107930. [Google Scholar] [CrossRef]
- Gill, K.S.; Lubbers, E.L.; Gill, B.S.; Raupp, W.J.; Cox, T.S. A genetic linkage map of Triticum tauschii (DD) and its relationship to the D genome of bread wheat (AABBDD). Genome 1991, 34, 362–374. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2018, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.-W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2, An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Michael, L.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Eickmeyer, K.; Huggins, P.; Pachter, L. On the optimality of the neighbor-joining algorithm. Algorithms Mol. Biol. 2008, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Roose, M.L. Patterns of Genotypic Diversity in Clonal Plant Species. Am. J. Bot. 1987, 74, 123–131. [Google Scholar] [CrossRef]
- Parker, K.C.; Hamrick, J.L. Genetic diversity and clonal structure in a columnar cactus, Lophocereus schottii. Am. J. Bot. 1992, 79, 86–96. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5, Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Foll, M.; Gaggiotti, O. A Genome-Scan Method to Identify Selected Loci Appropriate for Both Dominant and Codominant Markers: A Bayesian Perspective. Genetics 2008, 180, 977–993. [Google Scholar] [CrossRef]
- Eric, F.; Schoville, S.D.; Guillaume, B.; Olivier, F. Testing for Associations between Loci and Environmental Gradients Using Latent Factor Mixed Models. Mol. Biol. Evol. 2013, 30, 1687–1699. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schffer, A.A.; Zhang, J.; Zhang, Z.; Webb, M.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Consortium, U.P. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef]
- Lyu, Y.Z.; Dong, X.Y.; Huang, L.B.; Zheng, J.W.; Jiang, Z.P. SLAF-seq Uncovers the Genetic Diversity and Adaptation of Chinese Elm (Ulmus parvifolia) in Eastern China. Forests 2020, 11, 80. [Google Scholar] [CrossRef]
- Fang, H.; Liu, H.; Ma, R.; Liu, Y.; Li, J.; Yu, X.; Zhang, H.; Yang, Y.; Zhang, G. Genome-wide assessment of population structure and genetic diversity of Chinese Lou onion using specific length amplified fragment (SLAF) sequencing. PLoS ONE 2020, 15, e0231753. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Yan, H.; Liu, X.; Zang, W.; Zhang, A.; Zhou, S.; Huang, L.; Liu, J. Genome-wide association study of rust traits in orchardgrass using SLAF-seq technology. Hereditas 2017, 154, 5. [Google Scholar] [CrossRef][Green Version]
- Yang, J.; Zhang, C.; Zhao, N.; Zhang, L.; Hu, Z.; Chen, S.; Zhang, M. Chinese Root-type Mustard Provides Phylogenomic Insights into the Evolution of the Multi-use Diversified Allopolyploid Brassica juncea. Mol. Plant 2018, 11, 512–514. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tan, F.; Su, G.; Deng, S.; He, H.; Shi, S. Population genetic structure of three tree species in the mangrove genus Ceriops (Rhizophoraceae) from the Indo West Pacific. Genetica 2008, 133, 47–56. [Google Scholar] [CrossRef]
- Arnaud-Haond, S.; Teixeira, S.; Massa, S.I.; Billot, C.; Serråo, E.A. Genetic structure at range edge: Low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Mol. Ecol. 2010, 15, 3515–3525. [Google Scholar] [CrossRef] [PubMed]
- Tomimatsu, H.; Yamagishi, H.; Tanaka, I.; Sato, M.; Kondo, R.; Konno, Y. Consequences of forest fragmentation in an understory plant community: Extensive range expansion of native dwarf bamboo. Plant Species Biol. 2011, 26, 3–12. [Google Scholar] [CrossRef]
- Scholtz, R.; Polo, J.A.; Tanner, E.P.; Fuhlendorf, S.D. Grassland fragmentation and its influence on woody plant cover in the southern Great Plains, USA. Landsc. Ecol. 2018, 33, 1785–1797. [Google Scholar] [CrossRef]
- Kim, C.; Paterson, I.D.; Carla, L.; Hill, M.P. Expansive reed populations—Alien invasion or disturbed wetlands? Aob Plants 2018, 10, ply014. [Google Scholar]
- Hao, B.; Wu, H.; Shi, Q.; Liu, G.; Xing, W. Facilitation and competition among foundation species of submerged macrophytes threatened by severe eutrophication and implications for restoration. Ecol. Eng. 2013, 60, 76–80. [Google Scholar] [CrossRef]
- Valéry, L.; Radureau, A.; Lefeuvre, J.C. Spread of the native grass Elymus athericus in salt marshes of Mont-Saint-Michel bay as an unusual case of coastal eutrophication. J. Coast. Conserv. 2016, 21, 1–13. [Google Scholar] [CrossRef]
- Hangelbroek, H.H.; Ouborg, N.J.; Santamaría, L.; Schwenk, K. Clonal diversity and structure within a population of the pondweed Potamogeton pectinatus foraged by Bewick’s swans. Mol. Ecol. 2002, 11, 2137–2150. [Google Scholar] [CrossRef]
- Bona, A.; Kulesza, U.; Jadwiszczak, K.A. Clonal diversity, gene flow and seed production in endangered populations of Betula humilis Schrk. Tree Genet. Genomes 2019, 15, 1–12. [Google Scholar] [CrossRef]
- Mikael, H.; Richard, L. Seed dispersal and fine-scale genetic structuring in the asexual Nigritella miniata (Orchidaceae) in the Alps. Bot. J. Linn. Soc. 2019, 190, 83–100. [Google Scholar]
- Brzosko, E.; Wróblewska, A.; Ratkiewicz, M. Spatial genetic structure and clonal diversity of island populations of lady’s slipper (Cypripedium calceolus) from the Biebrza National Park (Northeast Poland). Mol. Ecol. 2002, 11, 2499–2509. [Google Scholar] [CrossRef] [PubMed]
- Torimaru, T.; Tomaru, N.; Nishimura, N.; Yamamoto, S. Clonal diversity and genetic differentiation in Ilex leucoclada M. patches in an old-growth beech forest. Mol. Ecol. 2010, 12, 809–818. [Google Scholar] [CrossRef]
- Jacquemyn, H.; Brys, R.; Honnay, O.; Hermy, M.; Ruiz, I.R. Local forest environment largely affects below-ground growth, clonal diversity and fine-scale spatial genetic structure in the temperate deciduous forest herb Paris quadrifolia. Mol. Ecol. 2005, 14, 4479–4488. [Google Scholar] [CrossRef]
- Tang, S.Q.; Yuan, L.; Geng, Y.P.; Zhang, G.R.; Li, W.; Yang, Z. Clonal and spatial genetic structure in natural populations of Luohanguo (Siraitia grosvenorii), an economic species endemic to South China, as revealed by RAPD markers. Biochem. Syst. Ecol. 2007, 35, 557–565. [Google Scholar] [CrossRef]
- Peng, Y.L.; Macek, P.; Macková, J.; Romoleroux, K.; Hensen, I. Clonal Diversity and Fine-scale Genetic Structure in a High Andean Treeline Population. Biotropica 2015, 47, 59–65. [Google Scholar] [CrossRef]
- Leger, E.A.; Espeland, E.K.; Merrill, K.R.; Meyer, S.E. Genetic variation and local adaptation at a cheatgrass (Bromus tectorum) invasion edge in western Nevada. Mol. Ecol. 2009, 18, 4366–4379. [Google Scholar] [CrossRef] [PubMed]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef]
- Wang, G.; Long, Y.; Thomma, B.; Wit, P.D.; Angenent, G.C.; Fiers, M. Functional Analyses of the CLAVATA2-Like Proteins and Their Domains That Contribute to CLAVATA2 Specificity. Plant Physiol. 2010, 152, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Ohe, M.; Scoccianti, V.; Bagni, N.; Tassoni, A.; Matsuzaki, S. Putative occurrence of lysine decarboxylase isoforms in soybean (Glycine max) seedlings. Amino Acids 2009, 36, 65–70. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).