Patterns of Fine-Scale Spatial Genetic Structure and Pollen Dispersal in Giant Sequoia (Sequoiadendron giganteum)
Abstract
1. Introduction
2. Materials and Methods
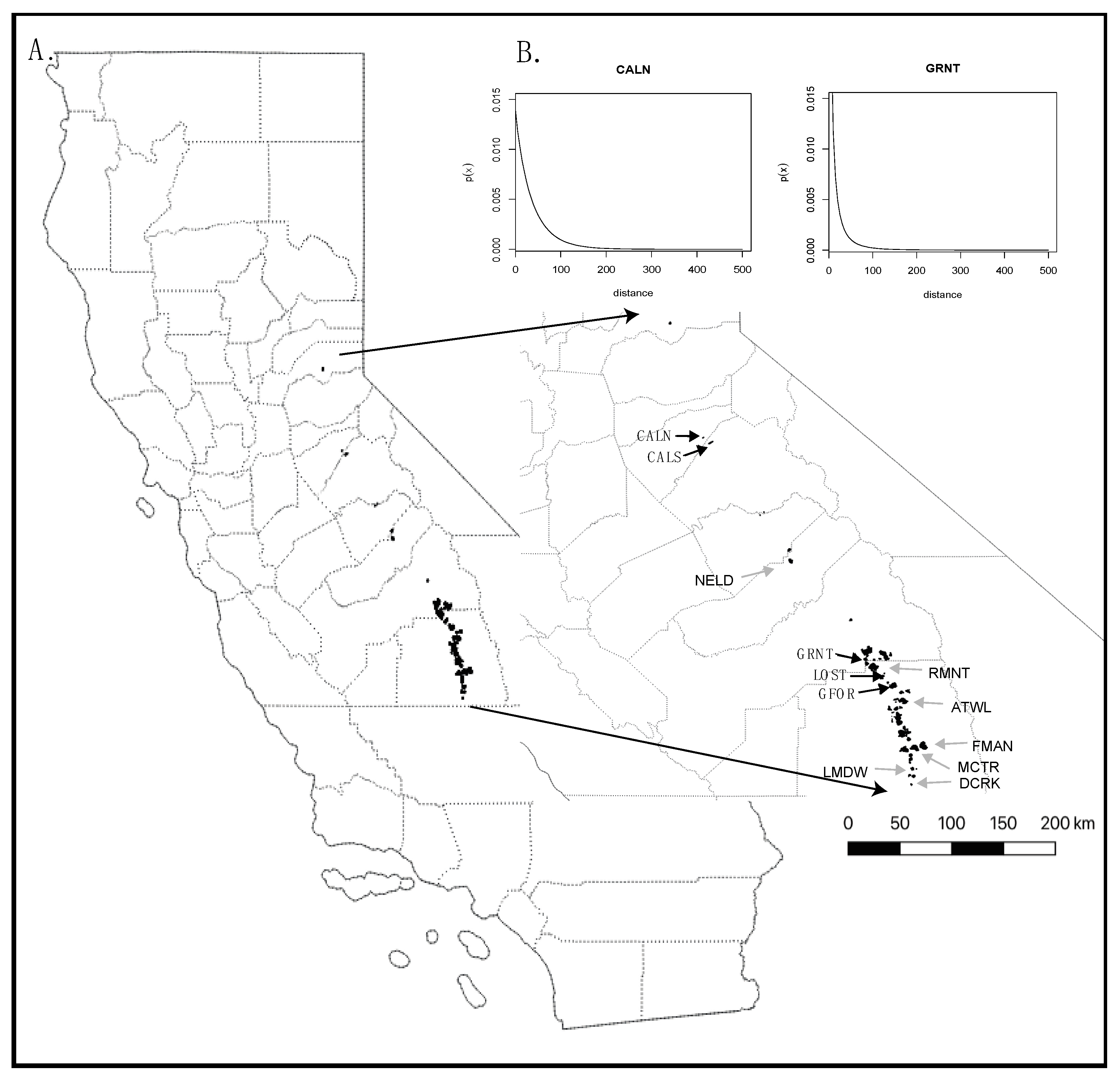
2.1. Sampling and Data Preparation
2.1.1. Fine-Scale Spatial Genetic Structure (FSGS)
2.1.2. Mating Parameters and Pollen Dispersal
2.1.3. Assigning Maternity
2.2. Data Analysis
2.2.1. Mating Parameters
2.2.2. Spatial Genetic Structure and Gene Dispersal
2.2.3. Pollen Dispersal Parameters
3. Results
3.1. Mating Parameters
3.2. Spatial Genetic Structure
3.3. Dispersal Dynamics
4. Discussion
4.1. Mating System and Pollen Pool Diversity
4.2. Evidence for Limited Dispersal
4.3. Evidence for Long-Distance Pollen Dispersal
4.4. Evidence for Demic Structure in Giant Sequoia Groves
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Seed Collection ID | Number of Seeds Genotyped |
| CALN 1 | 12 |
| CALN 2 | 22 |
| CALN 3 | 17 |
| CALN 4 | 12 |
| CALN 5 | 12 |
| CALN 6 | 12 |
| CALN 7 | 8 |
| CALN 8 | 22 |
| CALN 1 | 12 |
| CALS 1 | 29 |
| CALS 2 | 19 |
| CALS 3 | 16 |
| CALS 4 | 18 |
| CALS 5 | 21 |
| CALS 6 | 23 |
| CALS 7 | 15 |
| CALS 8 | 25 |
| CALS 9 | 25 |
| GRNT 1 | 13 |
| GRNT 2 | 29 |
| GRNT 3 | 27 |
| GRNT 4 | 20 |
| GRNT 5 | 20 |
| GRNT 6 | 31 |
| GRNT 7 | 18 |
| GRNT 8 | 17 |
| GRNT 9 | 11 |
| GRNT 10 | 13 |
| GRNT 11 | 13 |
| LOST 1 | 10 |
| LOST 2 | 23 |
| LOST 3 | 25 |
| LOST 4 | 24 |
| LOST 5 | 21 |
| LOST 6 | 26 |
| LOST 7 | 13 |
| LOST 8 | 10 |
| GFOR 1 | 15 |
| GFOR 2 | 20 |
| GFOR 3 | 20 |
| GFOR 4 | 9 |
| GFOR 5 | 30 |
| GFOR 6 | 22 |
| GFOR 7 | 28 |
| GFOR 8 | 21 |
| GFOR 9 | 29 |
| GFOR 10 | 20 |
| GFOR 11 | 35 |
| GFOR 12 | 18 |
| GFOR 13 | 13 |
| GFOR 14 | 34 |
| GFOR 15 | 28 |
| GFOR 16 | 28 |
| GFOR 17 | 30 |
| GFOR 18 | 10 |
Appendix B
| Population Code | Grove Size (ha) | Number of Spatial Groups | Distance Range within the First Distance Class (m) |
| CALN | 25 | 2 | 0–428 |
| CALS | 182 | 3 | 0–403 |
| NELD | 195 | 3 | 0–558 |
| GRNT | 163 | 3 | 0–293 |
| RMNT | 1466 | 6 | 0–415 |
| LOST | 21 | 2 | 0–138 |
| GFOR | 935 | 6 | 0–767 |
| ATWL | 542 | 5 | 0–236 |
| MCTR | 700 | 5 | 0–419 |
| FMAN | 580 | 5 | 0–456 |
| LMDW | 138 | 3 | 0–347 |
| DCRK | 21 | 2 | 0–269 |
References
- Nathan, R.; Schurr, F.M.; Spiegel, O.; Steinitz, O.; Trakhtenbrot, A.; Tsoar, A. Mechanisms of long-distance seed dispersal. Trends Ecol. Evol. 2008, 23, 638–647. [Google Scholar] [CrossRef]
- Kremer, A.; Ronce, O.; Robledo-Arnuncio, J.J.; Guillaume, F.; Bohrer, G.; Nathan, R.; Bridle, J.R.; Gomulkiewicz, R.; Klein, E.K.; Ritland, K.; et al. Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecol. Lett. 2012, 15, 378–392. [Google Scholar] [CrossRef]
- Ellstrand, N.C. Is gene flow the most important evolutionary force in plants? Am. J. Bot. 2014, 101, 737–753. [Google Scholar] [CrossRef]
- Robledo-Arnuncio, J.J.; Klein, E.K.; Muller-Landau, H.C.; Santamaria, L. Space, time and complexity in plant dispersal ecology. Mov. Ecol. 2014, 2, 16. [Google Scholar] [CrossRef]
- Vekemans, X.; Hardy, O.J. New insights from fine-scale spatial genetic structure analyses in plants populations. Mol. Ecol. 2004, 13, 921–935. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow and the Geographic Structure of Natural Populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Cain, M.L.; Milligan, B.G.; Strand, A.E. Long-distance seed dispersal in plant populations. Am. J. Bot. 2000, 87, 1217–1227. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Burke, J.M. The biological reality of species: Gene flow, selection, and collective evolution. Taxon 2001, 50, 47–67. [Google Scholar] [CrossRef]
- Heywood, J.S. Spatial analysis of genetic variation in plant populations. Annu. Rev. Ecol. Syst. 1991, 22, 335–355. [Google Scholar] [CrossRef]
- Wright, S. Isolation by distance. Genetics 1943, 28, 114–138. [Google Scholar]
- Ellstrand, N.C.; Elam, D.R. Population Genetic Consequences of Small Population Size: Implications for Plant Conservation. Annu. Rev. Ecol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]
- Krakowski, J.; Aitken, S.; El-Kassaby, Y. Inbreeding and conservation genetics in whitebark pine. Conserv. Genet. 2003, 4, 581–593. [Google Scholar] [CrossRef]
- Willi, Y.; Määttänen, K. The relative importance of factors determining genetic drift: Mating system, spatial genetic structure, habitat and census size in Arabidopsis lyrata. New Phytol. 2011, 189, 1200–1209. [Google Scholar] [CrossRef]
- Dow, B.D.; Ashley, M.V. Microsatellite analysis of seed dispersal and parentage of saplings in bur oak, Quercus Macrocarpa. Mol. Ecol. 1996, 5, 615–627. [Google Scholar] [CrossRef]
- Bacles, C.F.E.; Burczyk, J.; Lowe, A.J.; Ennos, R.A. Historical and contemporary mating patterns in remnant populations of the forest tree Fraxinus excelsior L. Evolution 2005, 59, 979–990. [Google Scholar] [CrossRef]
- De-Lucas, A.; Robledo-Arnuncio, J.; Hidalgo, E.; Gonzalez-Martinez, S.C. Mating system and pollen gene flow in Mediterranean maritime pine. Heredity 2008, 100, 390–399. [Google Scholar] [CrossRef]
- Chybicki, I.J.; Dzialuk, A. Bayesian approach reveals confounding effects of population size and seasonality on outcrossing rates in a fragmented subalpine conifer. Tree Genet. Genomes 2014, 10, 1723–1737. [Google Scholar] [CrossRef]
- O’Connell, L.M.; Mosseler, A.; Rajora, O.P. Extensive Long-Distance Pollen Dispersal in a Fragmented Landscape Maintains Genetic Diversity in White Spruce. J. Hered. 2007, 98, 640–645. [Google Scholar] [CrossRef]
- Colabella, F.; Gallo, L.A.; Moreno, A.C.; Marchelli, P. Extensive pollen flow in a natural fragmented population of patagonian cypress austrocedrus chilensis. Tree Genet. Genomes 2014, 10, 1519–1529. [Google Scholar] [CrossRef]
- Willard, D. A Guide to the Sierra Groves of California; Yosemite Conservancy: San Francisco, CA, USA, 2000. [Google Scholar]
- Hartesveldt, R.J.; Harvey, H.T.; Shellhammer, H.S.; Stecker, R.E. The Giant Sequoia of the Sierra Nevada; U.S. Department of the Interior, National Park Service: Washington, DC, USA, 1975.
- Rundel, P. Habitat Restriction in Giant Sequoia—Environmental Control of Grove Boundaries. Am. Midl. Nat. 1972, 87, 81–99. [Google Scholar] [CrossRef]
- Harvey, T.H.; Shellhammer, H.S.; Stecker, R.E. Giant Sequoia Ecology: Fire and Reproduction; Scientific Monograph Series No. 12; U.S. Department of the Interior, National Park Service: Washington, DC, USA, 1980.
- York, R.; Battles, J.; Heald, R. Edge effects in mixed conifer group selection openings: Tree height response to resource gradients. For. Ecol. Manag. 2003, 179, 107–121. [Google Scholar] [CrossRef]
- Shellhammer, H.S.; Shellhammer, T.H. Giant sequoia (Sequoiadendron giganteum Taxodiaceae) seedling survival and growth in the first four decades following managed fires. Madrono 2006, 53, 342–350. [Google Scholar] [CrossRef]
- York, R.; Thomas, Z.; Restaino, J. Influence of Ash Substrate Proximity on Growth and Survival of Planted Mixed-Conifer Seedlings. West. J. Appl. For. 2009, 24, 117–123. [Google Scholar] [CrossRef]
- Goss, M.; Swain, D.L.; Abatzoglou, J.T.; Sarhadi, A.; Kolden, C.A.; Williams, A.P.; Diffenbaugh, N.S. Climate change is increasing the likelihood of extreme autumn wildfire conditions across California. Environ. Res. Lett. 2020, 15, 094016. [Google Scholar] [CrossRef]
- Anderson, R.S. Modern pollen rain within and adjacent to two giant sequoia (Sequoiadendron giganteum) groves, Yosemite and Sequoia national parks, California. Can. J. For. Res. 1990, 20, 1289–1305. [Google Scholar] [CrossRef]
- DeSilva, R.; Dodd, R.S. Fragmented and isolated: Limited gene flow coupled with weak isolation by environment in the paleoendemic giant sequoia (Sequoiadendron giganteum). Am. J. Bot. 2020, 107, 45–55. [Google Scholar] [CrossRef]
- DeSilva, R.; Dodd, R.S. Development and characterization of microsatellite markers for giant sequoia, Sequoiadendron giganteum (Cupressaceae). Conserv. Genet. Resour. 2014, 6, 173–174. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- York, R.A.; Stephenson, N.L.; Meyer, M.; Hanna, S.; Moody, T.; Caprio, A.C.; Battles, J.J. A Natural Resource Condition Assessment for Sequoia and Kings Canyon National Parks: Appendix 11a—Giant Sequoias; Natural Resource Report NPS/SEKI/NRR—2013/665.11a; National Park Service: Fort Collins, CO, USA, 2013.
- Kalinowski, S.T.; Wagner, A.P.; Taper, M.L. ml-relate: A computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 2006, 6, 576–579. [Google Scholar] [CrossRef]
- Ritland, K. MLTR: Multilocus Mating System Program, Version 1.1; University of British Columbia: Vancouver, BC, Canada, 1996. [Google Scholar]
- Ritland, K. Extensions of models for the estimation of mating systems using n independent loci. Heredity 2002, 88, 221–228. [Google Scholar] [CrossRef]
- Smouse, P.E.; Dyer, R.J.; Westfall, R.D.; Sork, V.L. Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution 2001, 55, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Austerlitz, F.; Smouse, P.E. Two-generation analysis of pollen flow across a landscape; II. Relation between Φ(ft), pollen dispersal and interfemale distance. Genetics 2001, 157, 851–857. [Google Scholar]
- Loiselle, B.A.; Sork, V.L.; Nason, J.; Graham, C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 1995, 82, 1420–1425. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. Isolation by distance in a continuous population: Reconciliation between spatial autocorrelation and population genetics models. Heredity 1999, 83, 145–154. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. SPAGeDi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Fenster, C.B.; Vekemans, X.; Hardy, O.J. Quantifying gene flow from spatial genetic structure data in a metapopulation of Chamaecrista fasciculata (leguminosae). Evolution 2003, 57, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R. Effective population-size adult-population size ratios in wildlife—A review. Genet. Res. 1995, 66, 95–107. [Google Scholar] [CrossRef]
- Robledo-Arnuncio, J.J.; Austerlitz, F.; Smouse, P.E. POLDISP: A software package for indirect estimation of contemporary pollen dispersal. Mol. Ecol. Notes 2007, 7, 763–766. [Google Scholar] [CrossRef]
- Robledo-Arnuncio, J.J.; Austerlitz, F.; Smouse, P.E. A new method of estimating the pollen dispersal curve independently of effective density. Genetics 2006, 173, 1033–1045. [Google Scholar] [CrossRef]
- Austerlitz, F.; Dick, C.W.; Dutech, C.; Klein, E.K.; Oudu-Muratorio, S.; Smouse, P.; Sork, V.L. Using genetic markers to estimate the pollen dispersal curve. Mol. Ecol. 2004, 13, 937–954. [Google Scholar] [CrossRef]
- Marchelli, P.; Smouse, P.E.; Gallo, L.A. Short-distance pollen dispersal for an outcrossed, wind-pollinated southern beech (Nothofagus nervosa (Phil.) Dim. et Mil.). Tree Genet. Genomes 2012, 8, 1123–1134. [Google Scholar] [CrossRef]
- Moracho, E.; Moreno, G.; Jordano, P.; Hampe, A. Unusually limited pollen dispersal and connectivity of Pedunculate oak (Quercus robur) refugial populations at the species’ southern range margin. Mol. Ecol. 2016, 25, 3319–3331. [Google Scholar] [CrossRef] [PubMed]
- Burczyk, J.; Sandurska, E.; Lewandowski, A. Patterns of Effective Pollen Dispersal in Larch: Linking Levels of Background Pollination with Pollen Dispersal Kernels. Forests 2019, 10, 1139. [Google Scholar] [CrossRef]
- Burczyk, J.; Adams, W.T.; Shimizu, J.Y. Mating patterns and pollen dispersal in a natural knobcone pine (Pinus attenuata Lemmon.) stand. Heredity 1996, 77, 251–260. [Google Scholar] [CrossRef]
- Wasieliwska, M.; LKlemm, M.; Burczyk, J. Genetic diversity and mating system of Scots pine plus trees. Dendrobiology 2005, 53, 57–62. [Google Scholar]
- Sork, V.L.; Dyer, R.J.; Davis, F.W.; Smouse, P.E. Mating patterns in a savanna population of valley oak (Quercus lobata Neé). In Proceedings of the Fifth Symposium on Oak Woodlands: Oaks in California’s Changing Landscape, San Diego, CA, USA, 22–25 October 2001; Pacific SW Research Station, US Forest Service, USDA: San Diego, CA, USA, 2002; pp. 427–439. [Google Scholar]
- O’Connell, L.M.; Mosseler, A.; Rajora, O.P. Impacts of forest fragmentation on the mating system and genetic diversity of white spruce (Picea glauca) at the landscape level. Heredity 2006, 97, 418–426. [Google Scholar] [CrossRef]
- Bower, A.D.; Aitken, S.N. Mating system and inbreeding depression in whitebark pine (Pinus albicaulis Engelm.). Tree Genet. Genomes 2007, 3, 379–388. [Google Scholar] [CrossRef]
- Ledig, F.T.; Hodgskiss, P.D.; Johnson, D.R. Genic diversity, genetic structure, and mating system of Brewer spruce (Pinaceae), a relict of the Arcto-Tertiary forest. Am. J. Bot. 2005, 92, 1975–1986. [Google Scholar] [CrossRef]
- Mantovani, A.; Morellato, L.P.C.; Dos Reis, M.S. Internal Genetic Structure and Outcrossing Rate in a Natural Population of Araucaria angustifolia (Bert.) O. Kuntze. J. Hered. 2006, 97, 466–472. [Google Scholar] [CrossRef]
- Rajora, O.P.; Mosseler, A.; Major, J.E. Mating system and reproductive fitness traits of eastern white pine (Pinus strobus) in large, central versus small, isolated, marginal populations. Can. J. Bot. 2002, 80, 1173–1184. [Google Scholar] [CrossRef]
- El-Kassaby, Y.A.; Jaquish, B. Population density and mating pattern in western larch. J. Hered. 1996, 87, 438–443. [Google Scholar] [CrossRef][Green Version]
- Austerlitz, F.; Smouse, P.E. Two-generation analysis of pollen flow across a landscape. III. Impact of within-population structure. Genet. Res. 2001, 78, 271–278. [Google Scholar] [CrossRef] [PubMed]
- de-Lucas, A.I.; Gonzalez-Martinez, S.C.; Vendramin, G.G.; Hidalgo, E.; Heuertz, M. Spatial genetic structure in continuous and fragmented populations of Pinus pinaster Aiton. Mol. Ecol. 2009, 18, 4564–4576. [Google Scholar] [CrossRef]
- Aleksic, J.M.; Piotti, A.; Geburek, T.; Vendramin, G.G. Exploring and conserving a “microcosm”: Whole-population genetic characterization within a refugial area of the endemic, relict conifer Picea omorika. Conserv. Genet. 2017, 18, 777–788. [Google Scholar] [CrossRef]
- Eliades, N.H.; Fady, B.; Gailing, O.; Leinemann, L.; Finkeldey, R. Significant patterns of fine-scale spatial genetic structure in a narrow endemic wind-dispersed tree species, Cedrus brevifolia Henry. Tree Genet. Genomes 2018, 14, 15. [Google Scholar] [CrossRef]
- Mosca, E.; Di Pierro, E.A.; Budde, K.B.; Neale, D.B.; González-Martínez, S.C. Environmental effects on fine-scale spatial genetic structure in four Alpine keystone forest tree species. Mol. Ecol. 2018, 27, 647–658. [Google Scholar] [CrossRef]
- Gapare, W.J.; Aitken, S.N. Strong spatial genetic structure in peripheral but not core populations of Sitka spruce [Picea sitchensis (Bong.) Carr.]. Mol. Ecol. 2005, 14, 2659–2667. [Google Scholar] [CrossRef]
- Pluess, A.R.; Sork, V.L.; Dolan, B.; Davis, F.W.; Grivet, D.; Merg, K.; Papp, J.; Smouse, P.E. Short distance pollen movement in a wind-pollinated tree, Quercus lobata (Fagaceae). For. Ecol. Manag. 2009, 258, 735–744. [Google Scholar] [CrossRef]
- Crawford, T.J. The estimation of neighborhood parameters for plant populations. Heredity 1984, 52, 273–283. [Google Scholar] [CrossRef]
- Millerón, M.; López de Heredia, U.; Lorenzo, Z.; Perea, R.; Dounavi, A.; Alonso, J.; Gil, L.; Nanos, N. Effect of canopy closure on pollen dispersal in a wind-pollinated species (Fagus sylvatica L.). Plant Ecol. 2012, 213, 1715–1728. [Google Scholar]
- Latta, R.G.; Linhart, Y.B.; Fleck, D.; Elliot, M. Direct and indirect estimates of seed versus pollen movement within a population of Ponderosa pine. Evolution 1998, 52, 61–67. [Google Scholar] [CrossRef]
- Schuster, W.S.F.; Mitton, J.B. Paternity and gene dispersal in limber pine (Pinus flexilis James). Heredity 2000, 84, 348–361. [Google Scholar] [CrossRef]
- Heuertz, M.; Vekemans, X.; Hausman, J.F.; Palada, M.; Hardy, O.J. Estimating seed versus pollen dispersal from spatial genetic structure in the common ash. Mol. Ecol. 2003, 12, 2483–2495. [Google Scholar] [CrossRef]
- Chybicki, I.J.; Burczyk, J. Realized gene flow within mixed stands of Quercus robur L. and Q. petraea (Matt.) L. revealed at the stage of naturally established seedling. Mol. Ecol. 2010, 19, 2137–2151. [Google Scholar] [CrossRef]
- Sork, V.; Smouse, P.; Grivet, D.; Scofield, D. Impact of asymmetric male and female gamete dispersal on allelic diversity and spatial genetic structure in valley oak (Quercus lobata Nee). Evol. Ecol. 2015, 29, 927–945. [Google Scholar] [CrossRef]
- Ting, W.S. The Saccate Pollen Grains of Pinaceae Mainly of California. Grana 1965, 6, 270–289. [Google Scholar] [CrossRef]
- Bortenschlager, S. Aspects of pollen morphology in the cupressaceae. Grana 1990, 29, 129–138. [Google Scholar] [CrossRef]
- Schwendemann, A.B.; Wang, G.; Mertz, M.L.; McWilliams, R.T.; Thatcher, S.L.; Osborn, J.M. Aerodynamics of saccate pollen and its implications for wind pollination. Am. J. Bot. 2007, 94, 1371–1381. [Google Scholar] [CrossRef]
- Smouse, P.E.; Sork, V.L. Measuring pollen flow in forest trees: An exposition of alternative approaches. For. Ecol. Manag. 2004, 197, 21–38. [Google Scholar] [CrossRef]
- Robledo-Arnuncio, J.J. Wind pollination over mesoscale distances: An investigation with Scots pine. New Phytol. 2011, 190, 222–233. [Google Scholar] [CrossRef]
- Robledo-Arnuncio, J.J.; Gil, L. Patterns of pollen dispersal in a small population of Pinus sylvestris L. revealed by total-exclusion paternity analysis. Heredity 2005, 94, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Young, A.; Boyle, T.; Brown, T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 1996, 11, 413–418. [Google Scholar] [CrossRef]
- Whitlock, M.C.; Barton, N.H. The Effective Size of a Subdivided Population. Genetics 1997, 146, 427–441. [Google Scholar] [PubMed]
- Frankham, R. Inbreeding and extinction: A threshold effect. Conserv. Biol. 1995, 9, 792–799. [Google Scholar] [CrossRef]
| Population (ha) | Seed Collection Sites | Average Maternal Family Size | Census Density (m2) | Multi-Locus Outcrossing Rate: tm (SD) | Single-Locus Outcrossing Rate: ts (SD) | Bi-Parental Inbreeding: tm − ts | Nep |
|---|---|---|---|---|---|---|---|
| CALN (25) | 7 | 10 | 0.0005 | 0.92 (0.11) | 0.81 (0.09) | 0.117 (0.09) | 8.5 |
| CALS (182) | 9 | 11 | 0.0006 | 0.94 (0.12) | 0.76 (0.04) | 0.178 (0.11) | 8.4 |
| GRNT (163) | 11 | 14 | 0.0010 | 0.84 (0.03) | 0.66 (0.03) | 0.180 (0.03) | 4.7 |
| LOST (21) | 8 | 13 | 0.0018 | 0.88 (0.04) | 0.77 (0.04) | 0.112 (0.04) | 7.8 |
| GFOR (935) | 18 | 12 | 0.0010 | 0.95 (0.12) | 0.76 (0.02) | 0.189 (0.11) | 7.0 |
| AVERAGE | - | - | - | 0.91 | 0.75 | 0.155 | 7.5 |
| Population (ha) | Sp-Statistic | Sigma*2 (m) |
|---|---|---|
| CALN (25) | 0.0354 * | 252.6 |
| CALS (182) | 0.0444 ** | 374.0 |
| NELD (195) | 0.0417 ** | 153.6 |
| GRNT (163) | 0.0388 ** | _ |
| RMNT (1466) | 0.0235 * | 120.4 |
| LOST (21) | 0.0339 NS | _ |
| GFOR (935) | 0.0238 ** | 185.6 |
| ATWL (542) | 0.0292 NS | _ |
| MCTR (700) | 0.0414 ** | 264.2 |
| FMAN (580) | 0.0397 ** | 159.0 |
| LMDW (138) | 0.0308 * | 260.2 |
| DCRK (21) | 0.0303 * | 236.0 |
| Population | Scale (a) | Shape (b) | Average Distance, d (m) |
|---|---|---|---|
| (Ne/N = 0.5) | |||
| CALN | 34.62 | 0.93 | 80.7 |
| CALS | 98.54 | 25.94 | 64.6 |
| GRNT | 0.55 | 0.37 | 74.2 |
| LOST | 34.62 | 0.93 | 80.7 |
| GFOR | 0.004 | 0.22 | 203.0 |
| (Ne/N = 0.1) | |||
| CALN | 215.96 | 6.84 | 141.9 |
| CALS | 82.85 | 1.05 | 152.5 |
| GRNT | 4.75 | 0.47 | 129.0 |
| LOST | 1.05 | 0.35 | 213.5 |
| GFOR | 0.37 | 0.31 | 252.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeSilva, R.; Dodd, R.S. Patterns of Fine-Scale Spatial Genetic Structure and Pollen Dispersal in Giant Sequoia (Sequoiadendron giganteum). Forests 2021, 12, 61. https://doi.org/10.3390/f12010061
DeSilva R, Dodd RS. Patterns of Fine-Scale Spatial Genetic Structure and Pollen Dispersal in Giant Sequoia (Sequoiadendron giganteum). Forests. 2021; 12(1):61. https://doi.org/10.3390/f12010061
Chicago/Turabian StyleDeSilva, Rainbow, and Richard S. Dodd. 2021. "Patterns of Fine-Scale Spatial Genetic Structure and Pollen Dispersal in Giant Sequoia (Sequoiadendron giganteum)" Forests 12, no. 1: 61. https://doi.org/10.3390/f12010061
APA StyleDeSilva, R., & Dodd, R. S. (2021). Patterns of Fine-Scale Spatial Genetic Structure and Pollen Dispersal in Giant Sequoia (Sequoiadendron giganteum). Forests, 12(1), 61. https://doi.org/10.3390/f12010061






