Detection of Standing Deadwood from Aerial Imagery Products: Two Methods for Addressing the Bare Ground Misclassification Issue
Abstract
:1. Introduction
2. Materials and Methods
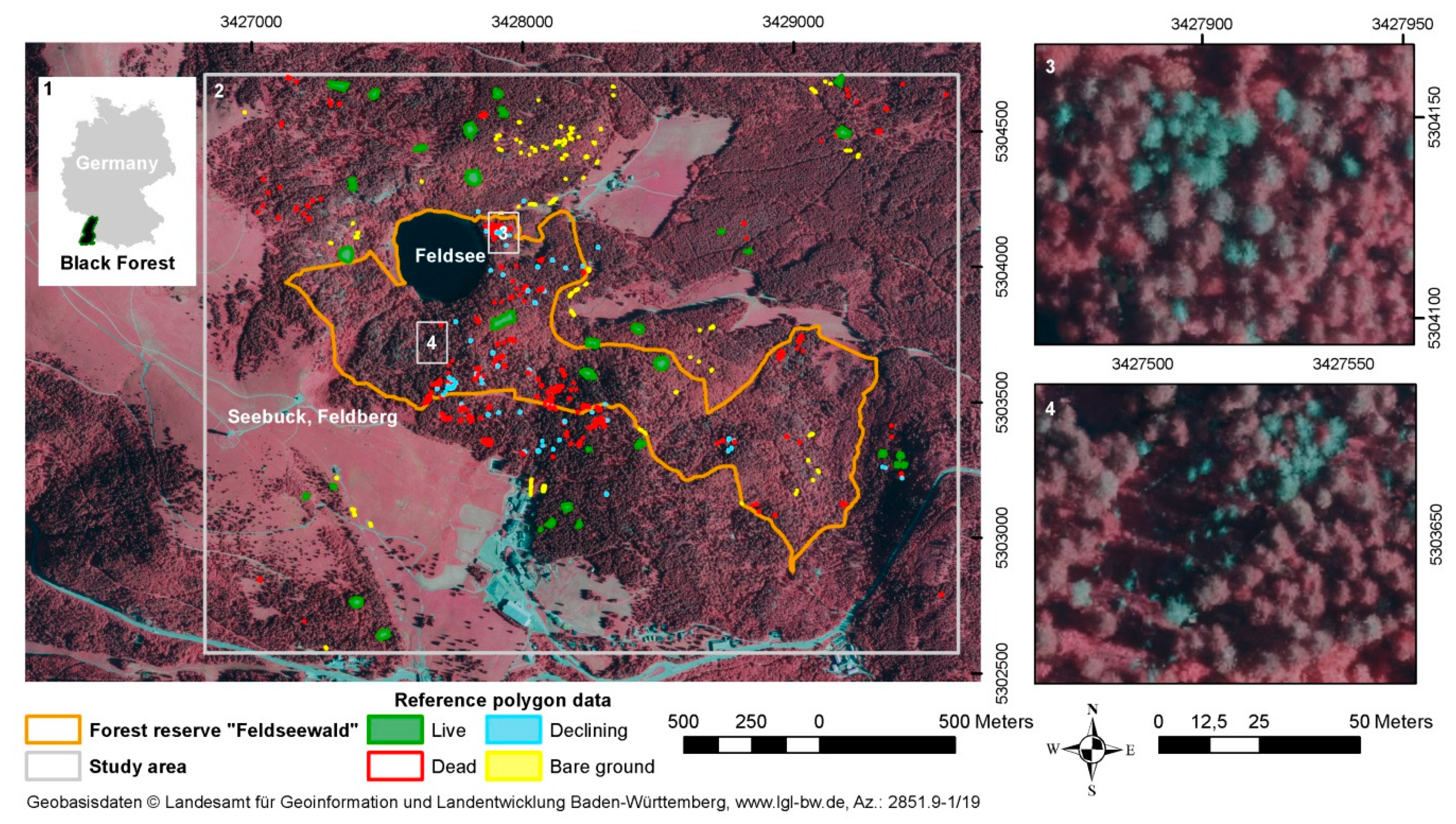
2.1. Study Site
2.2. Remote Sensing and GIS Data
2.3. Standing Deadwood Definition and Model Classes
2.4. Reference Polygons for Model Calibration
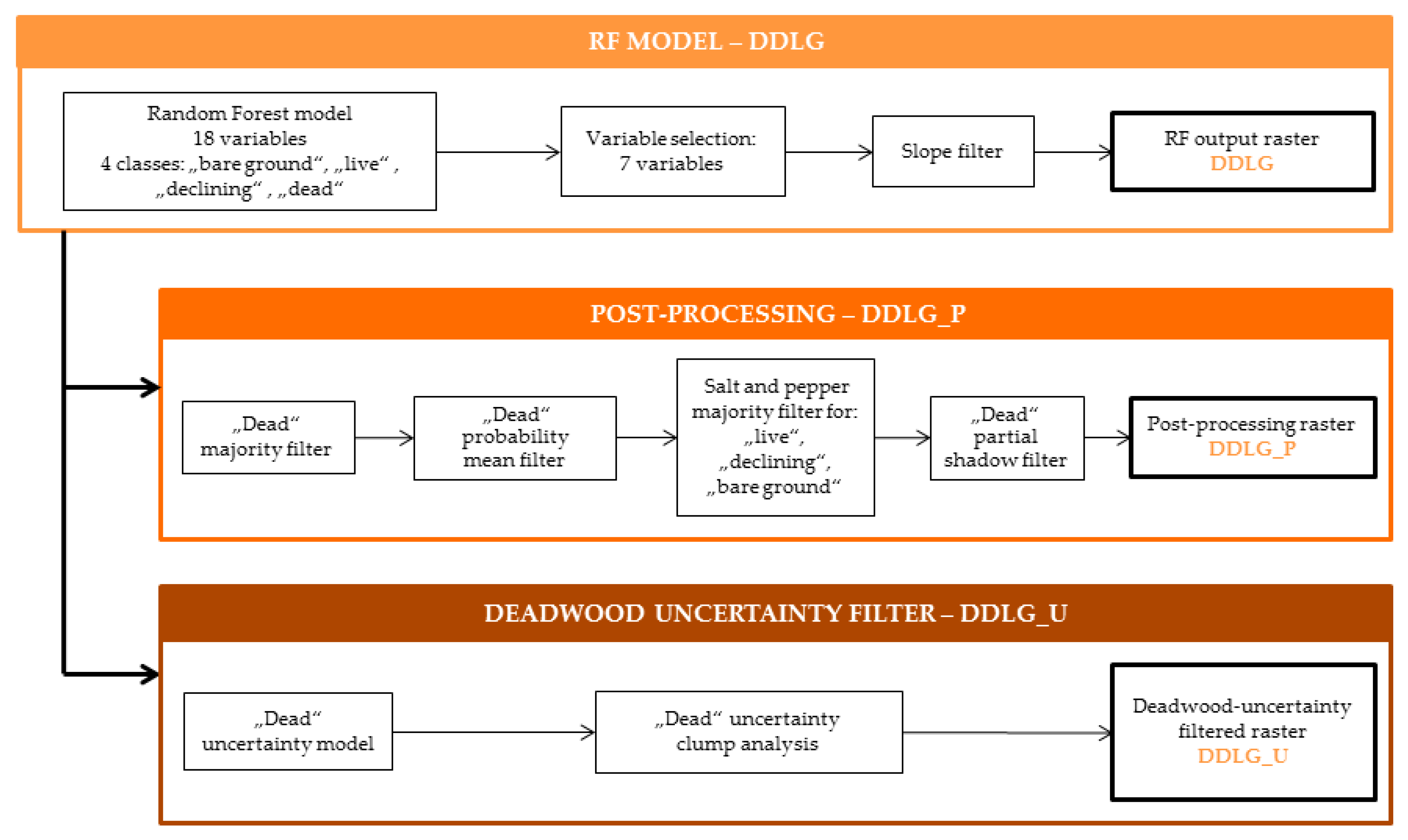
2.5. Deadwood Detection Method
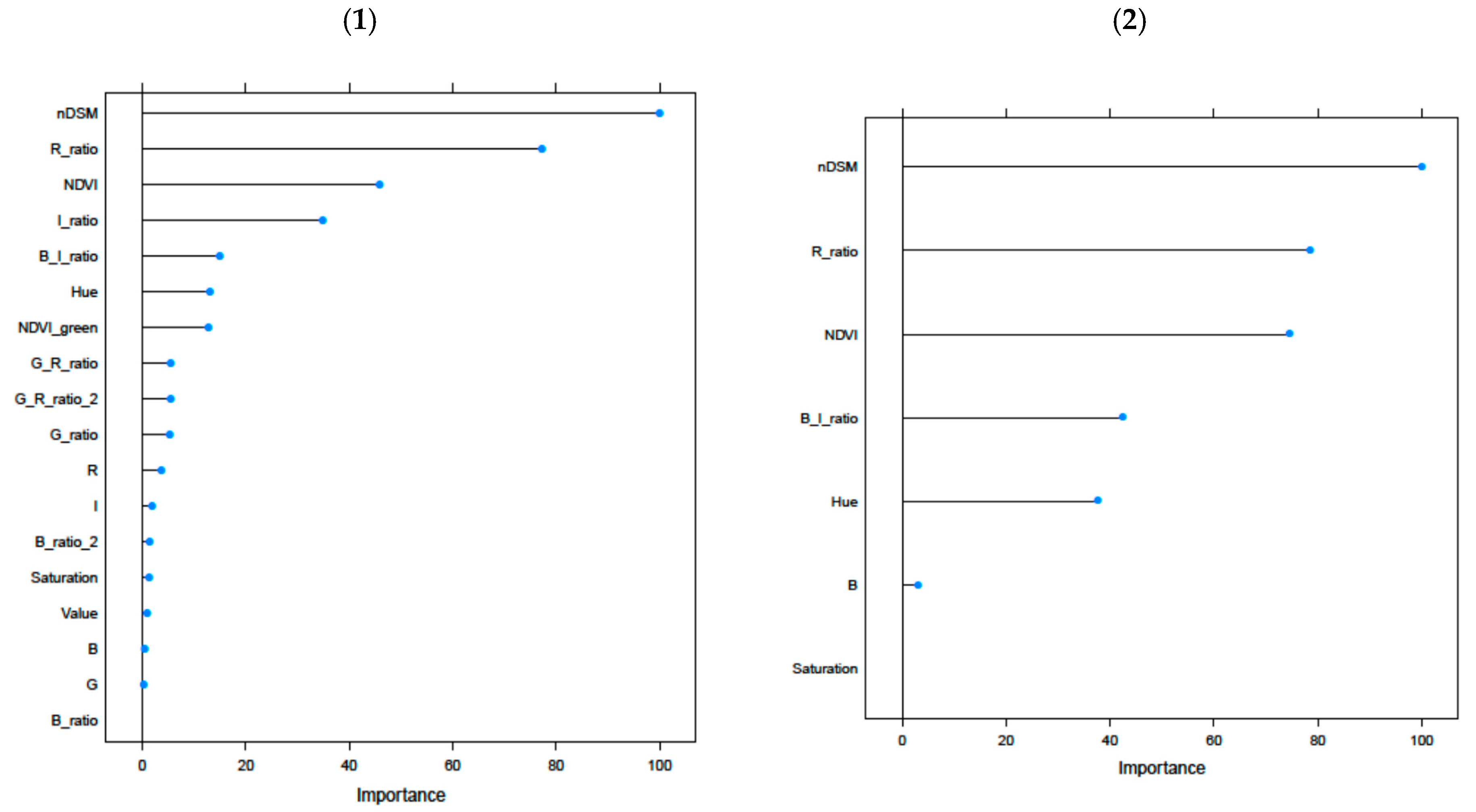
2.5.1. Random Forest Model (DDLG)
2.5.2. Post-Processing of RF Results (DDLG_P)
2.5.3. Deadwood Uncertainty Filter (DDLG_U)
2.6. Model Validation
2.6.1. Visual Assessment
2.6.2. “Pure Classes” Validation
2.6.3. Pixel-Based Validation Based on a Stratified Random Sample
2.6.4. Polygon-Based Deadwood Validation
3. Results
3.1. Random Forest Model and Pure Classes’ Validation
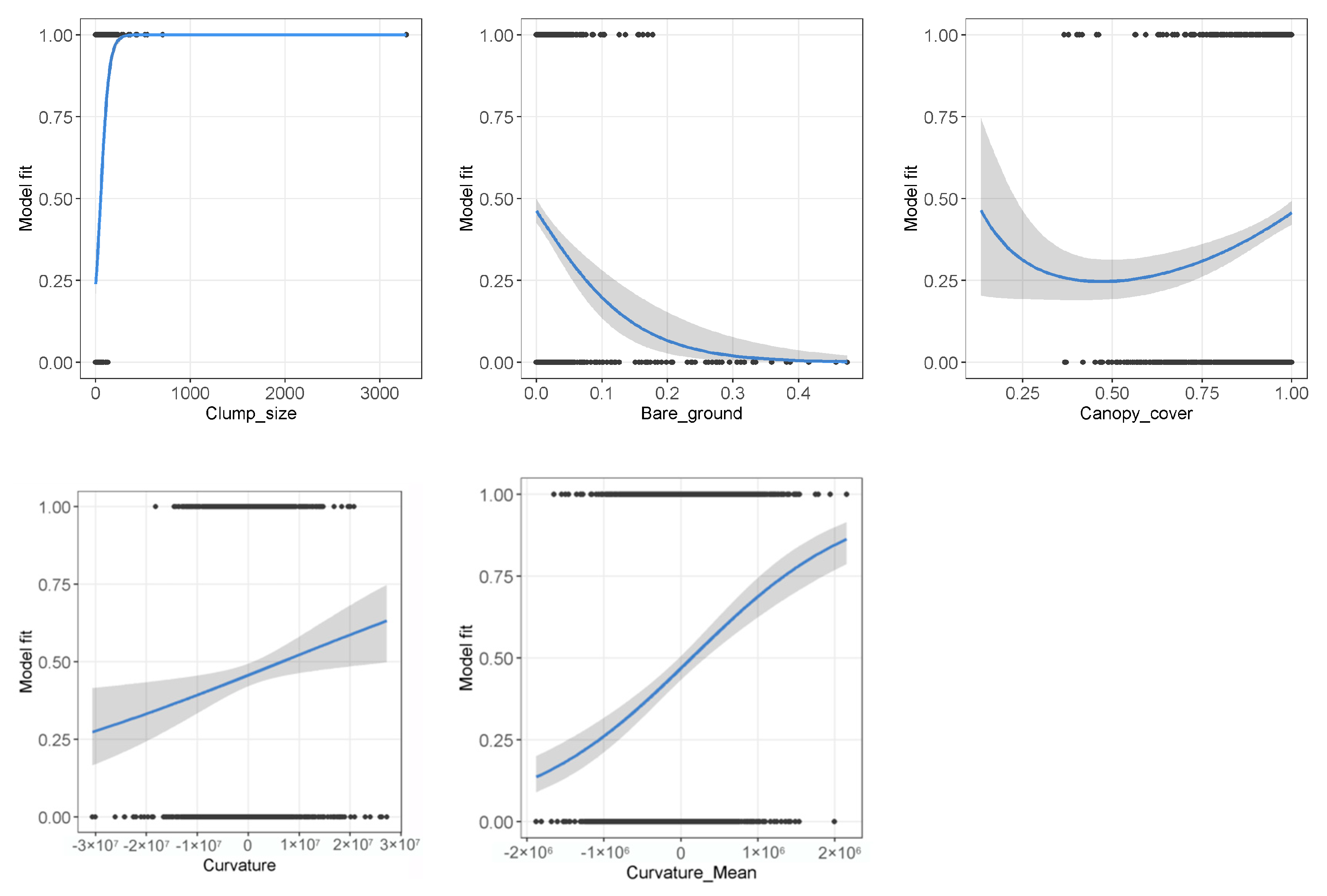
3.2. Uncertainty Model
3.3. Classification Results
3.4. Model Validation
3.4.1. Visual Assessment
3.4.2. Pixel-Based Validation
3.4.3. Polygon Based Deadwood Validation
4. Discussion
4.1. Deadwood Detection
4.2. Bare Ground Issue
4.3. Canopy Height Information
4.4. Deadwood Detection Algorithms
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| DDLG | Reference | Predicted Total | User’s Accuracy | |||||
|---|---|---|---|---|---|---|---|---|
| Bare Ground | Live | Declining | Dead | |||||
| 18 variables | Predicted | Bare ground | 1971 | 1 | 8 | 14 | 1994 | 0.99 |
| Live | 1 | 1940 | 112 | 4 | 2057 | 0.94 | ||
| Declining | 10 | 59 | 1792 | 145 | 2006 | 0.89 | ||
| Dead | 18 | 0 | 88 | 1837 | 1943 | 0.95 | ||
| Reference total | 2000 | 2000 | 2000 | 1986 | 7540 | |||
| Producer’s and overall accuracy | 0.99 | 0.97 | 0.90 | 0.92 | 0.94 | |||
| 7 variables | Predicted | Bare ground | 1989 | 0 | 12 | 22 | 2023 | 0.98 |
| Live | 0 | 1930 | 104 | 4 | 2038 | 0.95 | ||
| Declining | 4 | 66 | 1812 | 131 | 2013 | 0.90 | ||
| Dead | 7 | 4 | 72 | 1843 | 1926 | 0.96 | ||
| Reference total | 2000 | 2000 | 2000 | 2000 | 7574 | |||
| Producer’s and overall accuracy | 0.99 | 0.97 | 0.91 | 0.92 | 0.95 | |||
| PRES_Mean | PRES_SD | ABS_Mean | ABS_SD | p_l | AIC_l | p_q | AIC_q | |
|---|---|---|---|---|---|---|---|---|
| Clump_size | 634.17 | 1121.12 | 27.81 | 33.21 | 0.00 | 1791.67 | 0.00 | 1792.66 |
| Bare_ground | 0.01 | 0.02 | 0.02 | 0.06 | 0.00 | 2680.88 | 0.01 | 2678.66 |
| Canopy_cover | 0.97 | 0.08 | 0.91 | 0.13 | 0.00 | 2633.00 | 0.00 | 2601.77 |
| Curvature | 504,964.40 | 5,138,583.17 | −510,916.80 | 6,535,890.74 | 0.00 | 2761.7 | NA | NA |
| Curvature_mean | 101,697.81 | 546,521.45 | −169,517.63 | 520,512.28 | 0.00 | 2650.9 | NA | NA |
| Mean_Eucklidean_ Distance | 4945.16 | 2088.62 | 5078.54 | 3115.96 | 0.26 | 2775.3 | NA | NA |
| Reference | Total Predicted | User’s Accuracy | ||||||
|---|---|---|---|---|---|---|---|---|
| Bare Ground | Live | Declining | Dead | |||||
| DDLG | Predicted | Bare ground | 427 | 4 | 26 | 64 | 521 | 0.82 |
| Live | 8 | 590 | 163 | 3 | 764 | 0.77 | ||
| Declining | 32 | 148 | 423 | 30 | 633 | 0.67 | ||
| Dead | 283 | 7 | 138 | 653 | 1081 | 0.60 | ||
| Total reference | 750 | 749 | 750 | 750 | 2093 | |||
| Producer’s and overall accuracy | 0.57 | 0.79 | 0.56 | 0.87 | 0.70 | |||
| DDLG_P | Predicted | Bare ground | 492 | 7 | 51 | 110 | 660 | 0.75 |
| Live | 29 | 595 | 186 | 7 | 817 | 0.73 | ||
| Declining | 71 | 146 | 410 | 42 | 669 | 0.61 | ||
| Dead | 158 | 2 | 103 | 591 | 854 | 0.69 | ||
| Total reference | 750 | 750 | 750 | 750 | 2088 | |||
| Producer’s and overall accuracy | 0.66 | 0.79 | 0.55 | 0.79 | 0.70 | |||
| DDLG_U | Predicted | Bare ground | 612 | 8 | 67 | 119 | 806 | 0.76 |
| Live | 3 | 581 | 167 | 3 | 754 | 0.77 | ||
| Declining | 30 | 155 | 421 | 28 | 634 | 0.66 | ||
| Dead | 105 | 6 | 95 | 600 | 806 | 0.74 | ||
| Total reference | 750 | 750 | 750 | 750 | 2214 | |||
| Producer’s and overall accuracy | 0.82 | 0.77 | 0.56 | 0.80 | 0.74 | |||
| Reference Polygons | DDLG | DDLG_P | DDLG_U | |
|---|---|---|---|---|
| N | 315 | 323 | 238 | 285 |
| AREA SUM (m2) | 4295.7 | 6034.8 | 6024.5 | 6013.5 |
| % of reference polygons | 100% | 145% | 145% | 144% |
| AREA MEAN (m2) | 13.6 | 18.7 | 25.3 | 21.1 |
| AREA MEDIAN (m2) | 11.0 | 5.2 | 11.0 | 6.5 |
| AREA MAX (m2) | 65.9 | 813.8 | 811.0 | 813.8 |
| AREA SD (m2) | 12.1 | 56.6 | 64.6 | 59.8 |
References
- Thorn, S.; Müller, J.; Leverkus, A.B. Preventing European forest diebacks. Science 2019, 365. [Google Scholar] [CrossRef]
- Hahn, K.; Christensen, M. Dead Wood in European Forest Reserves—A reference for Forest Management. In EFI Proceedings No. 51. Monitoring and Indicators of Forest Biodiversity in Europe—From Ideas to Operationality; European Forest Institute: Joensuu, Finland, 2004; pp. 181–191. [Google Scholar]
- Schuck, A.; Meyer, P.; Menke, N.; Lier, M.; Lindner, M. Forest biodiversity indicator: Dead wood-a proposed approach towards operationalising the MCPFE indicator. In EFI Proceedings 51. Monitoring and Indicators of Forest Biodiversity in Europe—From Ideas to Operationality; Marchetti, M., Ed.; EFI: Joensuu, Finland, 2004; pp. 49–77. [Google Scholar]
- Paillet, Y.; Berges, L.; Hjalten, J.; Odor, P.; Avon, C.; Bernhardt-Romermann, M.; Bijlsma, R.J.; De Bruyn, L.; Fuhr, M.; Grandin, U.; et al. Biodiversity differences between managed and unmanaged forests: Meta-analysis of species richness in Europe. Conserv. Biol. 2010, 24, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Bußler, H.; Bense, U.; Brustel, H.; Flechtner, G.; Fowles, A.; Kahlen, M.; Möller, G.; Mühle, H.; Schmidl, J.; et al. Urwald relict species—Saproxylic beetles indicating structural qualities and habitat tradition. Wald. Online 2005, 2, 106–113. [Google Scholar]
- Seibold, S.; Brandl, R.; Buse, J.; Hothorn, T.; Schmidl, J.; Thorn, S.; Müller, J. Association of extinction risk of saproxylic beetles with ecological degradation of forests in Europe. Conserv. Biol. 2014, 29, 382–390. [Google Scholar] [CrossRef]
- Pechacek, P.; Krištín, A. Comparative diets of adult and young Threetoed Woodpeckers in a European alpine forest community. J. Wildl. Manag. 2004, 68, 683–693. [Google Scholar] [CrossRef]
- Kortmann, M.; Hurst, J.; Brinkmann, R.; Heurich, M.; Silveyra González, R.; Müller, J.; Thorn, S. Beauty and the beast: How a bat utilizes forests shaped by outbreaks of an insect pest. Anim. Conserv. 2018, 21, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Olchowik, J.; Hilszczanska, D.; Bzdyk, R.; Studnicki, M.; Malewski, T.; Borowski, Z. Effect of Deadwood on Ectomycorrhizal Colonisation of Old-Growth Oak Forests. Forests 2019, 10, 480. [Google Scholar] [CrossRef] [Green Version]
- Baldrian, P.; Zrůstová, P.; Tláskal, V.; Davidová, A.; Merhautová, V.; Vrška, T. Fungi associated with decomposing deadwood in a natural beech-dominated forest. Fungal Ecol. 2016, 23, 109–122. [Google Scholar] [CrossRef]
- Bader, P.; Jansson, S.; Jonsson, B.G. Wood-inhabiting fungi and substratum decline in selectively logged boreal spruce forests. Biol. Conserv. 1995, 72, 355–362. [Google Scholar] [CrossRef]
- Stighäll, K.; Roberge, J.-M.; Andersson, K.; Angelstam, P. Usefulness of biophysical proxy data for modelling habitat of an endangered forest species: The white-backed woodpecker Dendrocopos leucotos. Scand. J. For. Res. 2011, 26, 576–585. [Google Scholar] [CrossRef]
- Braunisch, V. Spacially Explicit Species-Habitat Models for Large-Scale Conservation Planning. Modelling Habitat Potential and Habitat Connectivity for Capercaillie (Tetrao urogallus). Ph.D. Thesis, Albert-Ludwigs-Universität, Freiburg im Breisgau, Germany, 2008. [Google Scholar]
- Kortmann, M.; Heurich, M.; Latifi, H.; Rösner, S.; Seidl, R.; Müller, J.; Thorn, S. Forest structure following natural disturbances and early succession provides habitat for two avian flagship species, capercaillie (Tetrao urogallus) and hazel grouse (Tetrastes bonasia). Biol. Conserv. 2018, 226. [Google Scholar] [CrossRef]
- Bouvet, A.; Paillet, Y.; Archaux, F.; Tillon, L.; Denis, P.; Gilg, O.; Gosselin, F. Effects of forest structure, management and landscape on bird and bat communities. Environ. Conserv. 2016, 43, 148–160. [Google Scholar] [CrossRef]
- Zielewska-Büttner, K.; Heurich, M.; Müller, J.; Braunisch, V. Remotely Sensed Single Tree Data Enable the Determination of Habitat Thresholds for the Three-Toed Woodpecker (Picoides tridactylus). Remote Sens. 2018, 10, 1972. [Google Scholar] [CrossRef] [Green Version]
- Balasso, M. Ecological Requirements of the Threetoed woodpecker (Picoides tridactylus L.) in Boreal Forests of Northern Sweden. Master’s Thesis, Swedish University of Agricultural Sciences, Umeå, Sweden, 2016. Available online: https://stud.epsilon.slu.se/8777/7/balasso_m_160204.pdf (accessed on 24 July 2020).
- Ackermann, J.; Adler, P.; Bauerhansl, C.; Brockamp, U.; Engels, F.; Franken, F.; Ginzler, C.; Gross, C.-P.; Hoffmann, K.; Jütte, K.; et al. Das digitale Luftbild. Ein Praxisleitfaden für Anwender im Forst- und Umweltbereich; Luftbildinterpreten, A.F., Ed.; Universitätsverlag Göttingen: Göttingen, Germany, 2012; Volume 7, p. 79. [Google Scholar]
- AFL. Luftbildinterpretationsschlüssel—Bestimmungsschlüssel für die Beschreibung von strukturreichen Waldbeständen im Color-Infrarot-Luftbild; Troyke, A., Habermann, R., Wolff, B., Gärtner, M., Engels, F., Brockamp, U., Hoffmann, K., Scherrer, H.-U., Kenneweg, H., Kleinschmit, B., et al., Eds.; Landesforstpräsidium (LFP) Freistaat Sachsen: Pirna, Germany, 2003; p. 48. [Google Scholar]
- Wulder, M.A.; Dymond, C.C.; White, J.C.; Leckie, D.G.; Carroll, A.L. Surveying mountain pine beetle damage of forests: A review of remote sensing opportunities. For. Ecol. Manag. 2006, 221, 27–41. [Google Scholar] [CrossRef]
- Heurich, M.; Krzystek, P.; Polakowsky, F.; Latifi, H.; Krauss, A.; Müller, J. Erste Waldinventur auf Basis von Lidardaten und digitalen Luftbildern im Nationalpark Bayerischer Wald. Forstl. Forsch. München 2015, 214, 101–113. [Google Scholar]
- Hildebrandt, G. Fernerkundung und Luftbildmessung für Forstwirtschaft, Vegetationskartierung und Landschaftsökologie; Wichmann Verlag: Heidelberg, Germany, 1996. [Google Scholar]
- Adamczyk, J.; Bedkowski, K. Digital analysis of relationships between crown colours on aerial photographs and trees health status. Rocz. Geomatyki 2006, 4, 47–54. [Google Scholar]
- Kenneweg, H. Auswertung von Farbluftbildern für die Abgrenzung von Schädigungen an Waldbeständen. Bildmess. U. Luftbildwes. 1970, 38, 283–290. [Google Scholar]
- Fassnacht, F.E.; Latifi, H.; Gosh, A.; Joshi, P.K.; Koch, B. Assessing the potential of hyperspectral imagery to map bark beetle-induced tree mortality. Remote Sens. Environ. 2014, 140, 533–548. [Google Scholar] [CrossRef]
- Adamczyk, J.; Osberger, A. Red-edge vegetation indices for detecting and assessing disturbances in Norway spruce dominated mountain forests. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 90–99. [Google Scholar] [CrossRef]
- ENVI. Vegetation Indices. Available online: http://www.harrisgeospatial.com/docs/VegetationIndices.html (accessed on 1 December 2019).
- Waser, L.T.; Küchler, M.; Jütte, K.; Stampfer, T. Evaluating the Potential of WorldView-2 Data to Classify Tree Species and Different Levels of Ash Mortality. Remote Sens. 2014, 6, 4515–4545. [Google Scholar] [CrossRef] [Green Version]
- Fassnacht, F.E. Assessing the Potential of Imaging Spectroscopy Data to Map Tree Species Composition and Bark Beetle-Related Tree Mortality. Ph.D. Thesis, Faculty of Environment and Natural Resources, Albert-Ludwigs-University, Freiburg, Germany, 2013. [Google Scholar]
- Heurich, M.; Fahse, L.; Reinelt, A. Die Buchdruckermassenvermehrung im Nationalpark Bayerischer Wald. In Waldentwicklung im Bergwald nach Windwurf und Borkenkäferbefall; Nationalparkverwaltung Bayerischer Wald: Grafenau, Germany, 2001; Volume 16, pp. 9–48. [Google Scholar]
- Zielewska, K. Ips Typographus. Ein Katalysator für einen Waldstrukturenwandel. Wsg Baden-Württemberg 2012, 15, 19–42. [Google Scholar]
- European Commission. Remote Sensing Applications for Forest Health Status Assessment. In European Union Scheme on the Protection of Forests Against Atmospheric Pollution, 2nd ed.; Office for Official Publications of the European Communities: Luxembourg, 2000; p. 216. [Google Scholar]
- Ahrens, W.; Brockamp, U.; Pisoke, T. Zur Erfassung von Waldstrukturen im Luftbild; Arbeitsanleitung für Waldschutzgebiete Baden-Württemberg; Forstliche Versuchs- und Forschungsanstalt Baden-Württemberg: Freiburg im Breisgau, Germany, 2004; Volume 5, p. 54. [Google Scholar]
- Bütler, R.; Schlaepfer, R. Spruce snag quantification by coupling colour infrared aerial photos and a GIS. For. Ecol. Manag. 2004, 195, 325–339. [Google Scholar] [CrossRef]
- Martinuzzi, S.; Vierling, L.A.; Gould, W.A.; Falkowski, M.J.; Evans, J.S.; Hudak, A.T.; Vierling, K.T. Mapping snags and understory shrubs for a LiDAR-based assessment of wildlife habitat suitability. Remote Sens. Environ. 2009, 113, 2533–2546. [Google Scholar] [CrossRef] [Green Version]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Pasher, J.; King, D.J. Mapping dead wood distribution in a temperate hardwood forest using high resolution airborne imagery. For. Ecol. Manag. 2009, 258, 1536–1548. [Google Scholar] [CrossRef]
- Kamińska, A.; Lisiewicz, M.; Stereńczak, K.; Kraszewski, B.; Sadkowski, R. Species-related single dead tree detection using multi-temporal ALS data and CIR imagery. Remote Sens. Environ. 2018, 219, 31–43. [Google Scholar] [CrossRef]
- Polewski, P.; Yao, W.; Heurich, M.; Krzystek, P.; Stilla, U. Active learning approach to detecting standing dead trees from ALS point clouds combined with aerial infrared imagery. In Proceedings of the CVPR Workshops, Boston, MA, USA, 7–12 June 2015; pp. 10–18. [Google Scholar]
- Krzystek, P.; Serebryanyk, A.; Schnörr, C.; Červenka, J.; Heurich, M. Large-Scale Mapping of Tree Species and Dead Trees in Šumava National Park and Bavarian Forest National Park Using Lidar and Multispectral Imagery. Remote Sens. 2020, 12, 661. [Google Scholar] [CrossRef] [Green Version]
- Maltamo, M.; Kallio, E.; Bollandsås, O.M.; Næsset, E.; Gobakken, T.; Pesonen, A. Assessing Dead Wood by Airborne Laser Scanning. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 375–395. [Google Scholar] [CrossRef]
- Marchi, N.; Pirotti, F.; Lingua, E. Airborne and Terrestrial Laser Scanning Data for the Assessment of Standing and Lying Deadwood: Current Situation and New Perspectives. Remote Sens. 2018, 10, 1356. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, L.; Salas, C.; Østgård, T.; Lien, V.; Gobakken, T.; Næsset, E. Predicting the occurrence of large-diameter trees using airborne laser scanning. Can. J. For. Res. 2016, 46, 461–469. [Google Scholar] [CrossRef] [Green Version]
- Yao, W.; Krzystek, P.; Heurich, M. Identifying standing dead trees in forest areas based on 3D single tree detection from full waveform Lidar data. In Proceedings of the ISPRS Annals of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Melbourne, Australia, 25 August–1 September 2012; pp. 359–364. [Google Scholar]
- Amiri, N.; Krzystek, P.; Heurich, M.; Skidmore, A.K. Classification of Tree Species as Well as Standing Dead Trees Using Triple Wavelength ALS in a Temperate Forest. Remote Sens. 2019, 11, 2614. [Google Scholar] [CrossRef] [Green Version]
- Casas, Á.; García, M.; Siegel, R.B.; Koltunov, A.; Ramírez, C.; Ustin, S. Burned forest characterization at single-tree level with airborne laser scanning for assessing wildlife habitat. Remote Sens. Environ. 2016, 175, 231–241. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, J.; Adler, P.; Aufreiter, C.; Bauerhansl, C.; Bucher, T.; Franz, S.; Engels, F.; Ginzler, C.; Hoffmann, K.; Jütte, K.; et al. Oberflächenmodelle aus Luftbildern für forstliche Anwendungen. Leitfaden AFL 2020. Wsl Ber. 2020, 87, 60. [Google Scholar]
- White, J.; Tompalski, P.; Coops, N.; Wulder, M. Comparison of airborne laser scanning and digital stereo imagery for characterizing forest canopy gaps in coastal temperate rainforests. Remote Sens. Environ. 2018, 208, 1–14. [Google Scholar] [CrossRef]
- Zielewska-Büttner, K.; Adler, P.; Ehmann, M.; Braunisch, V. Automated Detection of Forest Gaps in Spruce Dominated Stands Using Canopy Height Models Derived from Stereo Aerial Imagery. Remote Sens. 2016, 8, 175. [Google Scholar] [CrossRef] [Green Version]
- Meddens, A.J.H.; Hicke, J.A.; Vierling, L.A. Evaluating the potential of multispectral imagery to map multiple stages of tree mortality. Remote Sens. Environ. 2011, 115, 1632–1642. [Google Scholar] [CrossRef]
- Beck, H.E.; Zimmermann, N.E.; McVicar, T.R.; Vergopolan, N.; Berg, A.; Wood, E.F. Present and future Köppen-Geiger climate classification maps at 1-km resolution. Sci. Data 2018, 5, 180214. [Google Scholar] [CrossRef] [Green Version]
- Deutscher Wetterdienst. Wetter und Klima aus einer Hand. Station: Feldbarg/Schwarzwald. Available online: https://www.dwd.de/EN/weather/weather_climate_local/baden-wuerttemberg/feldberg/_node.html (accessed on 17 July 2020).
- Landesamt für Geoinformation und Landentwicklung Baden-Württemberg. Geodaten. Available online: https://www.lgl-bw.de/unsere-themen/Produkte/Geodaten (accessed on 3 January 2020).
- Rothermel, M.; Wenzel, K.; Fritsch, D.; Haala, N. Sure: Photogrammetric surface reconstruction from imagery. In Proceedings of the LC3D Workshop, Berlin, Germany, 4–5 December 2012. [Google Scholar]
- Schumacher, J.; Rattay, M.; Kirchhöfer, M.; Adler, P.; Kändler, G. Combination of Multi-Temporal Sentinel 2 Images and Aerial Image Based Canopy Height Models for Timber Volume Modelling. Forests 2019, 10, 746. [Google Scholar] [CrossRef] [Green Version]
- Mathow, T.; ForstBW. Regierungspräsidium Freiburg, Forstdirektion. Referat 84 Forsteinrichtung und Forstliche Geoinformation, Freiburg, Germany. Personal communication, 2015.
- Landesamt für Geoinformation und Landentwicklung Baden-Württemberg. ATKIS®—Amtliches Topographisch-Kartographisches Informationssystem. Available online: https://www.lgl-bw.de/unsere-themen/Geoinformation/AFIS-ALKIS-ATKIS/ATKIS/index.html (accessed on 3 January 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Hijmans, J.R. Raster: Geographic Data Analysis and Modeling, R Package Version 3.0-12; 2020. Available online: https://rdrr.io/cran/raster/ (accessed on 26 June 2020).
- Bivand, R.; Keitt, T.; Rowlingson, B. Package ‘rgdal’. Bindings for the ’Geospatial’ Data Abstraction Library, Version 1.2-16; 2019. Available online: https://CRAN.R-project.org/package=rgdal (accessed on 5 May 2019).
- Thomas, J.W.; Anderson, R.G.; Black, H.J.; Bull, E.L.; Canutt, P.R.; Carter, B.E.; Cromack, K.J.; Hall, F.C.; Martin, R.E.; Maser, C.; et al. Wildlife Habitats in Managed Forests—The Blue Mountains of Oregon and Washington. Agriculture Handbook No. 553; Thomas, J.W., Ed.; U.S. Department of Agriculture, Forest Service: Washington, DC, USA, 1979.
- Fassnacht, F.E.; Latifi, H.; Koch, B. An angular vegetation index for imaging spectroscopy data—Preliminary results on forest damage detection in the Bavarian National Park, Germany. Int. J. Appl. Earth Obs. Geoinf. 2012, 19, 308–321. [Google Scholar] [CrossRef]
- Kelly, M.; Blanchard, S.D.; Kersten, E.; Koy, K. Terrestrial Remotely Sensed Imagery in Support of Public Health: New Avenues of Research Using Object-Based Image Analysis. Remote Sens. 2011, 3, 2321–2345. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; The R Core Team; et al. Package ’caret’. Classification and Regression Training, Version 6.0-84; 2019. Available online: https://CRAN.R-project.org/package=caret (accessed on 20 September 2019).
- Ganz, S. Automatische Klassifizierung von Nadelbäumen Basierend auf Luftbildern. Automatic Classification of Coniferous Tree Genera Based on Aerial Images. Master’s Thesis, Albert-Ludwigs-Universität Freiburg, Freiburg, Germany, 2016. [Google Scholar]
- Jackson, R.D.; Huete, A.R. Interpreting vegetation indices. Prev. Vet. Med. 1991, 11, 185–200. [Google Scholar] [CrossRef]
- Ahamed, T.; Tian, L.; Zhang, Y.; Ting, K.C. A review of remote sensing methods for biomass feedstock production. Biomass Bioenergy 2011, 35, 2455–2469. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.; Merzlyak, M.N. Use of green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Interj. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Dowle, M.; Srinivasan, A. data.table: Extension of ‘data.frame’, R Package Version 1.12.8; 2019. Available online: https://cran.r-project.org/web/packages/data.table/index.html (accessed on 20 September 2019).
- Izrailev, S. Tictoc: Functions for Timing R Scripts, as Well as Implementations of Stack and List Structures, R Package Version 1.0; 2014. Available online: https://CRAN.R-project.org/package=tictoc (accessed on 10 November 2019).
- Environmental Systems Resource Institute. ArcGIS Desktop 10.5.1; ESRI: Redlands, CA, USA, 2018. [Google Scholar]
- Irons, J.R.; Petersen, G.W. Texture transforms of remote sensing data. Remote Sens. Environ. 1981, 11, 359–370. [Google Scholar] [CrossRef]
- Hexagon Erdas Imagine. Copyright 1990~2019. All Rights Reserved; Hexagon Geospatial, Intergraph Corporation: Madison, AL, USA, 2020. [Google Scholar]
- Barton, K. MuMIn: Multi-Model Inference, R Package Version 1.43.15; 2019. Available online: https://CRAN.R-project.org/package=MuMIn (accessed on 10 January 2020).
- Ballings, M.; Van den Poel, D. Auc: Threshold Independent Performance Measures For Probabilistic Classifiers, R Package Version 0.3.0; 2013. Available online: https://CRAN.R-project.org/package=AUC (accessed on 30 March 2020).
- Hosmer, D.H.; Lemeshow, S. Applied Logistic Regression, 2nd ed.; John Wiley & Sons: New York, NY, USA, 2000. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Thiele, C. cutpointr: Determine and Evaluate Optimal Cutpoints in Binary Classification Tasks, R Package Version 1.0.1; 2019. Available online: https://CRAN.R-project.org/package=cutpointr (accessed on 5 December 2019).
- DAT/EM. Summit Evolution. Available online: https://www.datem.com/summit-evolution/ (accessed on 8 June 2020).
- Congalton, R.G. A Review of Assessing the Accuracy of Classifications of Remotely Sensed Data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Kantola, T.; Vastaranta, M.; Yu, X.; Lyytikainen-Saarenmaa, P.; Holopainen, M.; Talvitie, M.; Kaasalainen, S.; Solberg, S.; Hyyppa, J. Classification of Defoliated Trees Using Tree-Level Airborne Laser Scanning Data Combined with Aerial Images. Remote Sens. 2010, 2, 2665–2679. [Google Scholar] [CrossRef] [Green Version]
- Wing, B.M.; Ritchie, M.W.; Boston, K.; Cohen, W.B.; Olsen, M.J. Individual snag detection using neighborhood attribute filtered airborne lidar data. Remote Sens. Environ. 2015, 163, 165–179. [Google Scholar] [CrossRef]
- Polewski, P.; Yao, W.; Heurich, M.; Krzystek, P.; Stilla, U. Detection of single standing dead trees from aerial color infrared imagery by segmentation with shape and intensity priors. In Proceedings of the ISPRS Annals of the Photogrammetry, Remote Sensing and Spatial Information Sciences 2015, 2015 PIA15+HRIGI15—Joint ISPRS Conference, Munich, Germany, 25–27 March 2015; Volume II-3/W4, pp. 181–188. [Google Scholar] [CrossRef] [Green Version]
- Wulder, M.A.; White, J.C.; Bentz, B. Detection and mapping of mountain pine beetle red attack: Matching information needs with appropriate remotely sensed data. In Proceedings of the Joint 2004 Annual General Meeting and Convention of the Society of American Foresters and the Canadian Institute of Forestry, Edmonton, AB, Canada, 2–6 October 2004. [Google Scholar]
- Sterenczak, K.; Kraszewski, B.; Mielcarek, M.; Piasecka, Z. Inventory of standing dead trees in the surroundings of communication routes—The contribution of remote sensing to potential risk assessments. For. Ecol. Manag. 2017, 402, 76–91. [Google Scholar] [CrossRef]
- Hengl, T. Finding the right pixel size. Comput. Geosci. 2006, 32, 1283–1298. [Google Scholar] [CrossRef]
- Dalponte, M.; Reyes, F.; Kandare, K.; Gianelle, D. Delineation of Individual Tree Crowns from ALS and Hyperspectral data: A comparison among four methods. Eur. J. Remote Sens. 2015, 48, 365–382. [Google Scholar] [CrossRef] [Green Version]
- Pluto-Kossakowska, J.; Osinska-Skotak, K.; Sterenczak, K. Determining the spatial resolution of multispectral satellite images optimal to detect dead trees in forest areas. Sylwan 2017, 161, 395–404. [Google Scholar]
- Straub, C.; Stepper, C.; Seitz, R.; Waser, L.T. Potential of UltraCamX stereo images for estimating timber volume and basal area at the plot level in mixed European forests. Can. J. For. Res. 2013, 43, 731–741. [Google Scholar] [CrossRef]
- Zielewska-Büttner, K.; Adler, P.; Peteresen, M.; Braunisch, V. Parameters Influencing Forest Gap Detection Using Canopy Height Models Derived From Stereo Aerial Imagery. In Proceedings of the 3 Wissenschaftlich-Technische Jahrestagung der DGPF. Dreiländertagung der DGPF, der OVG und der SGP, Bern, Switzerland, 7–9 June 2016; pp. 405–416. [Google Scholar]
- Trier, Ø.D.; Salberg, A.-B.; Kermit, M.; Rudjord, Ø.; Gobakken, T.; Næsset, E.; Aarsten, D. Tree species classification in Norway from airborne hyperspectral and airborne laser scanning data. Eur. J. Remote Sens. 2018, 51, 336–351. [Google Scholar] [CrossRef]
- Liu, Z.; Peng, C.; Timothy, W.; Candau, J.-N.; Desrochers, A.; Kneeshaw, D. Application of machine-learning methods in forest ecology: Recent progress and future challenges. Environ. Rev. 2018, 26. [Google Scholar] [CrossRef] [Green Version]
- Valbuena, R.; Maltamo, M.; Packalen, P. Classification of forest development stages from national low-density lidar datasets: A comparison of machine learning methods. Rev. De Teledetec. 2016, 45, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Wegmann, M.; Leutner, B.; Dech, S.e. Remote Sensing and GIS for Ecologists: Using Open Source Software; Wegmann, M., Leutner, B., Dech, S., Eds.; Pelagic Publishing: Exeter, UK, 2016; p. 352. [Google Scholar]
- Paoletti, M.E.; Haut, J.M.; Plaza, J.; Plaza, A. Deep learning classifiers for hyperspectral imaging: A review. Isprs J. Photogramm. Remote Sens. 2019, 158, 279–317. [Google Scholar] [CrossRef]
- Hamdi, Z.M.; Brandmeier, M.; Straub, C. Forest Damage Assessment Using Deep Learning on High Resolution Remote Sensing Data. Remote Sens. 2019, 11, 1976. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Yao, W.; Heurich, M. Dead wood detection based on semantic segmentation of vhr aerial cir imagery using optimized fcn-densenet. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, XLII-2/W16, 127–133. [Google Scholar] [CrossRef] [Green Version]
- O’Shea, K.; Nash, R. An Introduction to Convolutional Neural Networks. arXiv 2015, arXiv:1511.08458v2. [Google Scholar]
- European-Space-Imaging. WorldView-3. Data Sheet; European Space Imaging: Munich, Germany, 2018. [Google Scholar]
- European-Space-Imaging. WorldView-4. Data Sheet; European Space Imaging: Munich, Germany, 2018. [Google Scholar]
- Coeurdev, L.; Gabriel-Robe, C. Pléiades Imagery-User Guid; Astrium GEO-Information Services: Toulouse, France, 2012; p. 118. [Google Scholar]
- Piermattei, L.; Marty, M.; Ginzler, C.; Pöchtrager, M.; Karel, W.; Ressl, C.; Pfeifer, N.; Hollaus, M. Pléiades satellite images for deriving forest metrics in the Alpine region. Int. J. Appl. Earth Obs. Geoinf. 2019, 80, 240–256. [Google Scholar] [CrossRef]

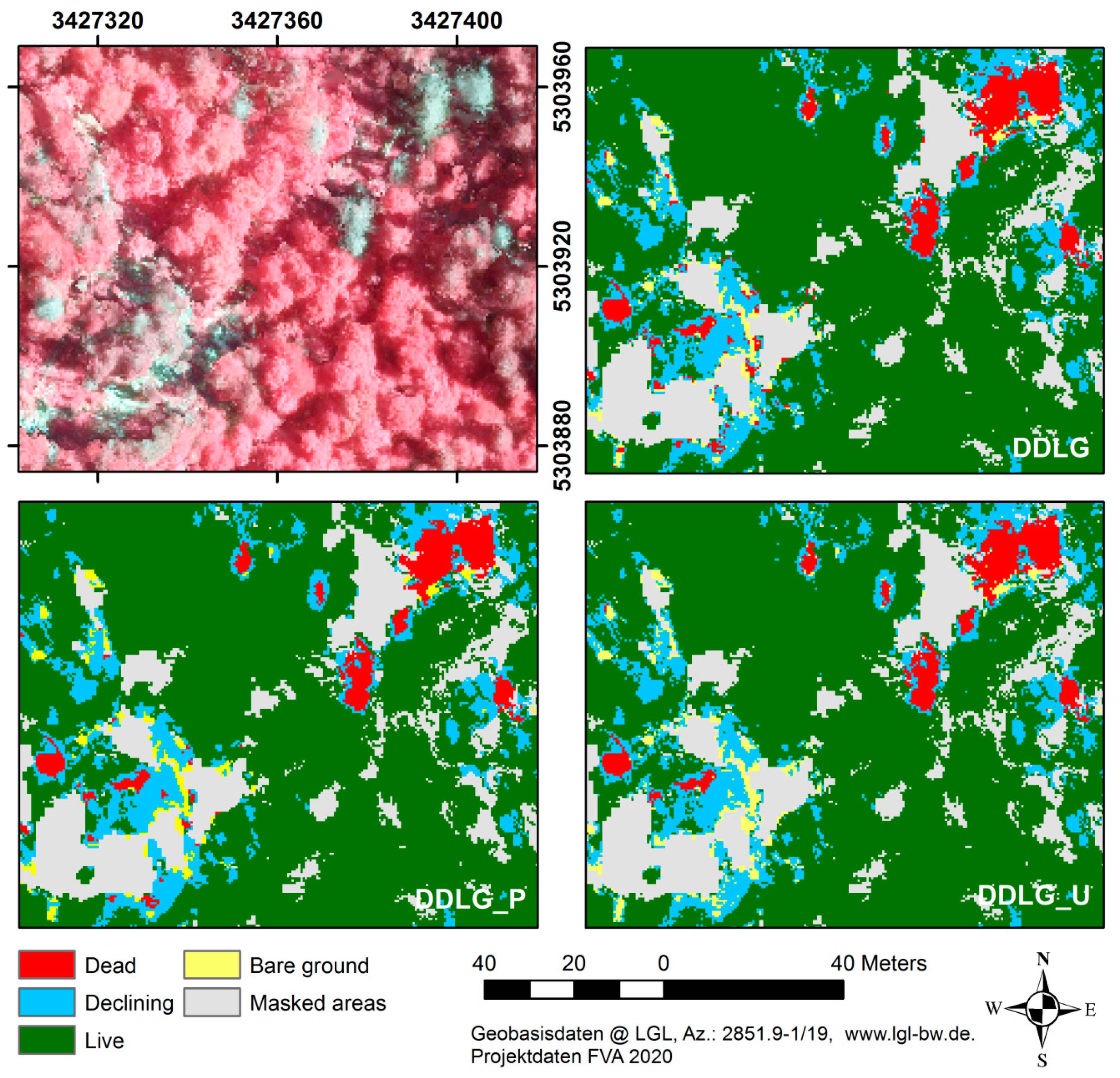
| Model Class | N | Polygon Area (m2) | No. of Pixels | |||||
|---|---|---|---|---|---|---|---|---|
| Sum | Min | Max | Mean | Median | SD | |||
| live | 33 | 24,789.7 | 1.2 | 2797 | 751.2 | 576.6 | 659.9 | 98,137 |
| dead | 315 | 4295.7 | 0.1 | 65.9 | 13.6 | 11 | 12.1 | 17,143 |
| declining | 64 | 1510.8 | 6.9 | 52.0 | 23.6 | 22.1 | 10.7 | 6048 |
| bare ground | 103 | 1208.9 | 0.5 | 178.7 | 11.7 | 4.2 | 22.3 | 4272 |
| Predictor Variables | Description or Formula | Reference |
|---|---|---|
| R, G, B, I | Red, green, blue, infrared bands | - |
| H, S, V | Hue, saturation, value calculated with “rgb2hsv” function in “grDevices” | R-package “grDevices” [58] |
| Vegetation_h | Vegetation height from CHM | - |
| R_ratio | R/(R + G + B + I) | Ganz [66] |
| G_ratio | G/(R + G + B + I) | |
| B_ratio | B/(R + G + B + I) | |
| I_ratio | I/(R + G + B +I) | |
| NDVI | (I − R)/(I + R) | Jackson and Huete [67] |
| NDVI_green | (I − G)/(I + G) | Ahamed et al. [68] |
| G_R_ratio | G/R | Waser et al. [28] |
| G_R_ratio_2 | (G − R)/(G + R) | Gitelson et al. [69] |
| B_ratio_2 | (R/B) × (G/B) × (I/B) | self-developed after Waser et al. [28] |
| B_I_ratio | B/I | self-developed |
| Variable | Description | Hypothesized Meaning | Unit |
|---|---|---|---|
| Clump_size | Size of the “dead” pixel clumps grouped with 8 neighbors (1 pixel = 0.25 m2) | Very small and very big clumps are more likely to be falsely classified | N |
| Bare_ground | Proportion of bare ground within a 11.5 × 11.5 m (23 × 23 pixels) moving window | Occurrence of “bare ground” next to “dead” pixels may indicate a possible misclassification of both classes | 0–1 |
| Canopy_cover | Proportion of pixels above 2 m vegetation height within a 11.5 × 11.5 m (23 × 23 pixels) moving window | Pixels with low canopy cover are likely to have false height values in transition areas between high and low vegetation | 0–1 |
| Curvature | Curvature values per pixel based on the I band | Form and direction of the I spectral signal may differ between “dead” and “bare ground” objects | Value (−∞)–∞ |
| Curvature_Mean | Mean curvature values within a 2.5 × 2.5 m (5 × 5 pixels) moving window based on the I band | Form and direction of the I spectral signal in a wider surrounding may show differences between “dead” and “bare ground” objects | Value (−∞)–∞ |
| Mean Euclidean distance (texture features) | Mean Euclidean distance values within a 2.5 × 2.5 m (5 × 5 pixels) moving window based on the I band | “Dead” and “bare ground” objects may show different texture characteristics in the I band | Value 0–∞ |
| DDLG Version | Accuracy Measure | Class | Overall Accuracy | Kappa | |||
|---|---|---|---|---|---|---|---|
| Bare Ground | Live | Declining | Dead | ||||
| 18 variables | Producer’s accuracy | 0.99 | 0.97 | 0.90 | 0.92 | 0.94 | 0.93 |
| User’s accuracy | 0.99 | 0.94 | 0.89 | 0.95 | |||
| 7 variables | Producer’s accuracy | 0.99 | 0.97 | 0.91 | 0.92 | 0.95 | 0.92 |
| User’s accuracy | 0.98 | 0.95 | 0.90 | 0.96 | |||
| Performance | Model Fit | 4 FOLDS Validation Max Sensitivity by Specificity = 0.70 (0.31) | ||
|---|---|---|---|---|
| metrics | Kappa maximum (0.39) | Max sensitivity by specificity = 0.70 (0.31) | Mean | Standard deviation |
| Sensitivity | 0.82 | 0.88 | 0.89 | 0.007 |
| Specificity | 0.77 | 0.70 | 0.72 | 0.009 |
| PPV | 0.78 | 0.75 | 0.76 | 0.005 |
| NPV | 0.81 | 0.85 | 0.87 | 0.007 |
| Overall accuracy | 0.80 | 0.79 | 0.80 | 0.003 |
| Cohen’s Kappa | 0.60 | 0.58 | 0.61 | 0.005 |
| AUC | 0.89 | 0.90 | 0.005 | |
| Class | DDLG | DDLG_P | DDLG_U | |||
|---|---|---|---|---|---|---|
| Pixel | % | Pixel | % | Pixel | % | |
| Live | 9,341,179 | 39.4% | 9,342,529 | 39.4% | 9,341,179 | 39.4% |
| Dead | 119,790 | 0.5% | 105,661 | 0.4% | 102,815 | 0.4% |
| Declining | 1,274,349 | 5.4% | 1,237,793 | 5.2% | 1,274,349 | 5.4% |
| Bare ground | 35,690 | 0.2% | 36,265 | 0.2% | 52,665 | 0.2% |
| NA | 12,933,862 | 54.6% | 12,982,622 | 54.8% | 12,933,862 | 54.6% |
| Total | 23,704,870 | 100.0% | 23,704,870 | 100.0% | 23,704,870 | 100.0% |
| DDLG | DDLG_P | DDLG_U | ||||
|---|---|---|---|---|---|---|
| Pixel | m2 | Pixel | m2 | Pixel | m2 | |
| N | 9868 | 3482 | 3139 | |||
| SUM | 119,790.00 | 29,947.50 | 105,661.00 | 26,415.25 | 102,815.00 | 25,703.75 |
| MEAN | 12.14 | 3.04 | 30.34 | 7.59 | 32.75 | 8.19 |
| MEDIAN | 2.00 | 0.50 | 10.00 | 2.50 | 12.00 | 3.00 |
| MIN | 1.00 | 0.25 | 1.00 | 0.25 | 1.00 | 0.25 |
| MAX | 3281.00 | 820.25 | 3258.00 | 814.50 | 3281.00 | 820.25 |
| SD | 53.82 | 13.45 | 87.30 | 21.82 | 91.98 | 22.99 |
| Model Scenario | Accuracy Measure | Class | Overall Accuracy | Kappa | |||
|---|---|---|---|---|---|---|---|
| Bare Ground | Live | Declining | Dead | ||||
| DDLG | User’s accuracy | 0.82 | 0.77 | 0.67 | 0.60 | 0.70 | 0.60 |
| Producer’s accuracy | 0.57 | 0.79 | 0.56 | 0.87 | |||
| DDLG_P | User’s accuracy | 0.75 | 0.73 | 0.61 | 0.69 | 0.70 | 0.59 |
| Producer’s accuracy | 0.66 | 0.79 | 0.55 | 0.79 | |||
| DDLG_U | User’s accuracy | 0.76 | 0.77 | 0.66 | 0.74 | 0.74 | 0.65 |
| Producer’s accuracy | 0.82 | 0.77 | 0.56 | 0.80 | |||
| Polygon Data | DDLG | DDLG_P | DDLG_PU | |||
|---|---|---|---|---|---|---|
| Intersected | Not Intersected | Intersected | Not Intersected | Intersected | Not Intersected | |
| N | 303 | 12 | 287 | 22 | 291 | 24 |
| N (% of reference) | 96% | 4% | 91% | 7% | 92% | 8% |
| AREA SUM (m2) | 3868.5 | 7.5 | 3844.7 | 23.1 | 3855.2 | 22.3 |
| AREA (% of reference) | 90% | 0% | 90% | 1% | 90% | 1% |
| AREA MEAN (m2) | 12.8 | 0.6 | 13.4 | 0.8 | 13.2 | 0.9 |
| AREA MEDIAN (m2) | 10.4 | 0.1 | 10.6 | 0.7 | 10.6 | 0.5 |
| AREA MAX (m2) | 64.9 | 4.3 | 65.1 | 4.3 | 64.9 | 4.3 |
| AREA SD (m2) | 11.5 | 1.2 | 11.5 | 0.9 | 11.5 | 1.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zielewska-Büttner, K.; Adler, P.; Kolbe, S.; Beck, R.; Ganter, L.M.; Koch, B.; Braunisch, V. Detection of Standing Deadwood from Aerial Imagery Products: Two Methods for Addressing the Bare Ground Misclassification Issue. Forests 2020, 11, 801. https://doi.org/10.3390/f11080801
Zielewska-Büttner K, Adler P, Kolbe S, Beck R, Ganter LM, Koch B, Braunisch V. Detection of Standing Deadwood from Aerial Imagery Products: Two Methods for Addressing the Bare Ground Misclassification Issue. Forests. 2020; 11(8):801. https://doi.org/10.3390/f11080801
Chicago/Turabian StyleZielewska-Büttner, Katarzyna, Petra Adler, Sven Kolbe, Ruben Beck, Lisa Maria Ganter, Barbara Koch, and Veronika Braunisch. 2020. "Detection of Standing Deadwood from Aerial Imagery Products: Two Methods for Addressing the Bare Ground Misclassification Issue" Forests 11, no. 8: 801. https://doi.org/10.3390/f11080801





