Genetic Diversity and Range Dynamics of Helleborus odorus subsp. cyclophyllus under Different Climate Change Scenarios
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Genetic Analysis
2.2.1. DNA Extraction
2.2.2. ISSR Genotyping
2.2.3. Statistical Analysis
2.3. Species Occurrence Data
2.4. Environmental Data
- Three Global Circulation Models (GCMs) that are rendered more suitable and realistic for the study area’s future climate.
- Four different IPCC scenarios from the Representative Concentration Pathways (RCP) family.
- Seven soil variables providing predicted values for the surface soil layer at varying depths.
- Elevation data from the CGIAR-CSI data-portal [43] were aggregated and resampled to match the resolution of the other environmental variables. Additional topographical variables (slope, aspect, heat load index, topographic position index and terrain ruggedness index) were computed based on elevation data using functions from the ‘raster’ 2.6.7 package [44] and ‘spatialEco’ 1.2–0 package [45].
- Geological data from the Geological Map of Greece [46].
2.5. Species Distribution Models
2.5.1. Model Parameterization and Evaluation
2.5.2. Model Projections
2.5.3. Area Range Change
2.6. Bioclimatic Congruence and Consistency
2.7. Generalized Dissimilarity Modelling
3. Results
3.1. Molecular Analysis
3.2. Species Distribution Modelling
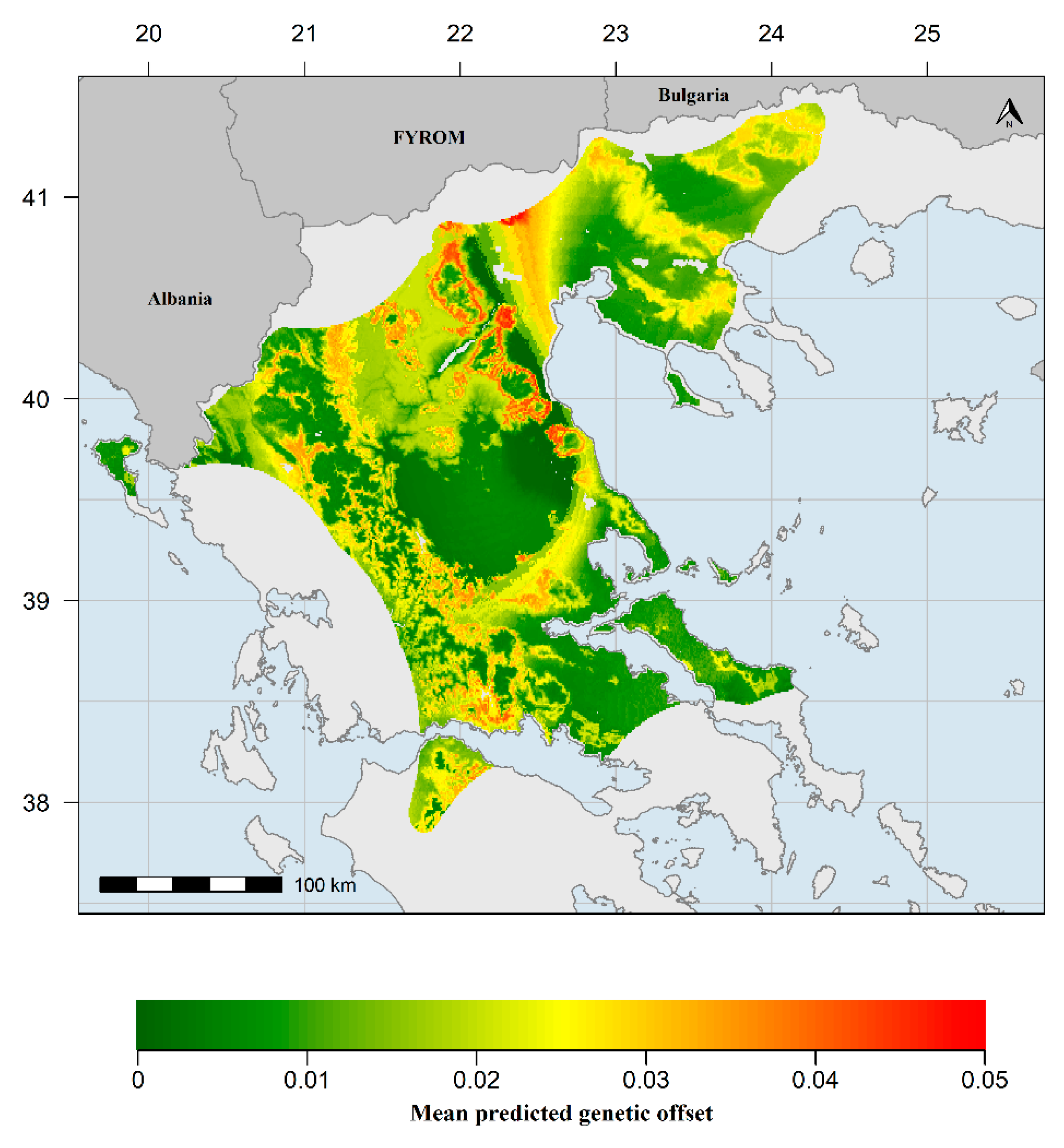
3.3. Generalized Dissimilarity Modelling
4. Discussion
4.1. Genetic Diversity and Structure
4.2. Climate Change Vulnerability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rahbek, C.; Borregaard, M.K.; Antonelli, A.; Colwell, R.K.; Holt, B.G.; Nogues-Bravo, D.; Rasmussen, C.M.; Richardson, K.; Rosing, M.T.; Whittaker, R.J.; et al. Building mountain biodiversity: Geological and evolutionary processes. Science 2019, 365, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Rahbek, C.; Gotelli, N.J.; Colwell, R.K.; Entsminger, G.L.; Rangel, T.F.; Graves, G.R. Predicting continental-scale patterns of bird species richness with spatially explicit models. Proc. R. Soc. B: Boil. Sci. 2006, 274, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Steinbauer, M.; Field, R.; Grytnes, J.A.; Trigas, P.; Ah-Peng, C.; Attorre, F.; Birks, H.J.B.; Borges, P.A.V.; Cardoso, P.; Chou, C.H.; et al. Topography-driven isolation, speciation and a global increase of endemism with elevation. Glob. Ecol. Biogeogr. 2016, 25, 1097–1107. [Google Scholar] [CrossRef]
- Thuiller, W.; Lavorel, S.; Araujo, M.B.; Sykes, M.T.; Prentice, I.C. Climate change threats to plant diversity in Europe. Proc. Natl. Acad. Sci. USA 2005, 102, 8245–8250. [Google Scholar] [CrossRef]
- Kier, G.; Kreft, H.; Lee, T.M.; Jetz, W.; Ibisch, P.L.; Nowicki, C.; Mutke, J.; Barthlott, W. A global assessment of endemism and species richness across island and mainland regions. Proc. Natl. Acad. Sci. USA 2009, 106, 9322–9327. [Google Scholar] [CrossRef]
- Kryštufek, B.; Reed, J.M. Pattern and Process in Balkan Biodiversity—An Overview. In Balkan Biodiversity; Springer: Dordrecht, The Netherlands, 2004; pp. 1–8. [Google Scholar] [CrossRef]
- Tzedakis, P.C. The Balkans as Prime Glacial Refugial Territory of European Temperate Trees. In Balkan Biodiversity; Springer: Dordrecht, The Netherlands, 2004; pp. 49–68. [Google Scholar] [CrossRef]
- Frajman, B.; Oxelman, B. Reticulate phylogenetics and phytogeographical structure of Heliosperma (Sileneae, Caryophyllaceae) inferred from chloroplast and nuclear DNA sequences. Mol. Phylogenetics Evol. 2007, 43, 140–155. [Google Scholar] [CrossRef]
- Surina, B.; Schönswetter, P.; Schneeweiss, G.M. Quaternary range dynamics of ecologically divergent species (Edraianthus serpyllifolius and E. tenuifolius, Campanulaceae) within the Balkan refugium. J. Biogeogr. 2011, 38, 1381–1393. [Google Scholar] [CrossRef]
- Kutnjak, D.; Kuttner, M.; Niketić, M.; Dullinger, S.; Schönswetter, P.; Frajman, B. Escaping to the summits: Phylogeography and predicted range dynamics of Cerastium dinaricum, an endangered high mountain plant endemic to the western Balkan Peninsula. Mol. Phylogenetics Evol. 2014, 78, 365–374. [Google Scholar] [CrossRef]
- Pauli, H.; Gottfried, M.; Dullinger, S.; Abdaladze, O.; Akhalkatsi, M.; Alonso, J.L.B.; Coldea, G.; Dick, J.; Erschbamer, B.; Calzado, R.F.; et al. Recent Plant Diversity Changes on Europe’s Mountain Summits. Science 2012, 336, 353–355. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; McInnes, K.H. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2002. [Google Scholar]
- Meiners, J.; Winkelmann, T. Evaluation of reproductive barriers and realisation of interspecific hybridisations depending on genetic distances between species in the genus Helleborus. Plant Boil. 2012, 14, 576–585. [Google Scholar] [CrossRef]
- Euro+Med (2006-): Euro+Med PlantBase-the information resource for Euro-Mediterranean Plant Diversity. Available online: http://ww2.bgbm.org/EuroPlusMed/ (accessed on 2 May 2020).
- Balázs, V.L.; Filep, R.; Ambrus, T.; Kocsis, M.; Farkas, Á.; Stranczinger, S.; Papp, N. Ethnobotanical, historical and histological evaluation of Helleborus L. genetic resources used in veterinary and human ethnomedicine. Genet. Resour. Crop. Evol. 2020, 67, 781–797. [Google Scholar] [CrossRef]
- Čakar, J.; Parić, A.; Vidic, D.; Haverić, A.; Haverić, S.; Maksimović, M.; Bajrović, K. Antioxidant and antiproliferative activities of Helleborus odorus Waldst. & Kit, H. multifidus Vis. and H. hercegovinus Martinis. Nat. Prod. Res. 2011, 25, 1969–1974. [Google Scholar] [CrossRef]
- Banuls, L.M.Y.; Katz, A.; Miklos, W.; Cimmino, A.; Tal, D.M.; Ainbinder, E.; Zehl, M.; Urban, E.; Evidente, A.; Kopp, B.; et al. Hellebrin and its aglycone form hellebrigenin display similar in vitro growth inhibitory effects in cancer cells and binding profiles to the alpha subunits of the Na+/K+-ATPase. Mol. Cancer 2013, 12, 33. [Google Scholar] [CrossRef]
- Reed, D.H.; Frankham, R. Society for Conservation Biology Correlation between Fitness and Genetic Diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Szczecińska, M.; Sramkó, G.; Wołosz, K.; Sawicki, J. Genetic Diversity and Population Structure of the Rare and Endangered Plant Species Pulsatilla patens (L.) Mill in East Central Europe. PLoS ONE 2016, 11, e0151730. [Google Scholar] [CrossRef]
- Reddy, M.P.; Sarla, N.; Siddiq, E. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Mondini, L.; Noorani, A.; Pagnotta, M.A. Assessing Plant Genetic Diversity by Molecular Tools. Diversity 2009, 1, 19–35. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T.B.J.; Ye, Z.H.; Mao, J.X. POPGENE, the User-Friendly Shareware for Population Genetic Analysis; University of Alberta: Edmonton, AB, Canada, 1997. [Google Scholar]
- Vekemans, X.; Beauwens, T.; Lemaire, M.; Roldan-Ruiz, I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol. Ecol. 2002, 11, 139–151. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinform 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Lynch, M.; Milligan, B.G. Analysis of population genetic structure with RAPD markers. Mol. Ecol. 1994, 3, 91–99. [Google Scholar] [CrossRef]
- Hampl, V.; Pavlíček, A.; Flegr, J. Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with the freeware program FreeTree: Application to trichomonad parasites. Int. J. Syst. Evol. Microbiol. 2001, 51, 731–735. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree, v1.3.1; beast: UK, 2010. [Google Scholar]
- Huff, D.R.; Peakall, R.; Smouse, P.E. RAPD variation within and among natural populations of outcrossing buffalograss [Buchloë dactyloides (Nutt.) Engelm.]. Theor. Appl. Genet. 1993, 86, 927–934. [Google Scholar] [CrossRef]
- Vavrek, M.J. Fossil: Palaeoecological and palaeogeographical analysis tools. Palaeontol. Electron. 2011, 14, 16. [Google Scholar]
- Oksanen, A.J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Minchin, P.R.; Hara, R.B.O.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package, version 2.5.2; CRAN, R Core Team: Cary, CA, USA, 2018. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.; Vonholdt, B.M. Structure Harvester: A website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Remington, D.L.; Thornsberry, J.M.; Matsuoka, Y.; Wilson, L.M.; Whitt, S.R.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 2001, 98, 11479–11484. [Google Scholar] [CrossRef]
- Chamberlain, S.; Boettiger, C. R Python, and Ruby clients for GBIF species occurrence data. PeerJ Preprints 2017, 5, e3304v1. [Google Scholar] [CrossRef]
- Robertson, M.P.; Visser, V.; Hui, C. Biogeo: An R package for assessing and improving data quality of occurrence record datasets. Ecography 2016, 39, 394–401. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Clim. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Karger, D.N.; Conrad, O.; Böhner, J.; Kawohl, T.; Kreft, H.; Soria-Auza, R.W.; Zimmermann, N.E.; Linder, H.P.; Kessler, M. Climatologies at high resolution for the earth’s land surface areas. Sci. Data 2017, 4, 1–20. [Google Scholar] [CrossRef] [PubMed]
- CHELSA Database. Available online: http://chelsa-climate.org/ (accessed on 2 May 2020).
- Morales-Barbero, J.; Vega-Álvarez, J. Input matters matter: Bioclimatic consistency to map more reliable species distribution models. Methods Ecol. Evol. 2018, 10, 212–224. [Google Scholar] [CrossRef]
- Araujo, M.B.; Anderson, R.P.; Barbosa, A.M.; Beale, C.M.; Dormann, C.F.; Early, R.; Garcia, R.A.; Guisan, A.; Maiorano, L.; Naimi, B.; et al. Standards for distribution models in biodiversity assessments. Sci. Adv. 2019, 5, eaat4858. [Google Scholar] [CrossRef]
- Jarvis, A.; Reuter, H.I.; Nelson, A.; Guevara, E. Hole-filled SRTM for the Globe Version 3, from the CGIAR-CSI SRTM 90m Database 2006. Available online: http//srtm.csi.cgiar.org (accessed on 31 April 2020).
- Hijmans, R.; van Etten, J. Raster: Geographic Data Analysis and Modeling. R Package Version 2014, 517, 2–12. [Google Scholar]
- Evans, J.S. spatialEco: Spatial Analysis and Modelling Utilities, version1.3-1; CRAN, R Core Team: Cary, CA, USA, 2020. [Google Scholar]
- Bornovas, I.; Rondogianni-Tsiambaou, T. Geological Map of Greece Scale 1:500,000, 2nd ed.; Inst. of Geology and Mineral Exploration, Division of General Geology and Economic Geology: Athens, Greece, 1983. [Google Scholar]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Márquez, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2012, 36, 27–46. [Google Scholar] [CrossRef]
- Araujo, M.B.; New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef]
- Thuiller, W.; Lafourcade, B.; Engler, R.; Araujo, M.B. BIOMOD—A platform for ensemble forecasting of species distributions. Ecography 2009, 32, 369–373. [Google Scholar] [CrossRef]
- Ferrier, S.; Manion, G.; Elith, J.; Richardson, K. Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Divers. Distrib. 2007, 13, 252–264. [Google Scholar] [CrossRef]
- Fitzpatrick, M.C.; Keller, S.R. Ecological genomics meets community-level modelling of biodiversity: Mapping the genomic landscape of current and future environmental adaptation. Ecol. Lett. 2014, 18, 1–16. [Google Scholar] [CrossRef]
- Manion, G.; Lisk, M.; Ferrier, S.; Nieto-Lugible, D.; Mokany, K.; Fitzpatrick, M.C. gdm: Generalized Dissimilarity Modeling, version 1.3.7; CRAN, R Core Team: Cary, CA, USA, 2018. [Google Scholar]
- R Core Development Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Rahbek, C. The elevational gradient of species richness: A uniform pattern? Ecography 1995, 18, 200–205. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Sgrò, C.M. Climate change and evolutionary adaptation. Nature 2011, 470, 479–485. [Google Scholar] [CrossRef]
- Pauls, S.U.; Nowak, C.; Bálint, M.; Pfenninger, M. The impact of global climate change on genetic diversity within populations and species. Mol. Ecol. 2012, 22, 925–946. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.F.; Anderson, N.O. How many marker loci are necessary? Analysis of dominant marker data sets using two popular population genetic algorithms. Ecol. Evol. 2013, 3, 3455–3470. [Google Scholar] [CrossRef] [PubMed]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Hogbin, P.M.; Peakall, R. Evaluation of the Contribution of Genetic Research to the Management of the Endangered Plant Zieria prostrata. Conserv. Boil. 1999, 13, 514–522. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Elam, D.R. Population Genetic Consequences of Small Population Size: Implications for Plant Conservation. Annu. Rev. Ecol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]
- Slatkin, M.; Excoffier, L. Serial Founder Effects during Range Expansion: A Spatial Analog of Genetic Drift. Genetics 2012, 191, 171–181. [Google Scholar] [CrossRef]
- Grdiša, M.; Radosavljević, I.; Liber, Z.; Stefkov, G.; Ralli, P.; Chatzopoulou, P.S.; Carović-Stanko, K.; Satovic, Z. Divergent selection and genetic structure of Sideritis scardica populations from southern Balkan Peninsula as revealed by AFLP fingerprinting. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Larena, B.G.; Aguilar, J.F.; Feliner, G.N. Glacial-induced altitudinal migrations in Armeria (Plumbaginaceae) inferred from patterns of chloroplast DNA haplotype sharing. Mol. Ecol. 2002, 11, 1965–1974. [Google Scholar] [CrossRef]
- Kramp, K.; Huck, S.; Niketić, M.; Tomović, G.; Schmitt, T. Multiple glacial refugia and complex postglacial range shifts of the obligatory woodland plant Polygonatum verticillatum (Convallariaceae). Plant Boil. 2009, 11, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Strid, A. The mountain flora of Greece with special reference to the Anatolian element. Proc. R. Soc. Edinburgh. Sect. B. Boil. Sci. 1986, 89, 59–68. [Google Scholar] [CrossRef]
- Feidas, H.; Karagiannidis, A.; Keppas, S.; Vaitis, M.; Kontos, T.; Zanis, P.; Melas, D.; Anadranistakis, E. Modeling and mapping temperature and precipitation climate data in Greece using topographical and geographical parameters. Theor. Appl. Clim. 2013, 118, 133–146. [Google Scholar] [CrossRef]
- Parmesan, C. Ecological and Evolutionary Responses to Recent Climate Change. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 637–669. [Google Scholar] [CrossRef]
- McInerny, G.; Turner, J.; Wong, H.; Travis, J.M.; Benton, T.G. How range shifts induced by climate change affect neutral evolution. Proc. R. Soc. B Boil. Sci. 2009, 276, 1527–1534. [Google Scholar] [CrossRef]
- Riddle, B.R.; Dawson, M.N.; Hadly, E.A.; Hafner, D.J.; Hickerson, M.J.; Mantooth, S.J.; Yoder, A.D. The role of molecular genetics in sculpting the future of integrative biogeography. Prog. Phys. Geogr. Earth Environ. 2008, 32, 173–202. [Google Scholar] [CrossRef]
- Scoble, J.; Lowe, A. A case for incorporating phylogeography and landscape genetics into species distribution modelling approaches to improve climate adaptation and conservation planning. Divers. Distrib. 2010, 16, 343–353. [Google Scholar] [CrossRef]
- Eckert, C.G.; Samis, K.E.; Lougheed, S.C. Genetic variation across species’ geographical ranges: The central–marginal hypothesis and beyond. Mol. Ecol. 2008, 17, 1170–1188. [Google Scholar] [CrossRef]
- Sagarin, R.; Gaines, S.D. The ‘abundant centre’ distribution: To what extent is it a biogeographical rule? Ecol. Lett. 2002, 5, 137–147. [Google Scholar] [CrossRef]
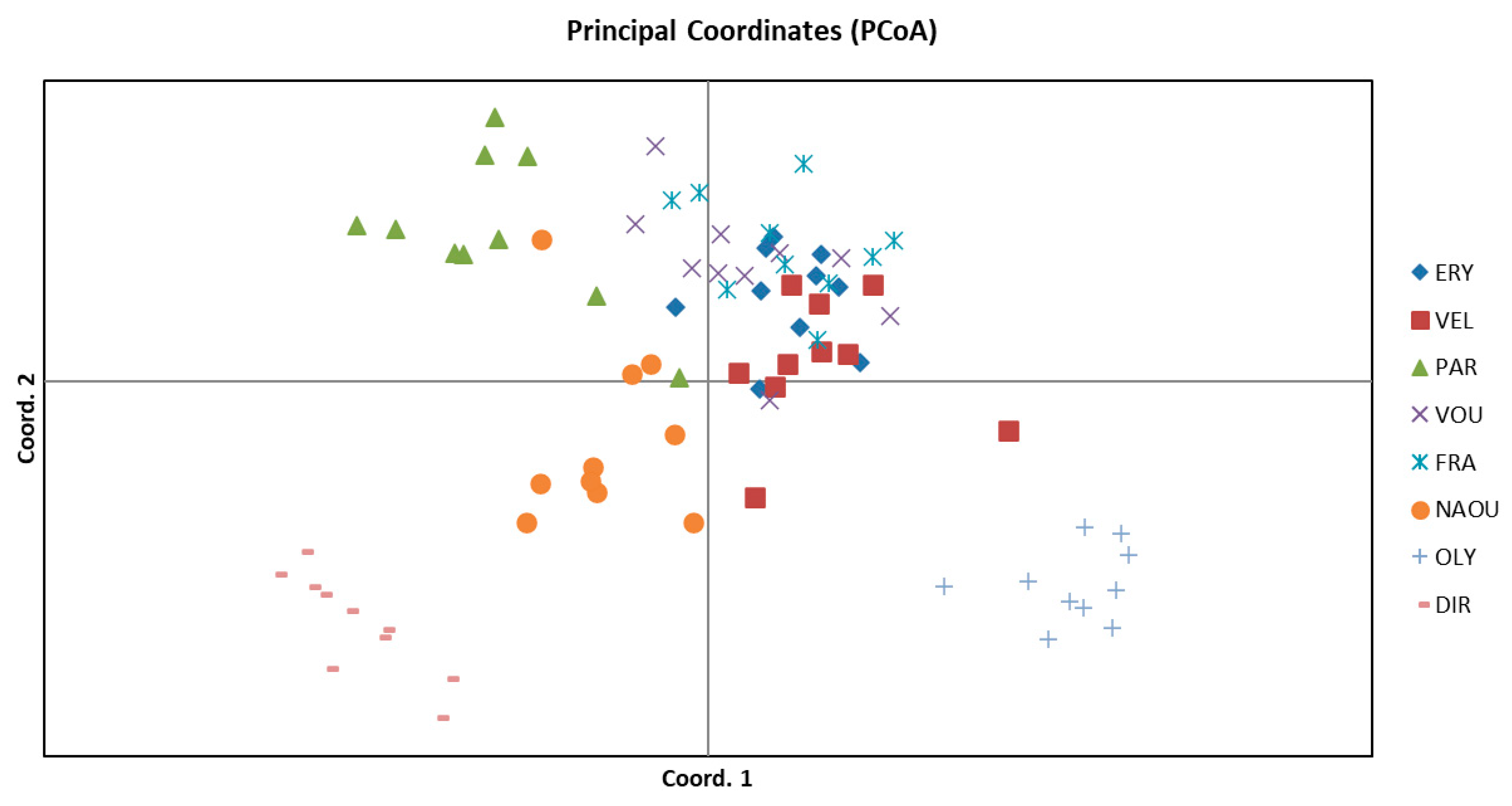
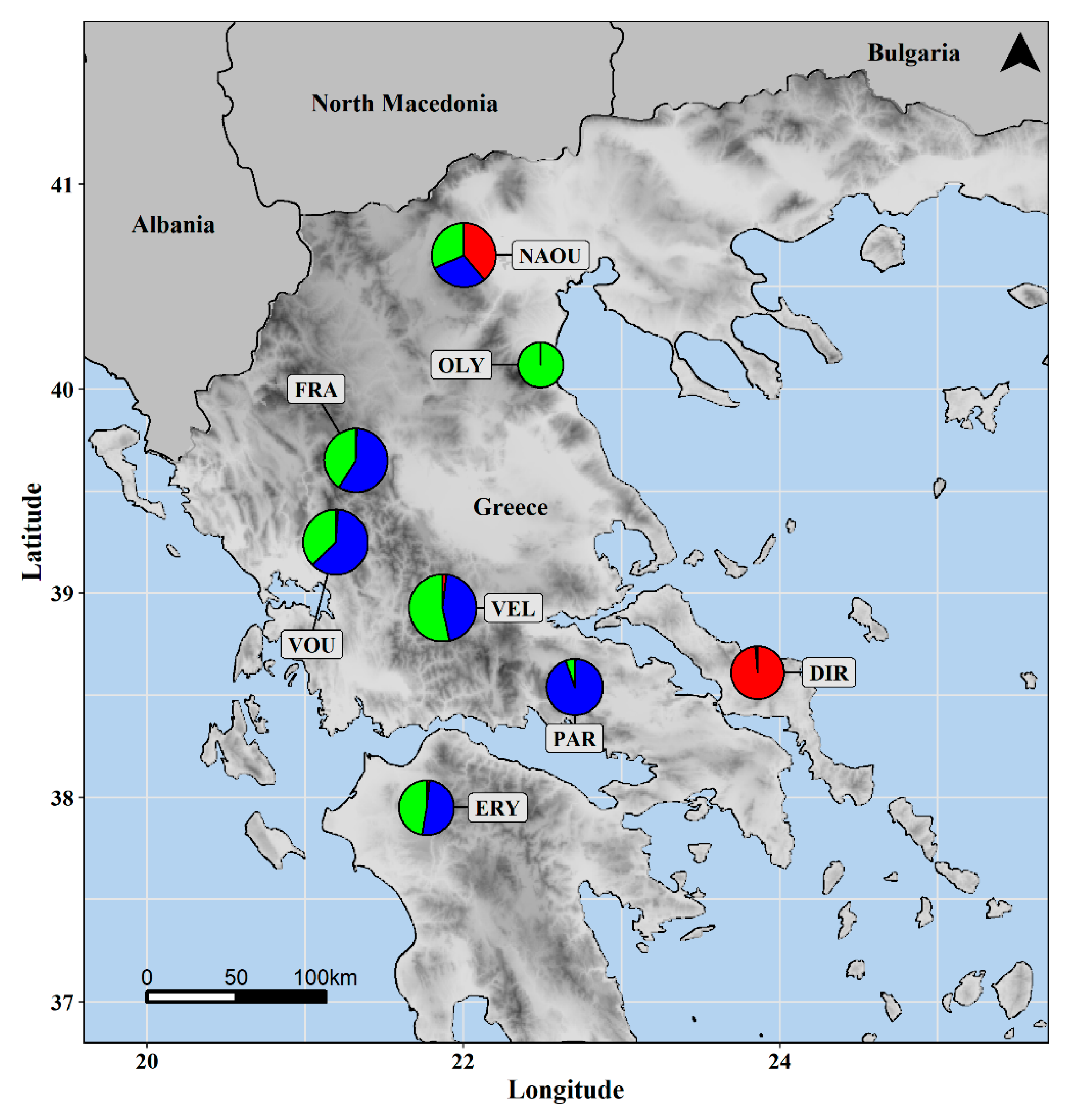
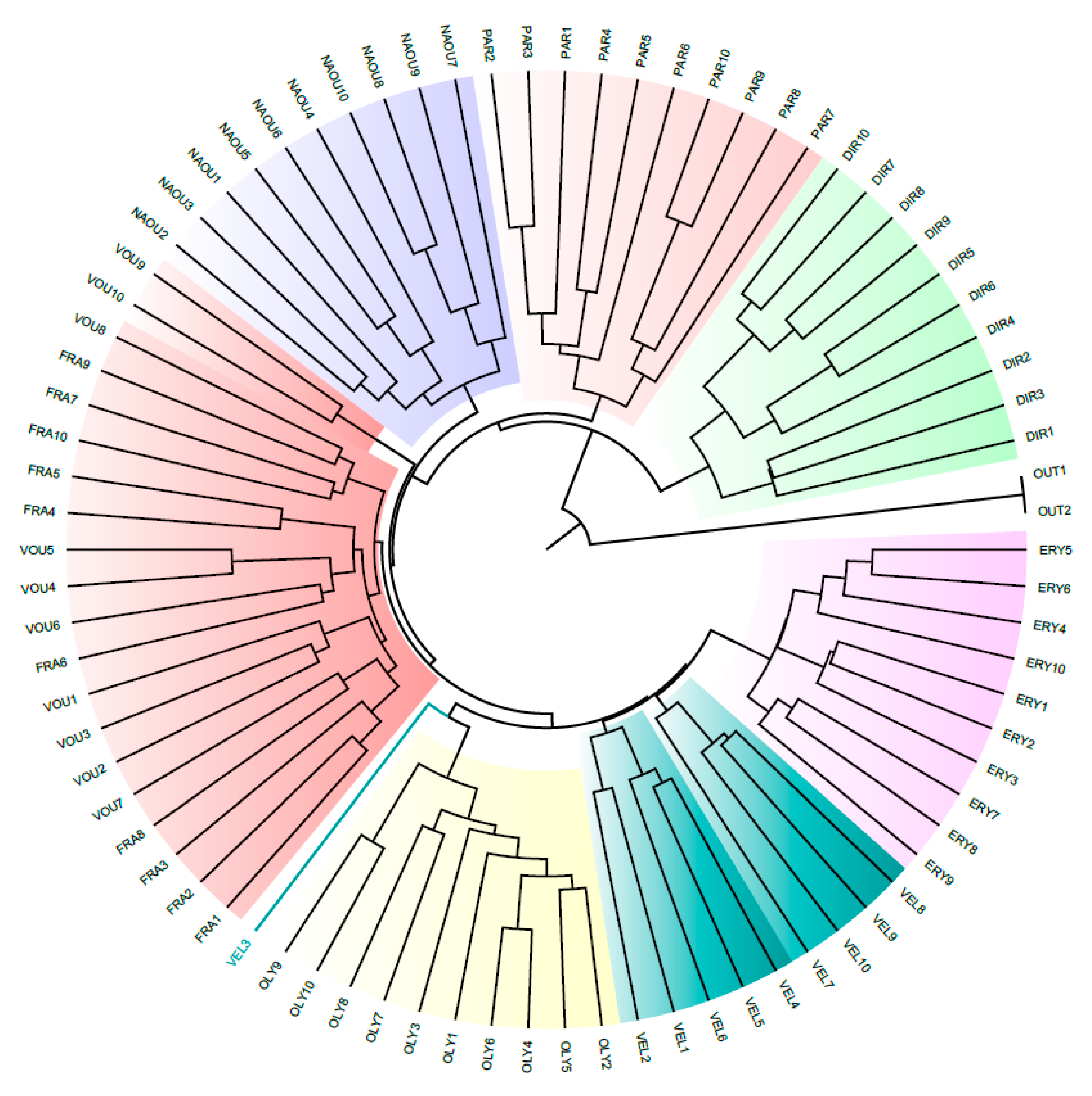
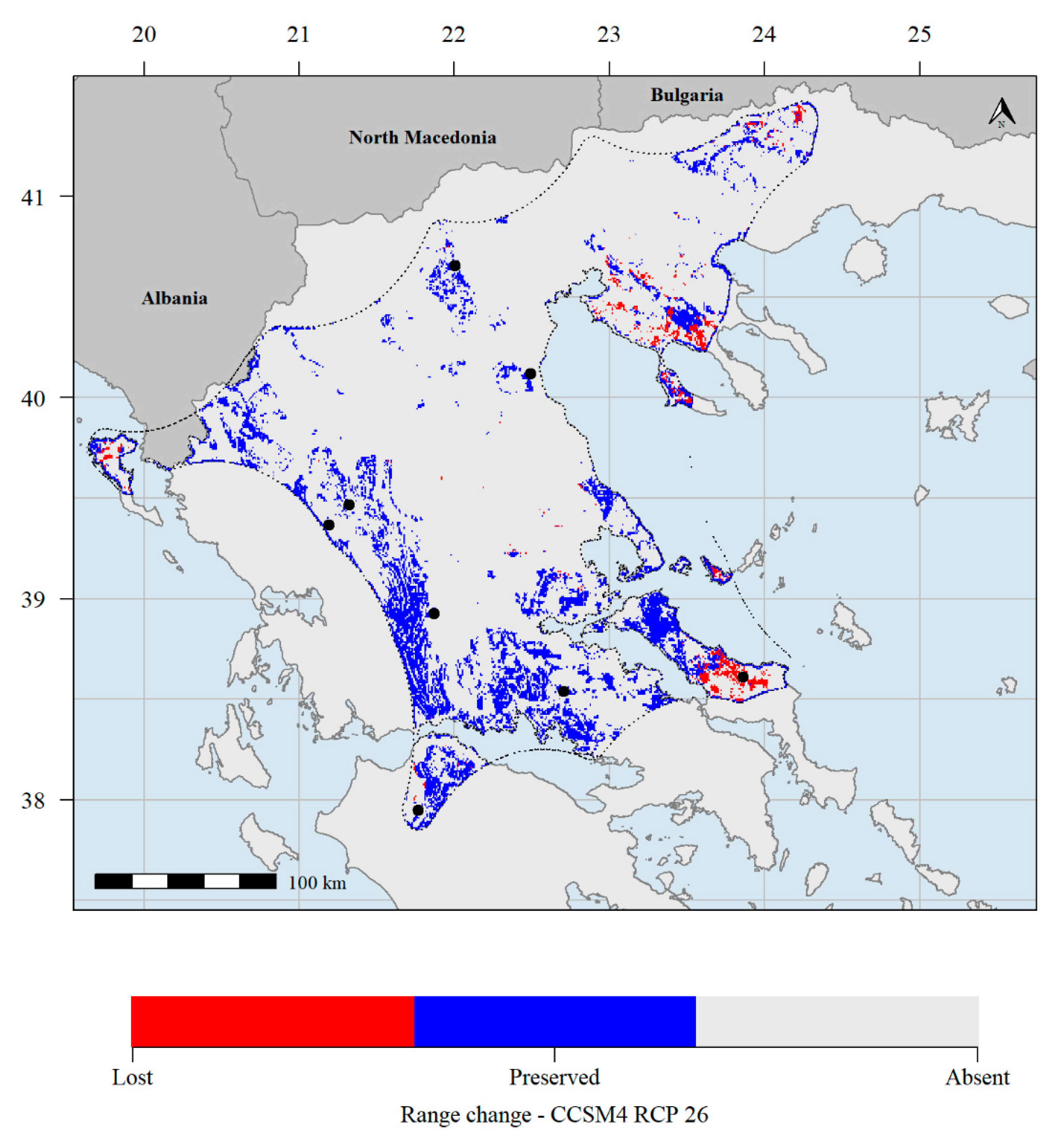
| Population | Population ID | Latitude | Longitude | Altitude | Part of Greece | Habitat |
|---|---|---|---|---|---|---|
| Erymanthos | ERY | 37.9504 | 21.7669 | 950 m | West | Forest openings with Abies cephalonica |
| Velouchi | VEL | 38.9285 | 21.8683 | 1400 m | West | Shrubs |
| Parnassos | PAR | 38.5397 | 22.7038 | 700 m | East | Forest openings with Abies cephalonica |
| Vourgareli | VOU | 39.3684 | 21.1919 | 750 m | West | Forest edges with Fagus sylvatica & Abies borisii-regis |
| Fragma Mesachoras | FRA | 39.4683 | 21.3199 | 850 m | West | Forest edges with Fagus sylvatica & Abies borisii-regis |
| Naousa | NAOU | 40.6548 | 22.0024 | 1050 m | East | Forest edges with Fagus |
| Olympos | OLY | 40.1178 | 22.4881 | 400 m | East | Forest openings with Quercus ilex & Q. rotundifolia |
| Dirfi | DIR | 38.6112 | 23.8602 | 1100 m | East | Forest openings with Abies cephalonica |
| Primer ID | Primer Sequence (5′–3′) | Tm (°C) | No of Bands Scored | Range (bp) |
|---|---|---|---|---|
| UBC-807 | (AG)₈T | 52 | 19 | 420–2000 |
| UBC-811 | (GA)₈T | 52 | 22 | 430–1980 |
| UBC-819 | (GT)₈A | 52 | 9 | 650–1190 |
| UBC-821 | (GT)₈T | 48.6 | 7 | 950–1780 |
| UBC-822 | (CT)₈A | 45.7 | 13 | 680–2000 |
| UBC-826 | (AC)₈C | 52 | 14 | 670–1800 |
| UBC-840 | (GA)₈YT | 45.7 | 17 | 470–1500 |
| UBC-845 | (CT)₈RG | 45.7 | 10 | 1000–2400 |
| UBC-869 | (GTT)₆ | 51 | 9 | 1100–2470 |
| UBC-880 | (GGAG)4 | 51 | 14 | 470–1560 |
| Population ID | N | No.B | PPB (%) | PrB | I | Hj–HWE | Hj–non HWE | Nm |
|---|---|---|---|---|---|---|---|---|
| ERY | 10 | 87 | 55.22 | 13 | 0.3167 ± 0.026 | 0.2719 ± 0.017 | 0.2317 ± 0.018 | |
| VEL | 10 | 115 | 76.87 | 12 | 0.3951 ± 0.022 | 0.3319 ± 0.013 | 0.3490 ± 0.015 | |
| PAR | 10 | 98 | 66.42 | 9 | 0.3368 ± 0.024 | 0.2781 ± 0.015 | 0.2910 ± 0.017 | |
| VOU | 10 | 112 | 75.37 | 11 | 0.3843 ± 0.023 | 0.3208 ± 0.014 | 0.3395 ± 0.016 | |
| FRA | 10 | 111 | 75.37 | 10 | 0.3788 ± 0.023 | 0.3131 ± 0.014 | 0.3274 ± 0.016 | |
| NAOU | 10 | 108 | 71.64 | 12 | 0.3745 ± 0.024 | 0.3171 ± 0.015 | 0.3214 ± 0.017 | |
| OLY | 10 | 84 | 43.28 | 26 | 0.2322 ± 0.025 | 0.2239 ± 0.016 | 0.2010 ± 0.018 | |
| DIR | 10 | 94 | 58.96 | 15 | 0.2983 ± 0.024 | 0.2613 ± 0.016 | 0.2592 ± 0.018 | |
| Mean | 0.3396 ± 0.009 | 0.2898 ± 0.013 | 0.2901 ± 0.019 | 1.3098 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fassou, G.; Kougioumoutzis, K.; Iatrou, G.; Trigas, P.; Papasotiropoulos, V. Genetic Diversity and Range Dynamics of Helleborus odorus subsp. cyclophyllus under Different Climate Change Scenarios. Forests 2020, 11, 620. https://doi.org/10.3390/f11060620
Fassou G, Kougioumoutzis K, Iatrou G, Trigas P, Papasotiropoulos V. Genetic Diversity and Range Dynamics of Helleborus odorus subsp. cyclophyllus under Different Climate Change Scenarios. Forests. 2020; 11(6):620. https://doi.org/10.3390/f11060620
Chicago/Turabian StyleFassou, Georgia, Konstantinos Kougioumoutzis, Gregoris Iatrou, Panayiotis Trigas, and Vasileios Papasotiropoulos. 2020. "Genetic Diversity and Range Dynamics of Helleborus odorus subsp. cyclophyllus under Different Climate Change Scenarios" Forests 11, no. 6: 620. https://doi.org/10.3390/f11060620
APA StyleFassou, G., Kougioumoutzis, K., Iatrou, G., Trigas, P., & Papasotiropoulos, V. (2020). Genetic Diversity and Range Dynamics of Helleborus odorus subsp. cyclophyllus under Different Climate Change Scenarios. Forests, 11(6), 620. https://doi.org/10.3390/f11060620








