Development and Characterization of Simple Sequence Repeat Markers for, and Genetic Diversity Analysis of Liquidambar formosana
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Transcriptome Assembly, SSR Mining, and Primer Design
2.3. DNA Extraction, PCR Amplification, and Detection of Polymorphisms
2.4. Genetic Diversity of Plus Trees of L. formosana, and Statistical Analysis
3. Results
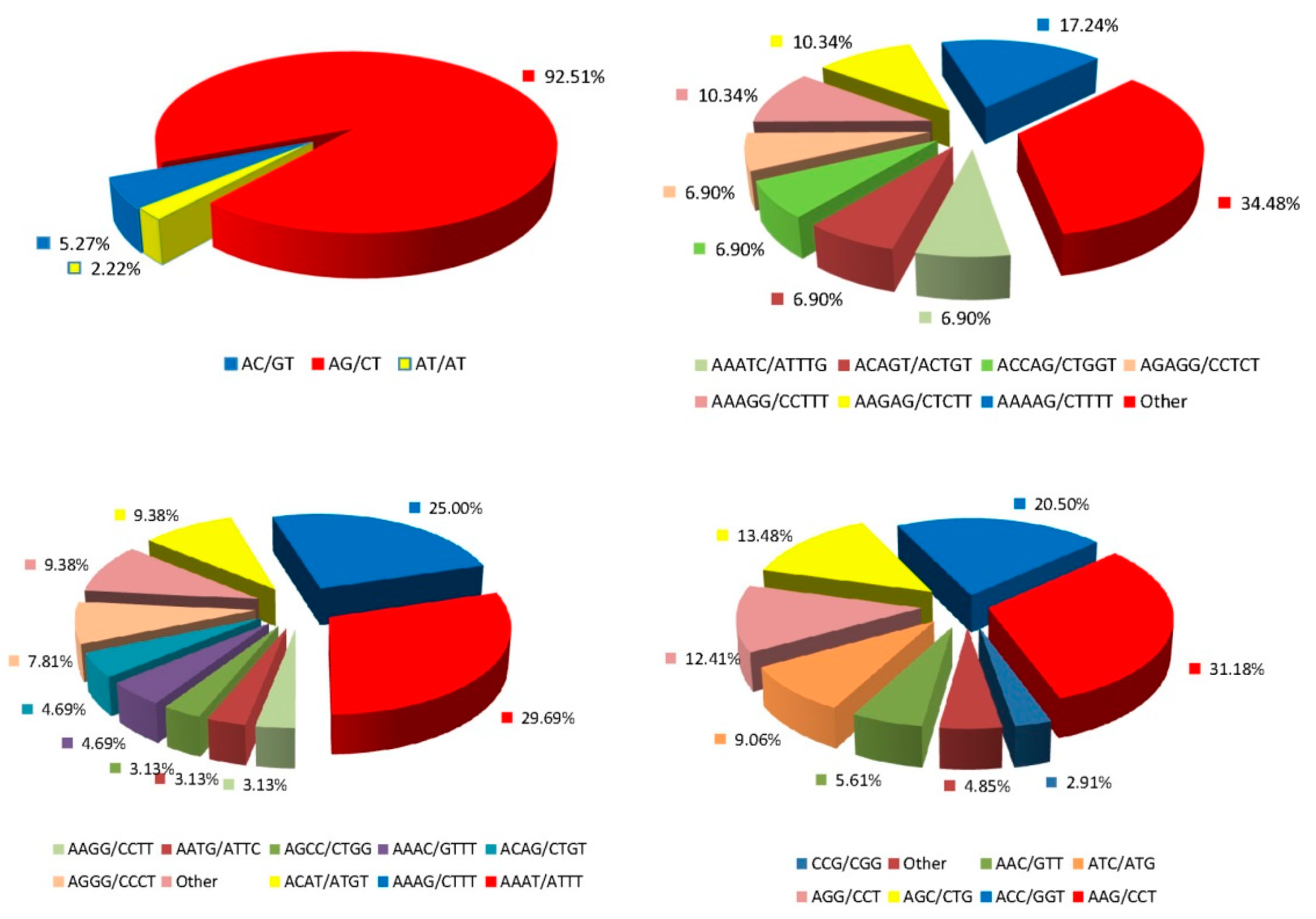
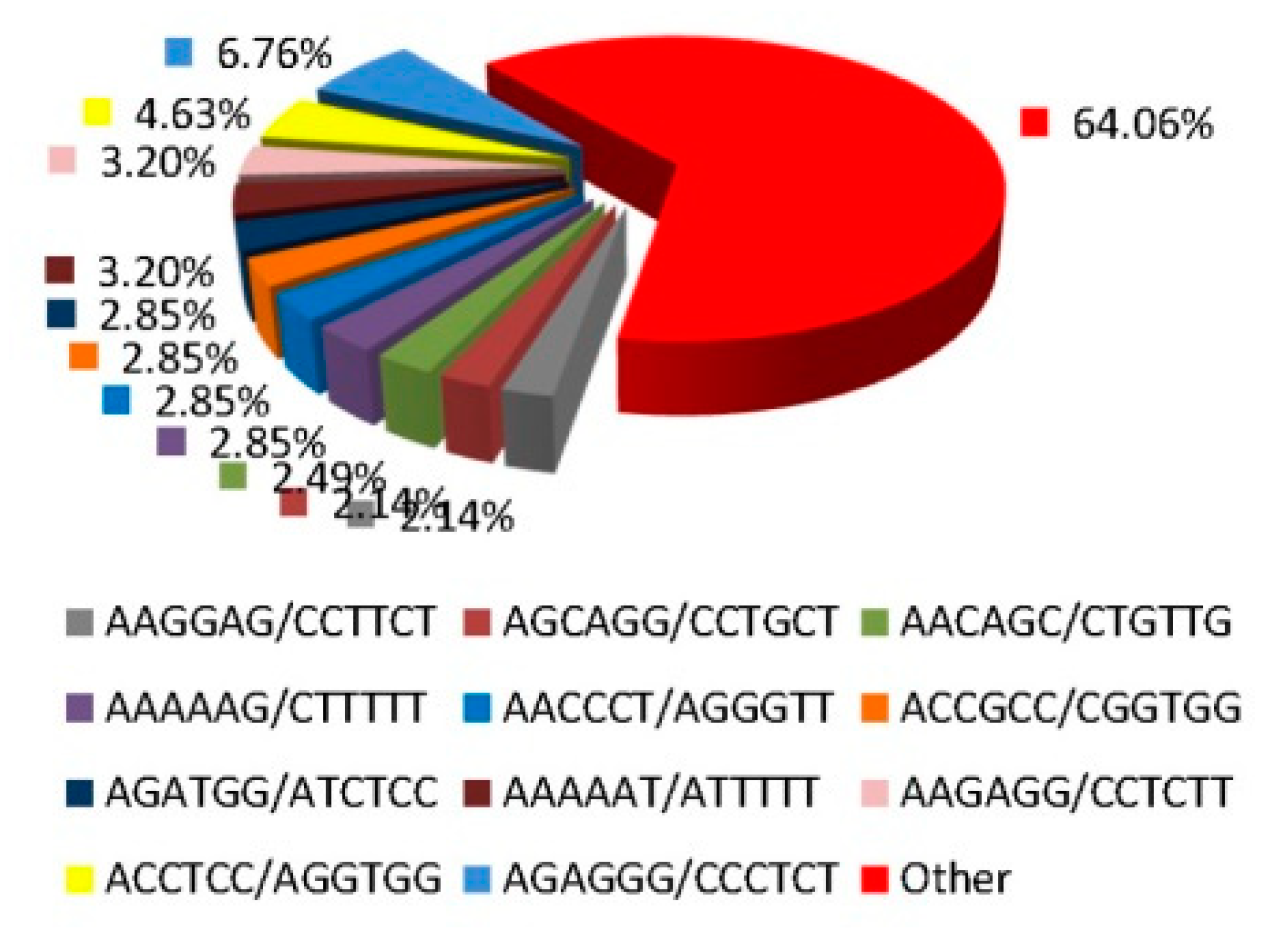
3.1. The Frequency and Distribution of SSR Loci
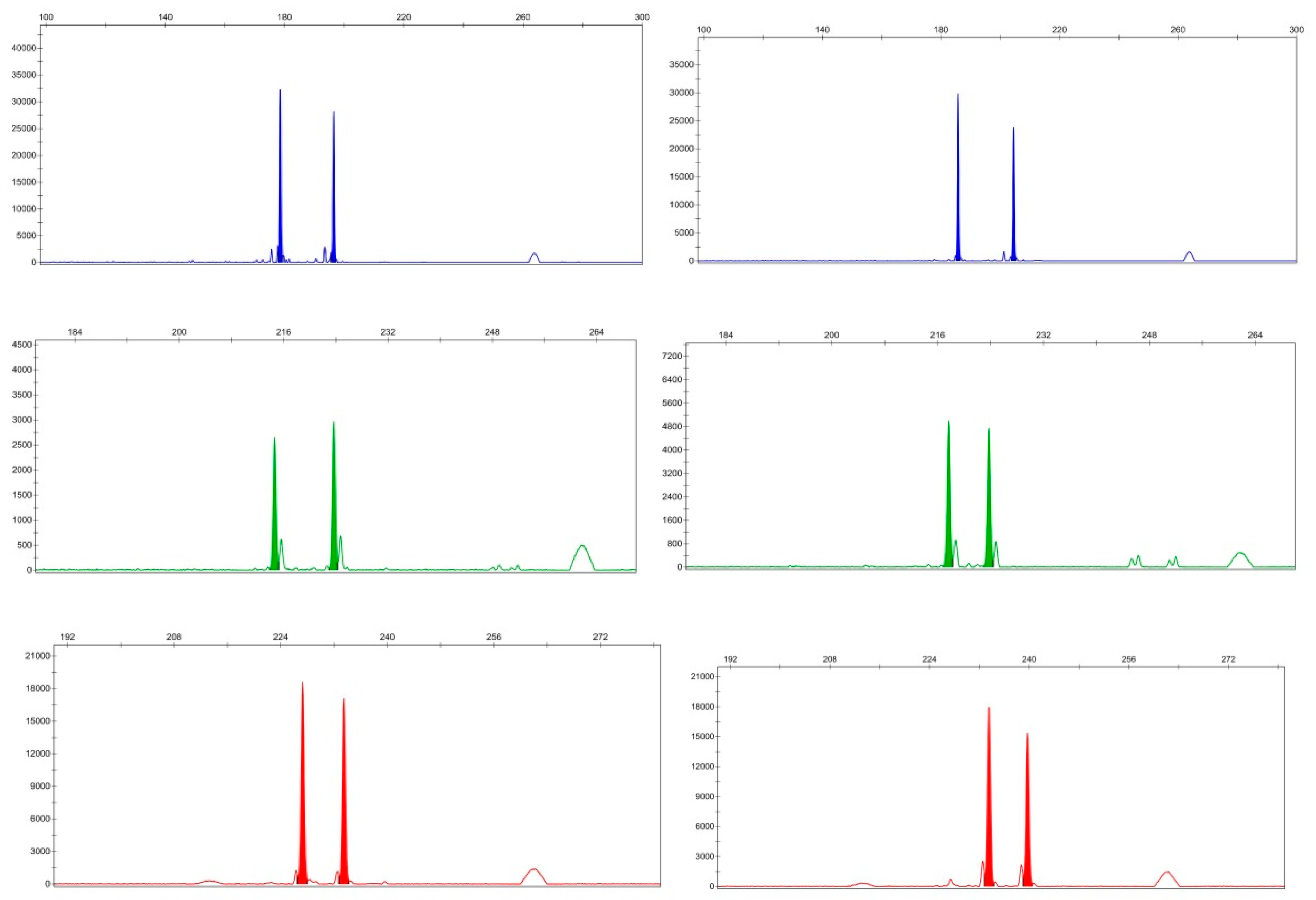
3.2. Synthesis, Screening, and Polymorphisms of EST-SSR Primers
3.3. Genetic Diversity of Plus Trees of L. Formosana
3.4. Cross-Transferability of EST-SSR Primers
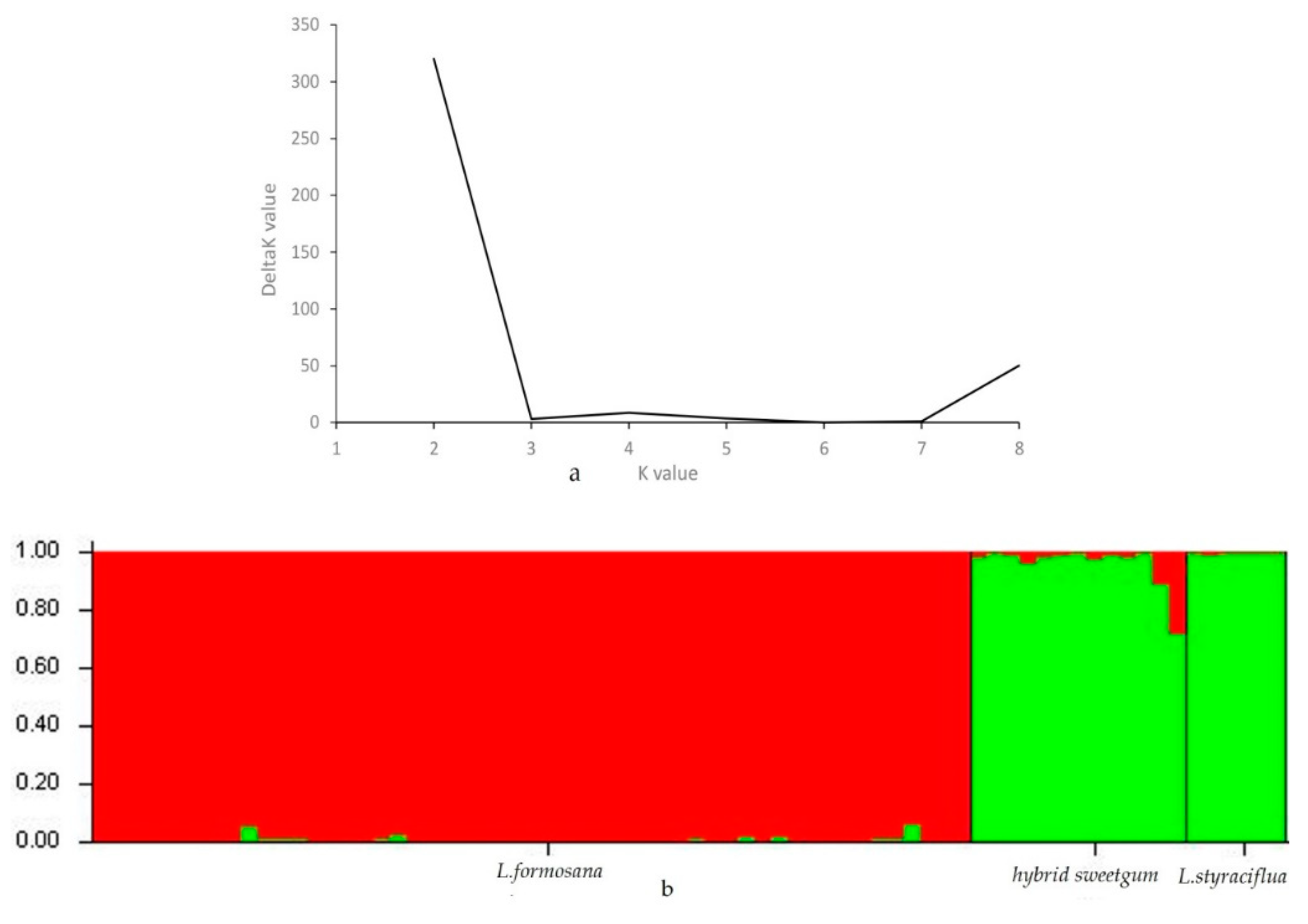
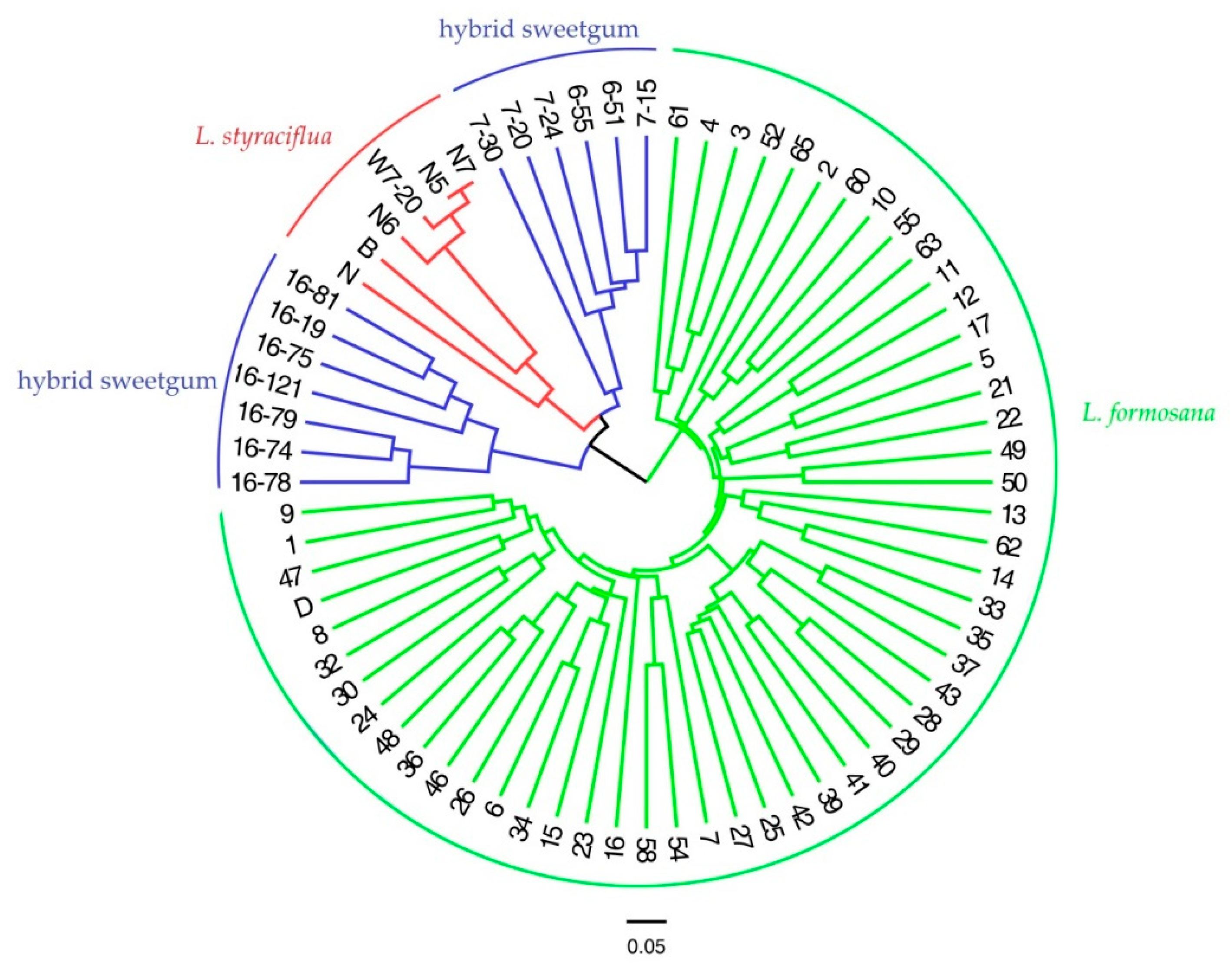
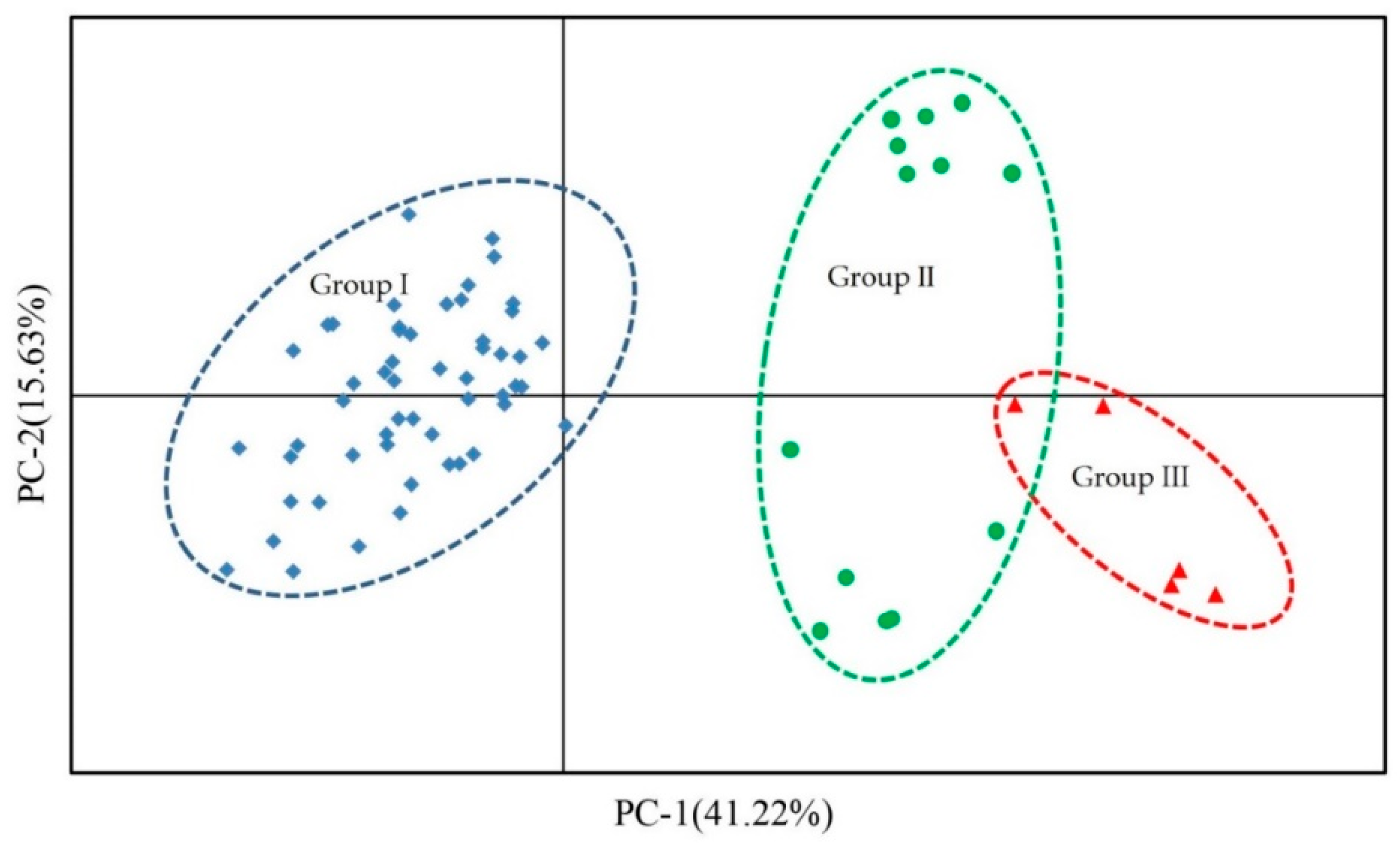
3.5. Population Genetic Structure Analysis, Cluster Analysis, and PCA of Three Species of Liquidambar
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhou, G.Y. Chinese sweetgum. J. Ningbo Univ. 1995, 2, 34–41. [Google Scholar]
- Zheng, Y.Q.; Pan, B.; Itohl, T. Chemical induction of traumatic gum ducts in Chinese sweetgum, Liquidambar formosana. IAWA J. 2015, 36, 58–68. [Google Scholar] [CrossRef]
- Merkle, S.A.; Neu, K.A.; Battle, P.J.; Bailey, R.L. Somatic embryogenesis and plantlet regeneration from immature and mature tissues of sweetgum (Liquidambar styraciflua). Plant Sci. 1998, 132, 169–178. [Google Scholar] [CrossRef]
- Kathleen, B.P.; Ickert-Bond, S.M.; Wen, J. Anatomically preserved Liquidambar (Altingiaceae) from the middle Miocene of Yakima Canyon, Washington state, USA, and its biogeographic implications. Am. J. Bot. 2004, 91, 499–509. [Google Scholar]
- Jiang, P.C. A survey of studies on Liquidambar formosana. Guangxi Med. J. 1998, 20, 241–242. [Google Scholar]
- Zeng, T.; Song, X. Study on the Methods of the Tapping from Chinese SweetGum. China For. Sci. Technol. 2010, 24, 84–87. [Google Scholar]
- Weng, L.L.; Jiang, J.D. The research progresses and prospects of local species Liquidamba formosana. J. Fujian For. Sci. Technol. 2007, 34, 184–189. [Google Scholar]
- Winstead, J.E. Populational Differences in Seed Germination and Stratification Requirements of Sweetgum. For. Sci. 1971, 17, 34–36. [Google Scholar]
- Suhaendi, H. Seed germination and seedling characteristics of twelve greenhouse grown provenances of L. Styraciflua. Buletin Penelitian Hutan 1989, 520, 1–18. [Google Scholar]
- Kariuki, J.G. Interim results of a provenancetrial of liquidambar styraciflua in Kenya. Kenya For. Res. Inst. 1989, 5, 1–17. [Google Scholar]
- He, G.P.; Chen, Y.T.; Tang, X.Y.; Shen, G.P.; Gao, S.M. A Study on Variations of Young Forest Growing Traits in Liquidambar formosana Geographic Provenances. Acta Agric. Univ. Jiangxiensis 2005, 27, 585–589. [Google Scholar]
- Shi, J.L.; Zhu, S.L.; Ma, K.X.; Zhu, J.P. Study on multi-objective breeding of Liquidambar formosana. For. Sci. Technol. 1997, 10, 13–14. [Google Scholar]
- Zhao, Y.; Wang, X.R.; Tang, R.H. The elite tree selection of Liquidambar formosana in Guizhou Province. J. Mt. Agric. Biol. 2009, 28, 530–534. [Google Scholar]
- Cornelius, J. The effectiveness of plus-tree selection for yield. For. Ecol. Manag. 1994, 67, 23–34. [Google Scholar] [CrossRef]
- Xu, Y.B.; Crouch, J.H. Marker-assisted selection in plant breeding: From publications to practice. Crop. Sci 2008, 48, 391–407. [Google Scholar] [CrossRef]
- Vendrame, W.A.; Holliday, C.P.; Merkle, S.A. Clonal propagation of hybrid sweetgum (Liquidambar styraciflua × L. formosana) by somatic embryogenesis. Plant Cell Rep. 2001, 20, 691–695. [Google Scholar] [CrossRef]
- Bi, X.Q.; Jin, X.Z.; Li, H.J. Genetic diversity in the natural populations of Liquidambar fornosana revealed by ISSR molecular markers. Bull. Bot. Res. 2010, 30, 120–125. [Google Scholar]
- Sun, R.; Lin, F.; Huang, P.; Zheng, Y. Moderate Genetic Diversity and Genetic Differentiation in the Relict Tree Liquidambar formosana Hance Revealed by Genic Simple Sequence Repeat Markers. Front. Plant Sci. 2016, 7, 1411. [Google Scholar] [CrossRef]
- Yin-Gang, L.I.; Zhou, X.P.; Liu, X.H.; Hua, Z.H.; Liu, W.; Liu, J.L.; Xie, J.Q. Preliminary Study on Growth of Styrax tonkinensis Plantation and Plus Tree Seletion. J. Zhejiang For. Sci. Technol. 2010, 30, 24–28. [Google Scholar]
- Kuang, H.; Richardson, T.; Carson, S.; Wilcox, P.; Bongarten, B. Genetic analysis of inbreeding depression in plus tree 850.55 of Pinus radiata D. Don. I. Genetic map with distorted markers. Theor. Appl. Genet. 1999, 98, 697–703. [Google Scholar] [CrossRef]
- Li, D.; Deng, Z.; Qin, B.; Liu, X.; Men, Z. De novoassembly and characterization of bark transcriptome using Illumina sequencing and development of EST-SSR markers in rubber tree (Hevea brasiliensis Muell. Arg.). BMC Genom. 2012, 13, 192. [Google Scholar] [CrossRef] [PubMed]
- Colburn, B.C.; Mehlenbacher, S.A.; Sathuvalli, V.R. Development and mapping of microsatellite markers from transcriptome sequences of European hazelnut (Corylus avellana L.) and use for germplasm characterization. Mol. Breed. 2017, 37, 16. [Google Scholar] [CrossRef]
- Aggarwal, R.K.; Hendre, P.S.; Varshney, R.K.; Bhat, P.R.; Krishnakumar, V.; Singh, L. Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor. Appl. Genet. 2007, 114, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Zalapa, J.E.; Cuevas, H.; Zhu, H.; Steffan, S.; Senalik, D.; Zeldin, E.; McCown, B.; Harbut, R.; Simon, P. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am. J. Bot. 2012, 99, 193–208. [Google Scholar] [CrossRef]
- Wei, W.; Wang, C.B.; Fang, L.; Chen, H.D.; Lin, W.; Wang, C.Y.; Zhang, X.D.; Wang, Y.H.; Kun-Bo, A.W. Development and Evaluation of New Non-Redundant EST-SSR Markers from Gossypium. Acta Agron. Sin. 2012, 38, 1443–1451. [Google Scholar]
- Xu, S.C.; Gong, Y.-M.; Mao, W.-H.; Hu, Q.-Z.; Zhang, G.-W.; Fu, W.; Xian, Q.-Q. Development and characterization of 41 novel EST-SSR markers for Pisum sativum (Leguminosae). Am. J. Bot. 2012, 99, e149–e153. [Google Scholar] [CrossRef]
- Yadong, Z.; Hu, X.; Zhang, Y.D.; Song, C.; Hu, Y.X.; Song, C.W. Identification of Populus Varieties from Hubei Province by EST-SSR Marker. Mol. Plant. Breed. 2009, 7, 105–109. [Google Scholar]
- Zhang, Y.; Wang, Z.W.; Qi, S.Z.; Wang, X.Q.; Zhao, J.; Zhang, J.F.; Li, B.l.; Zhang, Y.D.; Liu, X.Z.; Yuan, W. In vitro tetraploid induction from leaf and petiole explants of hybrid sweetgum (Liquidambar styraciflua × Liquidambar formosana). Forests 2017, 8, 264. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 27 April 2017). [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, C. Glaubitz convert: A user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol. Ecol. Resour. 2004, 4, 309–310. [Google Scholar]
- Popgene. Version 1.32. The User-Friendly Shareware for Population Genetic Analysismolecular Biology and Biotechnology Center, University of AlbertaEdmonton. Available online: http://www.ualberta.ca/~{}fyeh (accessed on 23 November 2017).
- Nagy, S.; Poczai, P.; Cernák, I.; Taller, J. PICcalc: An Online Program to Calculate Polymorphic Information Content for Molecular Genetic Studies. Biochem. Genet. 2012, 50, 670–672. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2005, 6, 288–295. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Q.; Huai, H.; Liu, A. Development of EST-SSR markers and investigation of genetic relatedness in tung tree. Tree Genet. Genomes 2012, 8, 933–940. [Google Scholar] [CrossRef]
- Wu, H.; Chen, D.; Li, J.; Yu, B.; Qiao, X. De Novo Characterization of Leaf Transcriptome Using 454 Sequencing and Development of EST-SSR Markers in Tea (Camellia sinensis). Plant. Mol. Biol. Rep. 2013, 31, 524–538. [Google Scholar] [CrossRef]
- Xiang, X.; Zhang, Z.; Wang, Z.; Zhang, X.; Wu, G. Transcriptome sequencing and development of EST-SSR markers in Pinus dabeshanensis, an endangered conifer endemic to China. Mol. Breed. 2015, 35, 158. [Google Scholar] [CrossRef]
- Dong, M.; Wang, Z.; He, Q.; Zhao, J.; Fan, Z.; Zhang, J. Development of EST-SSR markers in Larix principis-rupprechtii Mayr and evaluation of their polymorphism and cross-species amplification. Trees 2018, 32, 1559–1571. [Google Scholar] [CrossRef]
- Dong, L.; Sun, Y.H.; Zhao, J.; Zhang, Y.W.; Li, X.Y. Development and Application of EST-SSR Markers for DNA Fingerprinting and Genetic Diversity Analysis of the Main Cultivars of Black Locust (Robinia pseudoacacia L.) in China. Forests 2019, 10, 644. [Google Scholar] [CrossRef]
- Du, Q.; Wang, B.; Wei, Z.; Zhang, D.; Li, B. Genetic Diversity and Population Structure of Chinese White Poplar (Populus tomentosa) Revealed by SSR Markers. J. Hered. 2012, 103, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Nirapathpongporn, K.; Kongsawadworakul, P.; Viboonjun, U.; Teerawattanasuk, K.; Chrestin, H.; Segiun, M.; Clément-Dement, A.; Narangajavana, J. Development and mapping of functional expressed sequence tag-derived simple sequence repeat markers in a rubber tree RRIM600 × PB217 population. Mol. Breed. 2016, 36, 39. [Google Scholar] [CrossRef]
- Ge, Y.; Tan, L.; Wu, B.; Wang, T. Transcriptome Sequencing of Different Avocado Ecotypes: de novo Transcriptome Assembly, Annotation, Identification and Validation of EST-SSR Markers. Forests 2019, 10, 411. [Google Scholar] [CrossRef]
- Liu, F.M.; Hong, Z.; Yang, Z.J.; Zhang, N.N. De Novo Transcriptome Analysis of Dalbergia odorifera T. Chen (Fabaceae) and Transferability of SSR Markers Developed from the Transcriptome. Forests 2019, 10, 99. [Google Scholar]
- Guo, Q.; Wang, J.-X.; Su, L.-Z.; Lv, W.; Sun, Y.-H.; Li, Y. Development and evaluation of a novel set of EST-SSR markers based on transcriptome sequences of Black Locust (Robinia pseudoacacia L.). Genes 2017, 8, 177. [Google Scholar] [CrossRef]
- Cheng, Y.L.; Yang, Y.; Wang, Z.Y.; Qi, B.Y.; Yin, Y.L.; Li, H.G. Development and Characterization of EST-SSR Markers in Taxodium ‘zhongshansa’. Plant. Mol. Biol. Rep. 2015, 33, 1–11. [Google Scholar] [CrossRef]
- Jia, X.; Deng, Y.; Sun, X.; Liang, L.; Su, J. De novo assembly of the transcriptome of Neottopteris nidus using Illumina paired-end sequencing and development of EST-SSR markers. Mol. Breed. 2016, 36, 94. [Google Scholar] [CrossRef]
- Chen, L.Y.; Cao, Y.N.; Yuan, N.; Koh, N.; Wang, G.M.; Qiu, Y.X. Characterization of transcriptome and development of novel EST-SSR makers based on next-generation sequencing technology in Neolitsea sericea (Lauraceae) endemic to East Asian land-bridge islands. Mol. Breed. 2015, 35, 187. [Google Scholar] [CrossRef]
- Postolache, D.; Leonarduzzi, C.; Piotti, A.; Spanu, I.; Vendramin, G.G. Transcriptome versus Genomic Microsatellite Markers: Highly Informative Multiplexes for Genotyping Abies alba Mill. and Congeneric Species. Plant. Mol. Biol. Rep. 2013, 32. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Liu, S.; Liu, H.; Wu, A.; Hou, Y.; An, Y.; Wei, C. Construction of fingerprinting for tea plant (Camellia sinensis) accessions using new genomic SSR markers. Mol. Breed. 2017, 37, 93. [Google Scholar] [CrossRef]
- Durand, J.; Bodénès, C.; Chancerel, E.; Frigerio, J.-M.; Vendramin, G.; Sebastiani, F.; Buonamici, A.; Gailing, O.; Koelewijn, H.-P.; Villani, F.; et al. A fast and cost-effective approach to develop and map EST-SSR markers: oak as a case study. BMC Genom. 2010, 11, 570. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Wang, H.; Xia, Z.; Zou, M.; Lu, C.; Wang, W. Developmenrt of EST-SSR and genomic-SSR markers to assess genetic diversity in Jatropha Curcas L. BMC Res. Notes 2010, 3, 42. [Google Scholar] [CrossRef] [PubMed]
- Merkle, S.; Montello, P.; Kormanik, T.; Le, H. Propagation of novel hybrid sweetgum phenotypes for ornamental use via somatic embryogenesis. Propag. Ornam. Plant. 2010, 10, 220–226. [Google Scholar]
| Sequence Type | Total |
|---|---|
| Co-detected Unigenes Length (kbp) | 24,122 |
| Average Length of Unigenes (bp) | 699 |
| Number of SSR loci detected | 3284 |
| Number of Unigenes Containing SSR Loci | 2949 |
| Number of Unigenes with more than one locus | 298 |
| Number of complex SSR loci | 109 |
| Frequency of SSR loci | 1/7.3 kbp |
| Repeat Motif | Repeat Number | Percentage (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | >11 | Total | ||
| Dinucleotide | 344 | 298 | 209 | 590 | 1441 | 52.56 | |||||
| Trinucleotide | 427 | 234 | 104 | 80 | 43 | 7 | 32 | 927 | 33.8 | ||
| Tetranucleotide | 47 | 10 | 4 | 3 | 64 | 2.33 | |||||
| Pentanucleotid-e | 20 | 9 | 29 | 0.88 | |||||||
| Hexonucleotid-e | 205 | 57 | 11 | 6 | 1 | 1 | 281 | 10.58 | |||
| Total | 205 | 124 | 457 | 244 | 108 | 425 | 341 | 216 | 622 | 2742 | 100 |
| Percentage (%) | 7.48 | 4.52 | 16.67 | 8.9 | 3.94 | 15.5 | 12.44 | 7.88 | 22.68 | 100 | 52.56 |
| Code | Locus | Primer Sequence | Repeat Motif | Tm (°C) | Product Size (bp) | Dye |
|---|---|---|---|---|---|---|
| 1 | Liq_eSSR1 | F: TGTAAAACGACGGCCAGTCATCATCAGACACGGTTTGG | (TTAGGG)4 | 60 | 217 | HEX |
| R:GTCCAGCTCCGTACATCCAT | ||||||
| 2 | Liq_eSSR2 | F: TGTAAAACGACGGCCAGTATGGGGACGATGACTGTGTT | (ACGGCA)4 | 60 | 200 | ROX |
| R: CGTTGCAAATCTCATTGACG | ||||||
| 3 | Liq_eSSR4 | F: TGTAAAACGACGGCCAGTAGTGGTTGTTGCTGCTGTTG | (CTGGTG)7 | 60 | 206 | FAM |
| R: CTTATGCAGCCCCAAATGTT | ||||||
| 4 | Liq_eSSR7 1 | F: TGTAAAACGACGGCCAGTCAAGTAGTCCAGGGCGTCTC | (CTCTTC)4 | 60 | 192 | ROX |
| R: CCTAACGAGAAGGGAGGCTT | ||||||
| 5 | Liq_eSSR10 1 | F: TGTAAAACGACGGCCAGTAGACCACATGCTCGTTACCC | (CTAGCA)5 | 60 | 142 | FAM |
| R: TGCTGGAACTGGTGCTAGTG | ||||||
| 6 | Liq_eSSR13 | F: TGTAAAACGACGGCCAGTTCTGGGTCAGGTTCAGGTTC | (GGTTCA)4 | 60 | 24 | TAMRA |
| R: GCCATATCCCTCACCATGTC | ||||||
| 7 | Liq_eSSR16 | F: TGTAAAACGACGGCCAGTTGGCAGATCTAGGGGATTTG | (GAGGTG)4 | 60 | 203 | FAM |
| R: AACCTCCATCTCCACGATTG | ||||||
| 8 | Liq_eSSR17 1 | F: TGTAAAACGACGGCCAGTAGAGGTGAATGGGACACCAG | (GGATGA)4 | 60 | 206 | HEX |
| R: CTGCCAACCTGGGAATAGAA | ||||||
| 9 | Liq_eSSR25 1 | F: TGTAAAACGACGGCCAGTTCAACTTGATGGGGTCAGTG | (TATG)6 | 60 | 194 | ROX |
| R: GACTGCCCGTACCCAGAATA | ||||||
| 10 | Liq_eSSR28 | F: TGTAAAACGACGGCCAGTACATCAGAAGGAGCCGAAGA | (GGT)6 | 60 | 175 | HEX |
| R: CGGATTGGCAAGAGAAGAAG | ||||||
| 11 | Liq_eSSR30 1 | F: TGTAAAACGACGGCCAGTAGGCAGGGTTAGTCCCTTGT | (CTG)6 | 60 | 224 | ROX |
| R: GCTTGCGGACTTGTTTTCTC | ||||||
| 12 | Liq_eSSR33 1 | F: TGTAAAACGACGGCCAGTGAGGGAGATTCACCACAGGA | (GCA)8 | 60 | 212 | HEX |
| R: TCATCATCAAGTCCGCCATA | ||||||
| 13 | Liq_eSSR34 1 | F: TGTAAAACGACGGCCAGTGCTGCCTCCAAAGAAGTGTC | (GCT)7 | 60 | 199 | ROX |
| R: AGGGCTCCTCAGCTAGTTCC | ||||||
| 14 | Liq_eSSR35 1 | F: TGTAAAACGACGGCCAGTTTACCACCGTCGTCGTCAT | (TCC)8 | 60 | 165 | FAM |
| R: GAACCAAGAGAAGCACCAGC | ||||||
| 15 | Liq_eSSR42 | F: TGTAAAACGACGGCCAGTCACATCGGAAGCTCCTTCTC | (TC)10 | 60 | 201 | HEX |
| R: GCGACCCACAATACTAGGGA | ||||||
| 16 | Liq_eSSR43 | F: TGTAAAACGACGGCCAGTCCAAAGCTTCCTCCTCCTCT | (CT)12 | 60 | 24 | ROX |
| R: ACATTTCTTCCGCATTGACC | ||||||
| 17 | Liq_eSSR44 | F: TGTAAAACGACGGCCAGTCTTGGAATCACCTTCGCATT | (CA)9 | 60 | 198 | FAM |
| R: CGATCGAATTGTAAAGCGGT | ||||||
| 18 | Liq_eSSR46 | F: TGTAAAACGACGGCCAGTTCTCCATGGTCCTTTCTTGG | (AGAGA)6 | 60 | 162 | ROX |
| R: CCAAACAGGGTGGAGAGAAA | ||||||
| 19 | Liq_eSSR48 | F: TGTAAAACGACGGCCAGTTCGATGATGAGAACACTGGAA | (GATTT)6 | 60 | 163 | FAM |
| R: ATCCGATAATAACGTCCCCC | ||||||
| 20 | Liq_eSSR51 | F: TGTAAAACGACGGCCAGTGAGAATGGCAGAGGAAGCAC | (TGT)9 | 60 | 197 | ROX |
| R: AAGCCATGAAGAGAAGGCAA | ||||||
| 21 | Liq_eSSR55 1 | F: TGTAAAACGACGGCCAGTCCTTCTTTGGATCTCCACCA | (CTC)8 | 60 | 194 | ROX |
| R: TTGCACTTTGACTGAGACCG | ||||||
| 22 | Liq_eSSR59 1 | F: TGTAAAACGACGGCCAGTCAAACGAAAGATGAGCGACA | (CAA)6 | 60 | 201 | ROX |
| R: CCGTTTTCATGGACCACTCT | ||||||
| 23 | Liq_eSSR60 1 | F: TGTAAAACGACGGCCAGTGAGACCTGGTTGGACTTGGA | (GGA)6 | 60 | 216 | HEX |
| R: CGGAGAATGACCGGTCTAAA | ||||||
| 24 | Liq_eSSR62 1 | F: TGTAAAACGACGGCCAGTCCTCGAACTGTTTCTCCAGC | (AGG)6 | 60 | 202 | FAM |
| R: CGCTCCTTAAACCCTAACCC | ||||||
| 25 | Liq_eSSR64 | F: TGTAAAACGACGGCCAGTCGGTACATGTGATCAGCCAC | (GAA)7 | 60 | 169 | ROX |
| R: CTTCTGCTTCTCCAAATGGC | ||||||
| 26 | Liq_eSSR67 1 | F: TGTAAAACGACGGCCAGTCCAAGGAGAGAGTGACGAGG | (CCT)8 | 60 | 273 | ROX |
| R: ATTGATTTTTGCGGACTTGG | ||||||
| 27 | Liq_eSSR70 | F: TGTAAAACGACGGCCAGTTGGGGGAATGAGAGTTTGAC | (GTG)7 | 60 | 174 | HEX |
| R: ATCTCAGTGGGCTCTCCTCA | ||||||
| 28 | Liq_eSSR86 1 | F: TGTAAAACGACGGCCAGTCTGAAAATAAGGCGGGTGAA | (TC)9 | 60 | 172 | HEX |
| R: GTCTGGGTCATCTTCGTCGT | ||||||
| 29 | Liq_eSSR89 | F: TGTAAAACGACGGCCAGTATCGTCCAGGCTTATCATCG | (GTT)7gatc(GGCTGT)5 | 60 | 201 | FAM |
| R: GCCTCTGCTGACACTGACAA | ||||||
| 30 | Liq_eSSR90 1 | F: TGTAAAACGACGGCCAGTCCATTGTTCCCCCTACCTCT | (ACC)6acttctcctatcagcaaagccaccgcg(ACC)7 | 60 | 196 | ROX |
| R: TGCAAGAACGAGAGCAGAGA | ||||||
| 31 | Liq_eSSR92 | F: TGTAAAACGACGGCCAGTAGAGGACACCCAAGGGAACT | (CCCTTG)4gagagagaaagcgatag(GT)11atagcgagagagaa(AG)9 | 60 | 181 | HEX |
| R: CCATTGTTCCCCCTACCTCT | (ACC)6acttctcctatcagcaaagccaccgcg(ACC)7 | 60 | ||||
| 32 | Liq_eSSR98 1 | F: TGTAAAACGACGGCCAGTTCCCAATCTCAATCTCGACC | (CCTCAA)4 | 60 | 220 | ROX |
| R: CCTCCGTAATCCGAAGAACA |
| Locus | Na 1 | Ne 2 | Ho 3 | He 4 | I5 | PIC 6 | HW 7 |
|---|---|---|---|---|---|---|---|
| Liq_eSSR35 | 13 | 8.872 | 0.978 | 0.887 | 2.317 | 0.877 | * |
| Liq_eSSR86 | 10 | 5.25 | 0.925 | 0.81 | 1.882 | 0.785 | ns |
| Liq_eSSR67 | 7 | 4.86 | 0.906 | 0.794 | 1.712 | 0.765 | ns |
| Liq_eSSR90 | 7 | 3.959 | 0.698 | 0.747 | 1.565 | 0.71 | * |
| Liq_eSSR98 | 6 | 3.822 | 0.736 | 0.738 | 1.469 | 0.694 | ns |
| Liq_eSSR25 | 6 | 3.451 | 0.491 | 0.71 | 1.459 | 0.671 | * |
| Liq_eSSR10 | 5 | 3.216 | 0.736 | 0.689 | 1.3 | 0.632 | ns |
| Liq_eSSR30 | 7 | 2.814 | 0.711 | 0.645 | 1.339 | 0.608 | ns |
| LF39 8 | 6 | 2.681 | 0.577 | 0.627 | 1.337 | 0.599 | ns |
| LF17 8 | 4 | 2.825 | 0.774 | 0.646 | 1.097 | 0.572 | ns |
| Liq_eSSR33 | 4 | 2.688 | 0.962 | 0.628 | 1.133 | 0.568 | *** |
| Liq_eSSR62 | 5 | 2.636 | 0.642 | 0.621 | 1.137 | 0.558 | ns |
| LF37 8 | 6 | 2.452 | 0.604 | 0.592 | 1.149 | 0.545 | ns |
| Liq_eSSR60 | 4 | 2.465 | 0.547 | 0.594 | 1.094 | 0.543 | ns |
| Liq_eSSR59 | 3 | 2.485 | 0.64 | 0.598 | 0.998 | 0.528 | ns |
| LF15 8 | 5 | 2.016 | 0.566 | 0.504 | 1 | 0.47 | ns |
| Liq_eSSR17 | 3 | 2.158 | 0.679 | 0.537 | 0.883 | 0.458 | ns |
| Liq_eSSR55 | 7 | 1.683 | 0.457 | 0.406 | 0.911 | 0.388 | ns |
| Liq_eSSR7 | 4 | 1.751 | 0.188 | 0.429 | 0.755 | 0.372 | * |
| Liq_eSSR34 | 3 | 1.286 | 0.245 | 0.222 | 0.454 | 0.211 | ns |
| Mean | 5.75 | 3.169 | 0.653 | 0.621 | 1.25 | 0.578 | ns |
| Locus | hybrid Sweetgum | L. styraciflua | ||||
|---|---|---|---|---|---|---|
| PIC 1 | Na 2 | Transferab-ility 3 | PIC 1 | Na 2 | Transferab-ility 3 | |
| LF15 | 0.577 | 4 | ++ | 0.000 | 1 | + |
| LF17 | 0.492 | 4 | ++ | 0.476 | 3 | ++ |
| LF19 | 0.552 | 4 | ++ | 0.346 | 2 | ++ |
| LF37 | 0.419 | 3 | ++ | 0.240 | 2 | ++ |
| Liq_eSSR 7 | - | 0 | − | 0.000 | 1 | + |
| Liq_eSSR 10 4 | 0.519 | 4 | ++ | 0.240 | 2 | ++ |
| Liq_eSSR 17 | 0.335 | 2 | ++ | 0.000 | 1 | + |
| Liq_eSSR 25 4 | 0.640 | 4 | ++ | 0.536 | 3 | ++ |
| Liq_eSSR 30 4 | 0.580 | 3 | ++ | 0.346 | 2 | ++ |
| Liq_eSSR 33 | 0.496 | 4 | ++ | 0.000 | 1 | + |
| Liq_eSSR 34 4 | 0.666 | 4 | ++ | 0.476 | 3 | ++ |
| Liq_eSSR 35 4 | 0.445 | 4 | ++ | 0.620 | 4 | ++ |
| Liq_eSSR 55 4 | 0.558 | 4 | ++ | 0.460 | 3 | ++ |
| Liq_eSSR 59 | 0.205 | 4 | ++ | 0.000 | 1 | + |
| Liq_eSSR 60 4 | 0.686 | 6 | ++ | 0.643 | 5 | ++ |
| Liq_eSSR 62 4 | 0.575 | 4 | ++ | 0.346 | 2 | ++ |
| Liq_eSSR 67 4 | 0.555 | 3 | ++ | 0.240 | 2 | ++ |
| Liq_eSSR 86 4 | 0.645 | 5 | ++ | 0.393 | 4 | ++ |
| Liq_eSSR 90 4 | 0.746 | 6 | ++ | 0.643 | 5 | ++ |
| Liq_eSSR 98 4 | 0.417 | 3 | ++ | 0.476 | 4 | ++ |
| Mean | 0.505 | 3.72 | 0.324 | 2.55 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Dong, M.; Zhang, Y.; Qi, S.; Liu, X.; Zhang, J.; Zhao, J. Development and Characterization of Simple Sequence Repeat Markers for, and Genetic Diversity Analysis of Liquidambar formosana. Forests 2020, 11, 203. https://doi.org/10.3390/f11020203
Chen S, Dong M, Zhang Y, Qi S, Liu X, Zhang J, Zhao J. Development and Characterization of Simple Sequence Repeat Markers for, and Genetic Diversity Analysis of Liquidambar formosana. Forests. 2020; 11(2):203. https://doi.org/10.3390/f11020203
Chicago/Turabian StyleChen, Siyuan, Mingliang Dong, Yan Zhang, Shuaizheng Qi, Xuezeng Liu, Jinfeng Zhang, and Jian Zhao. 2020. "Development and Characterization of Simple Sequence Repeat Markers for, and Genetic Diversity Analysis of Liquidambar formosana" Forests 11, no. 2: 203. https://doi.org/10.3390/f11020203
APA StyleChen, S., Dong, M., Zhang, Y., Qi, S., Liu, X., Zhang, J., & Zhao, J. (2020). Development and Characterization of Simple Sequence Repeat Markers for, and Genetic Diversity Analysis of Liquidambar formosana. Forests, 11(2), 203. https://doi.org/10.3390/f11020203






