Identification of Grafting-Responsive MicroRNAs Associated with Growth Regulation in Pecan [Carya illinoinensis (Wangenh.) K. Koch]
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Small RNA Library Construction and Deep Sequencing
2.3. Identification of Conserved and Novel MiRNAs
2.4. Analysis of Differential Expressed MiRNAs
2.5. Target Gene Prediction and Enrichment Analysis (GO and KEGG) of Targets
2.6. Validation of MiRNAs Using qRT-PCR
3. Results
3.1. Overview of Sequencing Data
3.2. Identification of Conserved and Novel MiRNAs
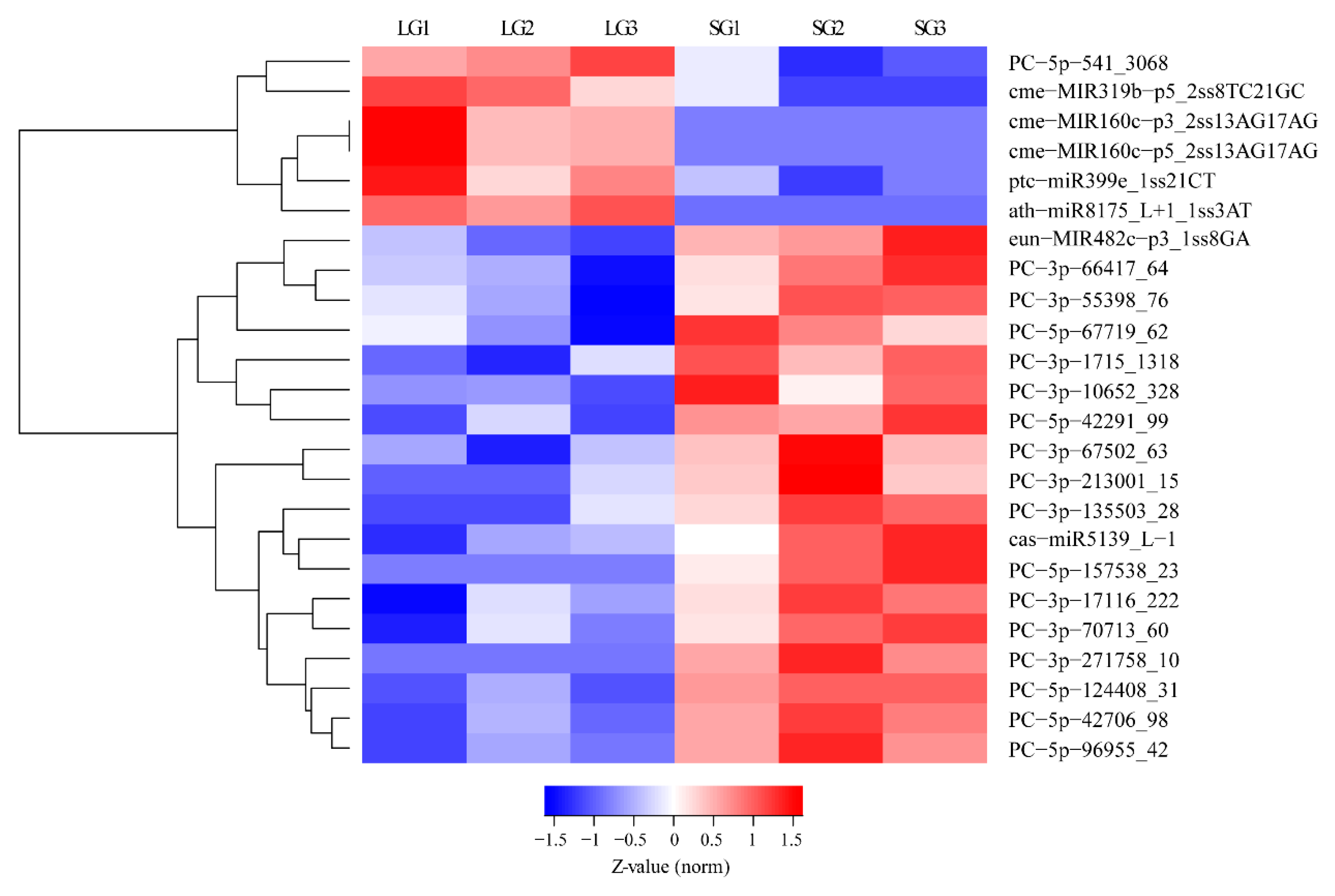
3.3. Differentially Expressed MiRNAs in Pecan Grafts with Different Growth Performance
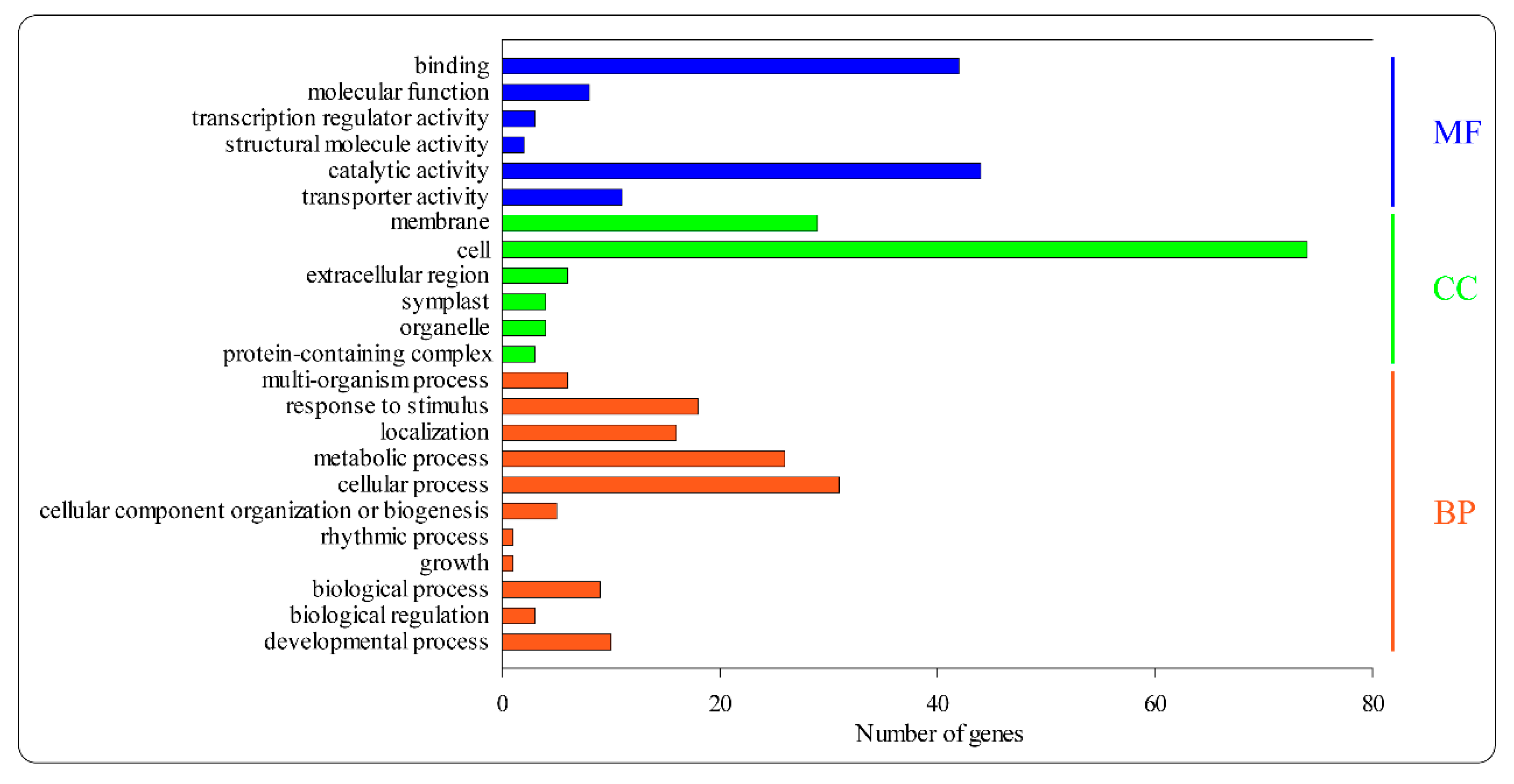
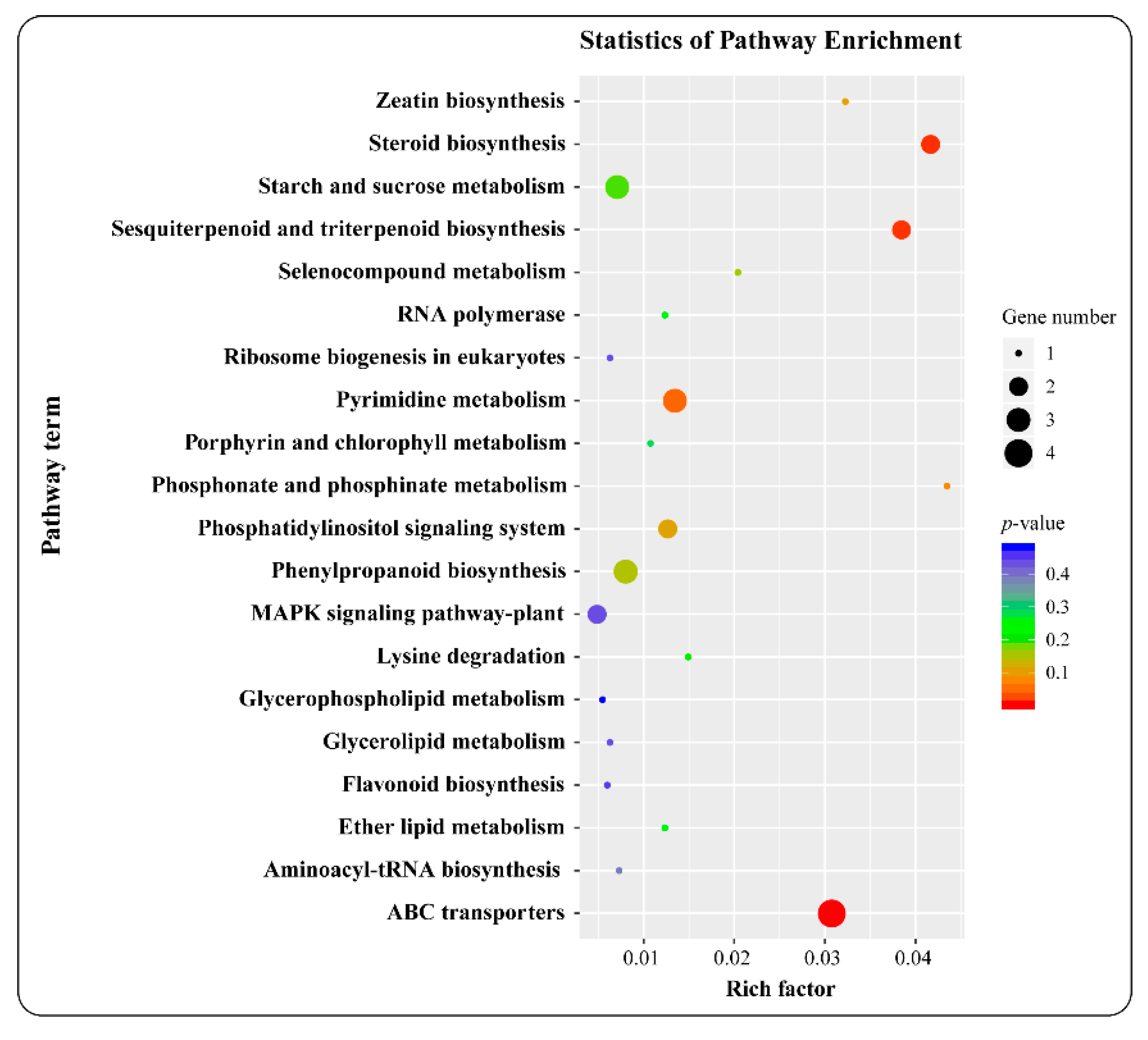
3.4. Go and KEGG Analysis of the Target Genes of Differentially Expressed MiRNAs
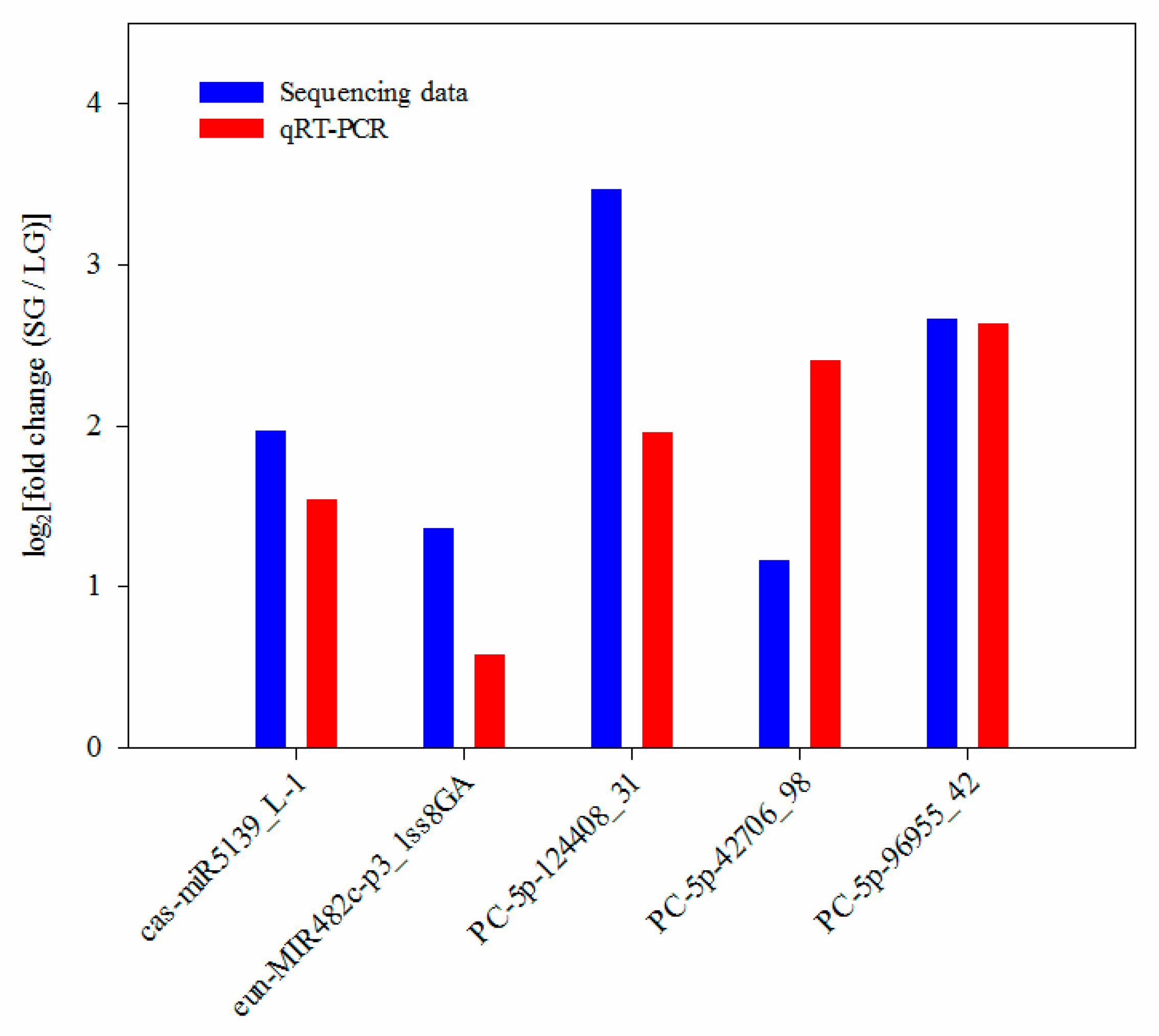
3.5. Expression Validation of Significantly Differentially Expressed MiRNAs Using qRT-PCR
4. Discussion
4.1. miRNA Population Identified in Pecan
4.2. Grafting-Responsive MiRNAs Involved in Graft Growth and Development
4.3. Long-Distance miRNA Transport in Grafts
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Harada, T. Grafting and RNA transport via phloem tissue in horticultural plants. Sci. Hortic. 2010, 125, 545–550. [Google Scholar] [CrossRef]
- Koepke, T.; Dhingra, A. Rootstock scion somatogenetic interactions in perennial composite plants. Plant Cell Rep. 2013, 32, 1321–1337. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.J.; Atkinson, C.J.; Bengough, A.G.; Else, M.A.; Fernández-Fernández, F.; Harrison, R.J.; Schmidt, S. Contributions of roots and rootstocks to sustainable, intensified crop production. J. Exp. Bot. 2013, 64, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Tworkoski, T.; Miller, S. Rootstock effect on growth of apple scions with different growth habits. Sci. Hortic. 2007, 111, 335–343. [Google Scholar] [CrossRef]
- North, M.; Cook, N. Effect of six rootstocks on ‘Forelle’ pear tree growth, production, fruit quality and leaf mineral content. In Proceedings of the XXVII International Horticultural Congress-IHC2006: International Symposium on Enhancing Economic and Environmental Sustainability of Fruit Production in a Global Economy, Seoul, Korea, 13–19 August 2006; Palmer, J.W., Ed.; Internation Society Horticultural Science: Leuven, Belgium, 2008; pp. 97–103. [Google Scholar]
- Sorce, C.; Massai, R.; Picciarelli, P.; Lorenzi, R. Hormonal relationships in xylem sap of grafted and ungrafted Prunus rootstocks. Sci. Hortic. 2002, 93, 333–342. [Google Scholar] [CrossRef]
- Fallahi, E.; Chun, I.-J.; Neilsen, G.H.; Colt, W.M. Effects of three rootstocks on photosynthesis, leaf mineral nutrition, and vegetative growth of “BC-2 Fuji” apple trees. J. Plant Nutr. 2001, 24, 827–834. [Google Scholar] [CrossRef]
- Basile, B.; Marsal, J.; DeJong, T.M. Daily shoot extension growth of peach trees growing on rootstocks that reduce scion growth is related to daily dynamics of stem water potential. Tree Physiol. 2003, 23, 695–704. [Google Scholar] [CrossRef]
- Cookson, S.J.; Ollat, N. Grafting with rootstocks induces extensive transcriptional re-programming in the shoot apical meristem of grapevine. BMC Plant Biol. 2013, 13, 147. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.J.; Rytter, J.; Detwiler, E.A.; Travis, J.W.; McNellis, T.W. Rootstock effects on gene expression patterns in apple tree scions. Plant Mol. Biol. 2003, 53, 493–511. [Google Scholar] [CrossRef]
- Prassinos, C.; Ko, J.-H.; Lang, G.; Iezzoni, A.F.; Han, K.-H. Rootstock-induced dwarfing in cherries is caused by differential cessation of terminal meristem growth and is triggered by rootstock-specific gene regulation. Tree Physiol. 2009, 29, 927–936. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Li, C.; Zhang, B. MicroRNAs in control of plant development. J. Cell Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005, 579, 5923–5931. [Google Scholar] [CrossRef]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef]
- Buhtz, A.; Pieritz, J.; Springer, F.; Kehr, J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 2010, 10, 64. [Google Scholar] [CrossRef]
- Martin, A.; Adam, H.; Díaz-Mendoza, M.; Żurczak, M.; González-Schain, N.D.; Suárez-López, P. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 2009, 136, 2873–2881. [Google Scholar] [CrossRef]
- Bhogale, S.; Mahajan, A.S.; Natarajan, B.; Rajabhoj, M.; Thulasiram, H.V.; Banerjee, A.K. MicroRNA156: A potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014, 164, 1011–1027. [Google Scholar] [CrossRef]
- Liu, N.; Yang, J.; Guo, S.; Xu, Y.; Zhang, M. Genome-wide identification and comparative analysis of conserved and novel microRNAs in grafted watermelon by high-throughput sequencing. PLoS ONE 2013, 8, e57359. [Google Scholar] [CrossRef]
- Khaldun, A.B.M.; Huang, W.J.; Lv, H.Y.; Liao, S.H.; Zeng, S.H.; Wang, Y. Comparative Profiling of miRNAs and Target Gene Identification in Distant-Grafting between Tomato and Lycium (Goji Berry). Front. Plant Sci. 2016, 7, 1475. [Google Scholar] [CrossRef]
- An, N.; Fan, S.; Yang, Y.; Chen, X.L.; Dong, F.; Wang, Y.B.; Xing, L.B.; Zhao, C.P.; Han, M.Y. Identification and characterization of miRNAs in self-Rooted and grafted Malus reveals critical networks associated with flowering. Int. J. Mol. Sci. 2018, 19, 2384. [Google Scholar] [CrossRef]
- Li, C.H.; Li, Y.S.; Bai, L.Q.; Zhang, T.Y.; He, C.X.; Yan, Y.; Yu, X.C. Grafting-responsive miRNAs in cucumber and pumpkin seedlings identified by high-throughput sequencing at whole genome level. Physiol. Plant 2014, 151, 406–422. [Google Scholar] [CrossRef] [PubMed]
- Ajamgard, F.; Rahemi, M.; Vahdati, K. Development of improved techniques for grafting of pecan. Sci. Hortic. 2016, 204, 65–69. [Google Scholar] [CrossRef]
- Zhang, R.; Peng, F.R.; Le, D.L.; Liu, Z.Z.; He, H.Y.; Liang, Y.W.; Tan, P.P.; Hao, M.Z.; Li, Y.R. Evaluation of Epicotyl Grafting on 25-to 55-day-old Pecan Seedlings. Horttechnology 2015, 25, 392–396. [Google Scholar] [CrossRef]
- Grauke, L.; Pratt, J. Pecan bud growth and freeze damage are influenced by rootstock. J. Am. Soc. Hortic. Sci. 1992, 117, 404–406. [Google Scholar] [CrossRef]
- Huang, Y.; Ren, H.T.; Xiong, J.L.; Gao, X.C.; Sun, X.H. Identification and characterization of known and novel microRNAs in three tissues of Chinese giant salamander base on deep sequencing approach. Genomics 2017, 109, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Wang, C.; Zhang, C.; Korir, N.K.; Yu, H.; Ma, Z.; Fang, J. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genom. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.; Feng, G.; Su, W.; Liu, Z.; Peng, F. Identification of miRNAs associated with graft union development in pecan [Carya illinoinensis (Wangenh.) K. Koch]. Forests 2018, 9, 472. [Google Scholar] [CrossRef]
- Huang, Y.; Xiao, L.; Zhang, Z.; Zhang, R.; Wang, Z.; Huang, C.; Huang, R.; Luan, Y.; Fan, T.; Wang, J. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. Gigascience 2019, 8, giz036. [Google Scholar] [CrossRef]
- Thompson, T.E.; Hunter, R.E. ‘Pawnee’ pecan. HortScience 1985, 20, 776. [Google Scholar]
- Zhang, R.Q.; Chen, J.H.; Xia, C.G.; Lv, F.D. Historical survey of the introduced pecan tree in China advances in science and technology. Hunan For. Sci. Technol. 2001, 28, 6–9. [Google Scholar]
- Sun, W.; Xu, X.H.; Wu, X.; Wang, Y.; Lu, X.; Sun, H.; Xie, X. Genome-wide identification of microRNAs and their targets in wild type and phyB mutant provides a key link between microRNAs and the phyB-mediated light signaling pathway in rice. Front. Plant Sci. 2015, 6, 372. [Google Scholar] [CrossRef] [PubMed]
- Ambady, S.; Wu, Z.; Dominko, T. Identification of novel microRNAs in Xenopus laevis metaphase II arrested eggs. Genesis 2012, 50, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shahid, M.; Wu, J.; Wang, L.; Liu, X.; Lu, Y. Comparative small RNA analysis of pollen development in autotetraploid and diploid rice. Int. J. Mol. Sci. 2016, 17, 499. [Google Scholar] [CrossRef] [PubMed]
- Qiang, J.; Bao, W.J.; Tao, F.Y.; He, J.; Li, X.H.; Xu, P.; Sun, L.Y. The expression profiles of miRNA–mRNA of early response in genetically improved farmed tilapia (Oreochromis niloticus) liver by acute heat stress. Sci. Rep. 2017, 7, 8705. [Google Scholar] [CrossRef] [PubMed]
- Lang, Q.-l.; Zhou, X.-c.; Zhang, X.-l.; Drabek, R.; Zuo, Z.-x.; Ren, Y.-l.; Li, T.-b.; Chen, J.-s.; Gao, X.-l. Microarray-based identification of tomato microRNAs and time course analysis of their response to Cucumber mosaic virus infection. J. Zhejiang Univ. Sci. B 2011, 12, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Yin, S.; Mao, J.; Liang, F.; Zhao, C.; Li, P.; Zhou, G.; Chen, S.; Tang, Z. Integrated analysis of mRNA-seq and miRNA-seq in the liver of Pelteobagrus vachelli in response to hypoxia. Sci. Rep. 2016, 6, 22907. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Webster, A. Vigour mechanisms in dwarfing rootstocks for temperate fruit trees. In Proceedings of the I International Symposium on Rootstocks for Deciduous Fruit Tree Species, Zaragoza, Spain, 11–14 June 2002; Sanchez, M.A., Webster, A.D., Eds.; International Society Horticultural Science: Leuven, Belgium, 2004; pp. 29–41. [Google Scholar]
- Sunkar, R.; Jagadeeswaran, G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008, 8, 37. [Google Scholar] [CrossRef]
- Wang, F.; Li, L.; Liu, L.; Li, H.; Zhang, Y.; Yao, Y.; Ni, Z.; Gao, J. High-throughput sequencing discovery of conserved and novel microRNAs in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol. Genet. Genom. 2012, 287, 555–563. [Google Scholar] [CrossRef]
- Axtell, M.J.; Bowman, J.L. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008, 13, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Gao, F.; Li, S.; Zhu, Y.; Yang, P. Identification of conserved and novel microRNAs from Liriodendron chinense floral tissues. PLoS ONE 2012, 7, e44696. [Google Scholar] [CrossRef] [PubMed][Green Version]
- An, W.; Gong, W.; He, S.; Pan, Z.; Sun, J.; Du, X. MicroRNA and mRNA expression profiling analysis revealed the regulation of plant height in Gossypium hirsutum. BMC Genom. 2015, 16, 886. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, A.; Zhang, X.; Hou, Y.; Shangguan, M.; Gajjeraman, P.; Li, Y.; Wei, C. Genome-wide identification of conserved and novel microRNAs in one bud and two tender leaves of tea plant (Camellia sinensis) by small RNA sequencing, microarray-based hybridization and genome survey scaffold sequences. BMC Plant Biol. 2017, 17, 212. [Google Scholar] [CrossRef]
- Pantaleo, V.; Szittya, G.; Moxon, S.; Miozzi, L.; Moulton, V.; Dalmay, T.; Burgyan, J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010, 62, 960–976. [Google Scholar]
- Cui, J.; Lu, W.; Lu, Z.; Ren, S.; Zhao, B.; Wang, L.; Teng, N.; Jin, B. Identification and analysis of microRNAs in the SAM and leaves of Populus tomentosa. Forests 2019, 10, 130. [Google Scholar] [CrossRef]
- Zhang, W.; Xie, Y.; Xu, L.; Wang, Y.; Zhu, X.; Wang, R.; Zhang, Y.; Muleke, E.M.; Liu, L. Identification of microRNAs and their target genes explores miRNA-mediated regulatory network of cytoplasmic male sterility occurrence during anther development in radish (Raphanus sativus L.). Front. Plant Sci. 2016, 7, 1054. [Google Scholar] [CrossRef]
- Tang, F.; Wei, H.; Zhao, S.; Wang, L.; Zheng, H.; Lu, M. Identification of microRNAs involved in regeneration of the secondary vascular system in Populus tomentosa Carr. Front. Plant Sci. 2016, 7, 724. [Google Scholar] [CrossRef]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef]
- Vanneste, S.; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef]
- Blakeslee, J.J.; Peer, W.A.; Murphy, A.S. Auxin transport. Curr. Opin. Plant Biol. 2005, 8, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Friml, J. Auxin transport—shaping the plant. Curr. Opin. Plant Biol. 2003, 6, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Kerr, I.D.; Bennett, M.J. New insight into the biochemical mechanisms regulating auxin transport in plants. Biochem. J. 2007, 401, 613–622. [Google Scholar] [CrossRef]
- Zažímalová, E.; Murphy, A.S.; Yang, H.; Hoyerová, K.; Hošek, P. Auxin transporters—why so many? Cold Spring Harb. Perspect. Biol. 2010, 2, a001552. [Google Scholar] [CrossRef]
- Kipreos, E.T.; Pagano, M. The F-box protein family. Genome Biol. 2000, 1, reviews3002.1–reviews3002.7. [Google Scholar] [CrossRef]
- Skowyra, D.; Craig, K.L.; Tyers, M.; Elledge, S.J.; Harper, J.W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 1997, 91, 209–219. [Google Scholar] [CrossRef]
- Harrison, C.; Katayama, S.; Dhut, S.; Chen, D.; Jones, N.; Bähler, J.; Toda, T. SCFPof1-ubiquitin and its target Zip1 transcription factor mediate cadmium response in fission yeast. EMBO J. 2005, 24, 599–610. [Google Scholar] [CrossRef]
- Rouillon, A.; Barbey, R.; Patton, E.E.; Tyers, M.; Thomas, D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCFMet30 complex. EMBO J. 2000, 19, 282–294. [Google Scholar] [CrossRef]
- Patton, E.E.; Peyraud, C.; Rouillon, A.; Surdin-Kerjan, Y.; Tyers, M.; Thomas, D. SCFMet30-mediated control of the transcriptional activator Met4 is required for the G1–S transition. EMBO J. 2000, 19, 1613–1624. [Google Scholar] [CrossRef]
- Aung, K.; Lin, S.-I.; Wu, C.-C.; Huang, Y.-T.; Su, C.-l.; Chiou, T.-J. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant. Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W.-R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant. Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Koizumi, R.; Shui, G.; Shimojima, M.; Wenk, M.R.; Ito, T.; Ohta, H. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. USA 2009, 106, 20978–20983. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y. Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 2013, 52, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-Z.; Ai, X.-Y.; Guo, W.-W.; Peng, S.-A.; Deng, X.-X.; Hu, C.-G. Identification of miRNAs and their target genes using deep sequencing and degradome analysis in trifoliate orange [Poncirus trifoliate (L.) Raf]. Mol. Biotechnol. 2012, 51, 44–57. [Google Scholar] [CrossRef]
- He, Q.; Zhu, S.; Zhang, B. MicroRNA–target gene responses to lead-induced stress in cotton (Gossypium hirsutum L.). Funct. Integr. Genom. 2014, 14, 507–515. [Google Scholar] [CrossRef]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef]
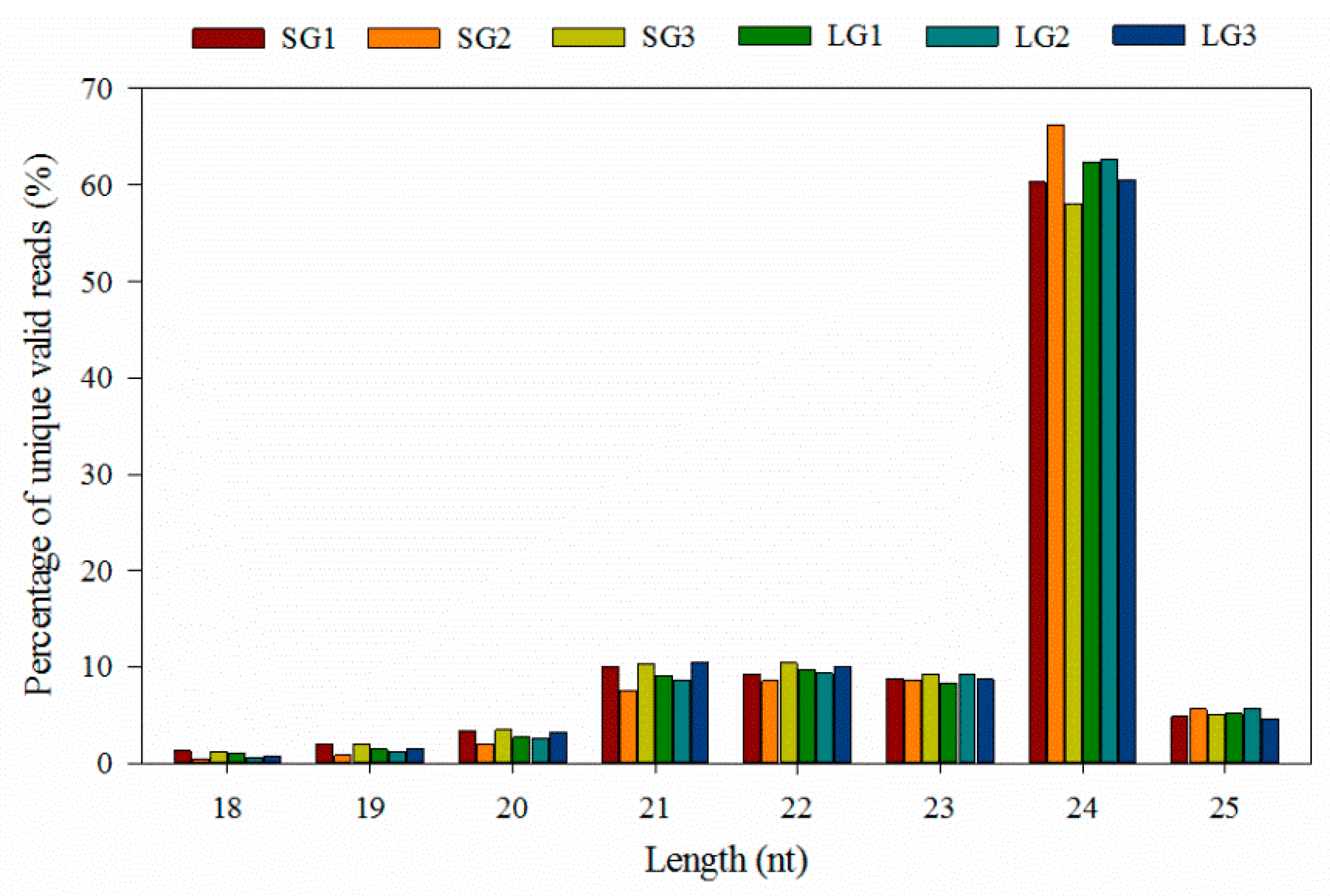
| SG1 | SG2 | SG3 | LG1 | LG2 | LG3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| lib | Total | Unique | Total | Unique | Total | Unique | Total | Unique | Total | Unique | Total | Unique |
| Raw reads | 21,056,396 | 3,677,206 | 13,652,899 | 3,103,481 | 10,785,140 | 2,648,359 | 11,708,837 | 2,438,848 | 15,829,590 | 5,570,725 | 18,327,912 | 5,805,385 |
| 3ADT&length filter | 2,360,700 | 816,006 | 809,523 | 370,969 | 2,002,022 | 554,824 | 1,490,397 | 544,606 | 908,635 | 607,526 | 1,167,251 | 577,406 |
| Junk reads | 42,994 | 16,383 | 25,905 | 15322 | 23,224 | 11,158 | 20,533 | 11,029 | 47,247 | 29,392 | 47,895 | 29,689 |
| mRNA | 921,861 | 21,309 | 341,968 | 9320 | 399,286 | 11,309 | 283,384 | 8922 | 699,798 | 12,479 | 801,490 | 16,101 |
| rRNA | 426,663 | 8809 | 183,608 | 4378 | 168,645 | 6050 | 150,534 | 4446 | 185,693 | 7249 | 218,823 | 6426 |
| tRNA | 125,670 | 1666 | 28,942 | 576 | 51,498 | 1092 | 34,216 | 637 | 45,548 | 1169 | 163,922 | 1346 |
| snoRNA | 5874 | 420 | 1375 | 145 | 1538 | 162 | 2371 | 210 | 2254 | 178 | 3233 | 240 |
| snRNA | 10,717 | 610 | 4350 | 195 | 4049 | 276 | 3428 | 187 | 6581 | 300 | 7934 | 396 |
| other Rfam RNA | 31,717 | 1189 | 7681 | 494 | 10,892 | 666 | 9844 | 646 | 8648 | 659 | 12,706 | 758 |
| Repeats | 12,009 | 445 | 3283 | 169 | 4976 | 228 | 3834 | 169 | 6201 | 271 | 10,994 | 389 |
| valid reads | 17,168,960 | 2,811,819 | 12,263,173 | 2,702,559 | 8,137,392 | 2,063,453 | 9,726,162 | 1,868,717 | 13,946,042 | 4,912,485 | 15,927,337 | 5,173,801 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Li, F.; Peng, F.; Tan, P.; Zhu, K.; Feng, G.; Mo, Z.; Li, Y. Identification of Grafting-Responsive MicroRNAs Associated with Growth Regulation in Pecan [Carya illinoinensis (Wangenh.) K. Koch]. Forests 2020, 11, 196. https://doi.org/10.3390/f11020196
Liu Z, Li F, Peng F, Tan P, Zhu K, Feng G, Mo Z, Li Y. Identification of Grafting-Responsive MicroRNAs Associated with Growth Regulation in Pecan [Carya illinoinensis (Wangenh.) K. Koch]. Forests. 2020; 11(2):196. https://doi.org/10.3390/f11020196
Chicago/Turabian StyleLiu, Zhuangzhuang, Fengda Li, Fangren Peng, Pengpeng Tan, Kaikai Zhu, Gang Feng, Zhenghai Mo, and Yongrong Li. 2020. "Identification of Grafting-Responsive MicroRNAs Associated with Growth Regulation in Pecan [Carya illinoinensis (Wangenh.) K. Koch]" Forests 11, no. 2: 196. https://doi.org/10.3390/f11020196
APA StyleLiu, Z., Li, F., Peng, F., Tan, P., Zhu, K., Feng, G., Mo, Z., & Li, Y. (2020). Identification of Grafting-Responsive MicroRNAs Associated with Growth Regulation in Pecan [Carya illinoinensis (Wangenh.) K. Koch]. Forests, 11(2), 196. https://doi.org/10.3390/f11020196




