Abstract
The purpose of this study was to characterize the endogenous cellulase gene MaCel1 of Monochamus alternatus, which is an important vector of Bursaphelenchus xylophilus, a pine wood nematode, which causes pine wilt disease (PWD). In this study, MaCel1 was cloned by rapid amplification of cDNA end (RACE), and its expression analyzed by RT-qPCR (real-time quantitative PCR detecting). A total of 1778 bp of cDNA was obtained. The encoding region of this gene was 1509 bp in length, encoding a protein containing 502 amino acids with a molecular weight of 58.66 kDa, and the isoelectric point of 5.46. Sequence similarity analysis showed that the amino acids sequence of MaCel1 had high similarity with the β-Glucosinolate of Anoplophora glabripennis and slightly lower similarity with other insect cellulase genes (GH1). The β-D-Glucosidase activity of MaCel1 was 256.02 ± 43.14 U/L with no β-Glucosinolate activity. MaCel1 gene was widely expressed in the intestine of M. alternatus. The expression level of MaCel1 gene in male (3.46) and female (3.51) adults was significantly higher than that in other developmental stages, and the lowest was in pupal stage (0.15). The results will help reveal the digestive mechanism of M. alternatus and lay the foundation for controlling PWD by controlling M. alternatus.
1. Introduction
Cellulose is a type of natural polymer compound widely present in plant tissues. It is the most abundant biomass resource on earth, and it is also an important industrial raw material and renewable energy resource [1]. Cellulose is the main nutrient in the food of wood borers and plays an important role in normal growth and development [2]. The digestive system of selected phytophagous insects has been examined as a potential resource for identification of novel cellulolytic enzymes with potential industrial applications [3]. Plants have rigid cells composed of abundant cellulose polymers strengthened by hydrogen bonds and van der waals forces. Phytophagous insects need cellulase, pectinase, lignan, xylanase, and xyloglucanase to digest plant materials. Insects have a complete and complex high-efficiency cellulose biocatalytic system that can digest and utilize a large amount of plant cellulose and convert cellulose into insects’own nutrition [4]. Traditionally, it is believed that insects cannot produce cellulase [5,6]. Instead, the cellulose in insects is decomposed by symbiotic microorganisms in the body [7]. However, recent studies have shown that insect cells and tissues produce cellulase to digest cellulose [8,9]. Cellulase genes, which were cloned in insects, such as Blattaria, Coleoptera, and Orthoptera [10,11,12,13,14,15,16,17,18,19,20], prove the existence of insect-endogenous cellulase. Insect cellulase belongs to the four main glycosyl hydrolase families (GHs), which are GH1, GH5, GH9, and GH45 [21,22]. Cellulase can be isolated from the intestine of the larvae of Psacothea hilaris. The purified cellulase can degrade carboxymethyl cellulose and insoluble cellulose (average degree of polymerization 34) [16]. A novel cellulase (β-1,4-endoglucanase, EC 3.2.1.4) cDNA belonging to the GH45 was cloned from the mulberry longicorn beetle, Apriona germari and Bateocera horsfieldi [17,23]. The AgEGase enzyme assay exhibited high activity only in midgut tissue, suggesting that the midgut is the prime site where large quantities of EGase are synthesized for degrading the absorbed cellulose from the diet. However, there are many studies on symbiotic microorganism in the cellulase of M. alternatus [7,24,25]. Despite emerging evidence of insect cellulase, there are few reports about endogenous cellulase in M. alternatus, which necessitates further study on endogenous cellulase of M. alternatus.
M. alternatus is a worldwide forest pest and an important vector for Bursaphelenchus xylophilus, a pine wood nematode, which causes pine wilt disease. Similar to other xylophagous insects, M. alternatus mainly digests cellulose to obtain nutrients throughout the growth and development process, and this process requires the participation of cellulase for better digestion of the plant material [26,27]. Pine wilt disease has many characteristics, such as high mortality and fast onset, is widespread, and is called pine cancer by scholars [28,29,30]. PWD causes huge damage to the forestry economy [31,32]. The huge tree damage is caused by adults and larvae [33,34]. In addition, M. alternatus carries the PWD virus, which is fatal to Pinus massoniana. In recent years, the research on controlling PWD by M. alternatus has been increasing and involves many aspects [32,33,35]. Thus, studying cellulase in M. alternatus is of great significance for elucidating the digestion physiology of beetles, developing and utilizing insect lignocellulase resources, and exploring new control strategies for beetles.
In this study, the MaCel1 gene of M. alternatus was cloned based on the transcriptome data of M. alternatus, which was obtained earlier in our research group. The bioinformatics of MaCel1 and the expression of MaCel1 in M. alternatus gene at different developmental stages were studied and analyzed. The results will provide a theoretical basis for revealing the function of MaCel1 gene of M. alternatus and laying a foundation for controlling PWD by controlling M. alternatus.
2. Materials and Methods
2.1. Test Materials
The infected trees of P. massoniana in Guantou Town, Lianjiang County, Fuzhou City, Fujian Province (26.150° N; 119.593° E) were used as experimental material. The infected trees were felled and sawn to a length of 1 m. The early larvae, late larvae and pupae of M. alternatus were collected in phloem and xylem. Then, the larvae were left to grow to the adult stage in the infected trees. Adults, both male and female, were collected and fed with fresh pine branches for later use.
2.2. RNA Isolation and cDNA Synthesis
We took 10 early larvae, late larvae, pupae, female and male adults of M. alternatus each and cut a small hole in their tail. The intestines were picked out with an insect needle under a stereo microscope. The total RNA was extracted according to the Trizol reagent instructions and stored at −80 °C. The first strand of cDNA was synthesized according to the instructions of BestarTM qPCR RT Kit and stored at −20 °C. The cDNA was used as a template for quantitative expression analysis. In addition, the first strand of cDNA was synthesized (Fermentas RevertAid First Strand cDNA Synthesis Kit) from fresh intestinal tissue of the late larvae of M. alternatus, which was used to amplify the middle fragment of the gene.
2.3. Amplification of the Intermediate Fragment of MaCel1
The M. alternatus cellulase gene was screened out by referring to the transcription data and annotation information. The primers were designed and synthesized by Shenggong Biological Engineering Co., Ltd. (Shanghai, China) (Table 1). The first strand of synthesized cDNA was used as a template for PCR amplification. The reaction mixture was as follows: PCR-Grade Water 15 μL, 2X Ex taq Buffer (takara) 25 μL, dNTP Mix (10 mM) 1 μL, Ex taq (takara) 1 μL, cDNA first strand 5 μL, and forward and reverse primers 1.5 μL each. PCR amplification of the cDNA of MaCel1 was performed at 94 °C for 2 min followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. The PCR products were electrophoresed on 1.0% agarose gel. After that, the target band was quickly cut out and recycled according to the instructions of the OMEGA kit. The purified PCR product was connected to plasmids pMD18T, which is a high-efficiency cloning vector. After transformation, the positive clones were sequenced, and sequence comparison analysis was performed to verify the transcriptome sequence.

Table 1.
Primers used in this study.
2.4. Full-Length RACE of MaCel1
The three specific 5′ RACE primers and two specific 3′ RACE primers were designed by Premier 5.0 software. The primers were synthesized by Shenggong Biological Engineering Co., Ltd. (Shanghai, China). and used to clone the 5′terminal and 3′terminal sequences of MaCel1 gene.
The total RNA of the M. alternatus intestine was used as the template and gene-specific primers (GSP1) (Table 1) as the primer. The first strand of the cDNA was synthesized by Superscript II RT enzyme. RNA was removed from the cDNA using RNase Mix. The dC-tailed cDNA was amplified by nested PCR with the first-round primers of GSP-2 (Table 1) and abridged anchor primer (AAP), followed by the second-round primers of GSP-3 (Table 1) and abridged universal amplification primer (AUAP) in the kit. The PCR product was linked to pMD18T and the positive clones were sequenced.
The total RNA of the M. alternatus intestine was used as the template to synthesize the cDNA of 3′RACE. The cDNA was synthesized by SMARTScribeTM reverse transcriptase and primer 3′CDS primer A. The cDNA was then used as a template of the first round of PCR and the primers were 3′977-1 (Table 1) and universal primer (UPM). The template of the second round PCR was cDNA with primers of 3′977-2 (Table 1) and UPM. The PCR was linked to pMD18T and the positive clones were sequenced by Shenggong Biological Engineering Co., Ltd. (Shanghai, China). Finally, the full-length cDNA sequence of MaCel1 gene was obtained by Vector NTI10.3.0. The clone sequence was compared with the cDNA sequence to confirm that the recombinant vector was successfully obtained.
2.5. Prokaryotic Expression and Purification of MaCel1
The full-length CDS (Met1-Leu502) were amplified using pMD8-T-Cel plasmid as the template. After recycling, the CDS was linked to 5′EcoRI-XhoI3′ of Pet-32a vector. The vector was connected and converted to DH5α competent cells. The recombinant plasmid pET32a-Cel was screened and identified. Then, pET32a-Cel was converted to Escherichia coli Rosetta (DE3) and identified. Rosetta (pET32a-MaCel1) was inoculated into Luria-Bertani (LB) medium and cultured overnight at 37 °C for 220 r/min. Isopropyl-beta-d-thiogalactopyranoside (IPTG) was added and the final concentration was 0.5 mmol/L at 18 °C for 10 h when OD600 was between 0.4 and 0.6. The recombinant cells were centrifuged for 10 min at 12000 r/min. Then the supernatants and precipitates were collected separately, and the solubility of the fusion protein was detected by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The inclusion bodies were obtained from the induction of Rosetta-pET32a-MaCel1. The inclusion bodies were denatured by adding urea. The soluble MaCel1 recombinant protein was obtained when the denatured inclusion bodies were added into the refolding buffer. The heteroprotein was removed by Ni-NTA (network terminal appliance) column. The MaCel1 protein was concentrated on a millipore 15 mL 10K (UFC910096) ultrafiltration centrifuge column.
2.6. Determination of Enzyme Activity of MaCel1
The activities of β-D-Glucosidase and β-Glucosinolate of MaCel1 protein were determined by 3,5-Dinitrosalicylic acid (DNS) colorimetric method with allyl glucoside and salicylic acid as the substrates. An enzyme activity unit was defined as the number of micromoles per minute per milligram of protein producing reducing sugars. The glucose standard curve was run and measured at 520 nm wavelength by enzyme-labeled instrument [36,37]. The DNS method was used to detect the glucose enzyme in the sample [36,37]. The method for determination of the β-D-Glucosidase activity was the same as that for determination of the glucosidase activity. The substrate was allyl glucoside [38,39].
2.7. Bioinformatics Analysis
DNAMAN software was used to predict the coding sequence of the target gene. The open reading frame (ORF) finder (www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to predict the ORF of MaCel1. The physical and chemical properties of MaCel1 encoded protein were predicted by Protparam (https://web.expasy.org/protparam/) while SignalP4.1 (http://www.cbs.dtu.dk/services/SignalP/) was used to predict the signal peptide encoding proteins.
The transmembrane structure and hydrophilicity of the protein encoded by MaCel1 gene were predicted by TMpredServer (https://embnet.vital-it.ch/software/TMPRED_form.html). The prediction of the secondary and tertiary structure of MaCel1 protein was achieved by using SOPMA (http://scop.mrc-lmb.cam.ac.uk/scop/). The amino acid sequences were compared by ClustalW2 software (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The sequences were downloaded from the NCBI and the phylogenetic tree was constructed with the LG+G evolutionary model and the maximum likelihood phylogenetic method by MEGA X software.
2.8. Quantitative Expression of MaCel1
RT-qPCR was used to analyze the expression of the MaCel1 gene in the intestinal tract of M. alternatus. The RT-qPCR was performed and analyzed using Bestar® SybrGreen qPCRmasterMix and a StepOnePlus ™ real-time fluorescence quantitative PCR instrument (Corbett Research, Australia). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (Table 1) was used as a reference gene to normalize expression data [37]. The RT-qPCR was performed in a final volume of 20 µL in a mixture containing 10 µL Bestar® SybrGreen qPCRmasterMix, 0.5 µL each of specific forward and reverse primers, 7 µL ddH2O, and 2 µL cDNA. The amplification program was 95 °C for 2 min followed by 40 cycles of 94 °C for 20 s, 58 °C for 20 s, and 72 °C for 30 s. The specificity of the amplification was controlled by melting curve analysis (from 65 to 95 °C). The relative expression of MaCel1 gene was calculated by the 2−∆∆Ct method, which is applicable when the amplification efficiency of the target and reference genes is approximately equal [40]. The data were analyzed by the one-way ANOVA using IBM SPSS software. Normality of the data was evaluated with the Kolmogorov–Smirnov test.
2.9. Prediction of Endogenous Cellulase in M. alternatus
Gene annotation was mainly based on the obtained sequencing information [28]. The gene sequence was compared with kyoto encyclopedia of genes and genomes (KEGG), cluster of orthologousgroup (COG), gene ontology (GO) and RefSeq non-redundant proteins (Nr) to obtain the corresponding functional annotation information, and the possible functions of the genes were inferred based on the results of each database [41]. The Blast comparison was conducted between the genes and each database and then filtered by Blast results. The sequence was annotated of the highest score comparison results (default identity ≥40%, coverage ≥40%). The expression of the lignocellulase-related genes was calculated by reads per kilobase per million mapped reads (RPKM) method [42]. The RPKM method could eliminate the influence of gene length and sequencing amount on the expression of the genes. Thus, the RPKM values could be directly used to differentiate the gene expression levels among different samples.
3. Results
3.1. Prediction of Endogenous Cellulase in M. alternatus
M. alternatus had 14 cellulase enzymes, including 3 exo-glucanase (GH48), 7 endoglucanase (GH5, GH9, GH45), and 4 β-Glucosidase (GH1, GH3), belonging to 6 glycosidase families (Table A1). There were two up-regulated genes, c79679 and c81097 (Figure 1), and six down-regulated genes, namely c89763, c39850, c77902, c94844, c71020, and c86570, in the late larvae compared with the early larvae. The absolute expression of the cellulase gene at different developmental stages was compared. The results show that the c39850 gene was found to be the highest in the early larvae (4445 RPMK), and c77902, c71020, and c86570 genes had more than 200 reads per kilobase per million mapped reads (RPMK). In the late larvae, the highest expression of c81097 gene was 83 RPMK, the lowest was 55 RPMK for c79679 gene, and the expression level of other genes did not exceed 20 RPMK.
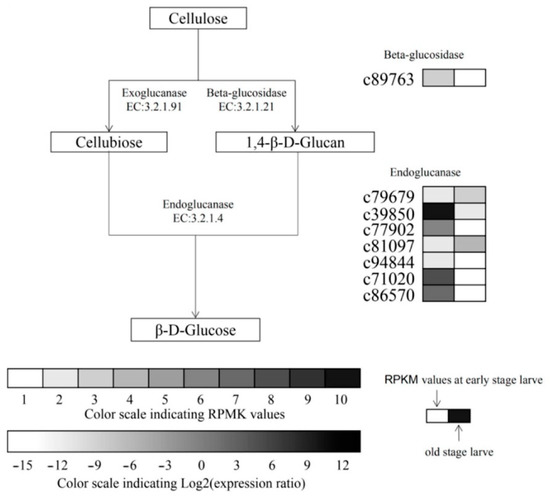
Figure 1.
Differential transcriptional levels of cellulolytic genes in different developmental stages in M. alternatus. The expression pattern of each transcriptome was shown on two grids, the RPKM values of the early larvae on the left and the relative log2 (expression ratio) of the later larvae on the right. The absolute expression range of early larvae is displayed with 10 different gray scale levels, and the RPKM values are 0–10, 10–20, 20–50, 50–100, 100–200, 200–400, 400–1000, 1000–2000, 2000–3000 and 3000–4000, respectively represented by scale 1–10.
3.2. Cloning and Sequence Analysis of MaCel1
According to the verification results of 5′RACE and 3′RACE based on the transcriptome sequence, the cDNA with a total length of 1778 bp was obtained. The 5′terminal non-coding region was 183 bp, and the 3′terminal non-coding region was 86 bp. We named the gene MaCel1 (KY073339). Sequence analysis revealed an ORF of 1509 bp, which encoded a predicted protein of 504 amino acids with a calculated molecular weight of 58.66 kDa and theoretical pI value of 5.46. The molecular formula of the protein was C2685H4005N687O771S13, and the instability coefficient was 41.66, which suggested that the protein was an unstable protein. The average hydrophilicity of MaCel1 gene was −0.449. MaCel1 gene coding protein contained the hydrophobic regions, which indicated that the MaCel1 coding protein was a hydrophilic protein. Using TMpred server online software to predict the transmembrane structure of the protein encoded by the MaCel1 gene, the results showed one transmembrane domain and three suspected transmembrane domains. The results of secondary structure of the protein was 37.85% α-helix related protein function, 7.17% β-sheet related protein function, and 15.14% the stretched chain between α-helix and irregular curl. The main skeleton of the protein in the tertiary structure was α-helix (Figure 2).

Figure 2.
Prediction of MaCel1 protein tertiary structure.
3.3. Similarity Analysis of MaCel1
The amino acid sequences of the MaCel1 gene and other insect Cel genes obtained from Blast were compared using DNAMAN V6 software (Figure 3). The phylogenetic analysis of the Cel genes from 9 insect species, including the MaCel1 gene, was performed using MEGA X (Figure 4). The results showed that the MaCel1 gene was most closely related to the β-Glucosinolate of A. glabripennis while Harpegnathos saltator was the outgroup species.

Figure 3.
The amino acid sequences of MaCel1 genes and some insect cellulase genes. MaCel1 protein source and GenBank accession number: MaCel1: M. alternatus (ATY36713); AnMyr1: A. glabripennis (XP018565386); PsGH1: Phyllotreta striolata (AHZ59658); AtMyr1: Aethina tumida (XP019880470); TcMyr1: Tribolium castaneum (XP966332); PaCel: Periplaneta Americana (AIA09348); ClMyr1: Chrysomela lapponica (XP966332); PpMyr1: Fireflies: Photinus pyralis (XP966332); LsGH1: Laodelphax striatellus (RZF43906); NkCel: Neotermes koshunensis (BAB91145).

Figure 4.
MaCel1 phylogenetic tree based on amino acid sequence.
3.4. Prokaryotic Expression of MaCel1 Gene and Determination of Enzyme Activity of MaCel1
SDS-PAGE analysis showed that a specific protein band of about 76 kD was expressed in the E. coli containing MaCel1 induced by IPTG (Figure 5A), which fitted the estimated size. The renatured protein was obtained by purification (Figure 5B). The results of enzyme activity showed that the β-D-Glucosidase activity was 256.02 ± 43.14 U/L, but no activity of the β-Glucosinolate was observed.
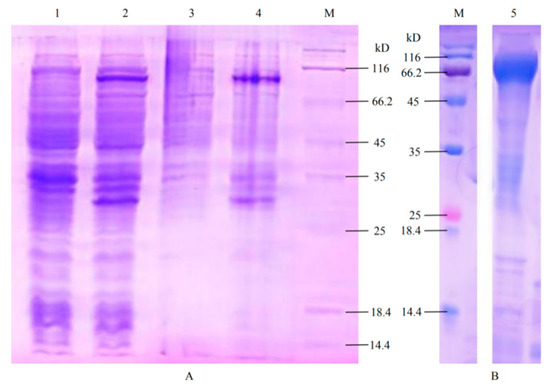
Figure 5.
SDS-PAGE analysis of recombinant protein of MaCel1 in M. alternatus. (A): Induced expression of recombinant vector pET32a-MaCel1; (M): Standard for protein molecular weight; (1): Expression of pET32a-MaCel1 without IPTG induction; (2): The expression product of pET32a-MaCel1 after IPTG induction; (3): Expression product of pET32a-MaCel1 in supernatant; (4): Expression products of pET32a-MaCel1 in inclusion bodies; (B): Renaturation of MaCel1 purified protein; (5): MaCel1 renatured protein.
3.5. Expression Analysis of MaCel1
In order to study the relative expression of cellulase in M. alternatus at different development stages, RT-qPCR analysis was carried out on the early larvae, the late larvae, pupae, female and male adults. There was a significant difference in the relative expression level of the MaCel1 gene among different developmental stages (ANOVA, F = 20.822, df = 4, p = 0.002). The relative expression of MaCel1 was the highest in both male and female adults, followed by the early larvae, while it was the lowest in late larvae and pupal stage (Figure 6).

Figure 6.
Expression profiles of MaCel1 in different developmental stages of M. alternatus. EL: early larvae; LL: late larvae; PP: pupae; MA: male adult; FA: female adult. The data in the graph were mean ± standard error, and the different letters on the column represent the significant difference in the relative expression in different developmental stages.
4. Discussion
Insects that feed on plant materials, such as termites and longhorn beetles, can use cellulose as a carbon source. Thus, cellulase is very important to digest the cellulose that the insects eat. We obtained the endogenous cellulase gene c39850 sequence based on the transcriptome data of M. alternatus at different developmental stages and named it MaCel1. The amino acid sequence of MaCel1 gene and β-Glucosidase gene of A. glabripennis had the highest similarity; suggesting that these two insects have similar evolutionary timing. The enzyme activity of the β-D-Glucosidase of the MaCel1 was 256.02 ± 43.14 U/L, which indicated that MaCel1 protein had certain β-D-Glucosidase activity. It is suggested that MaCel1 may be a new member of the insect cellulase gene and its catalytic properties should be further investigated. There are different types of cellulase in different types of insects [43]. The β-D-Glucosidase activity of the MaCel1 was higher than that of the M. alternatus digestive tract previously reported, which might be caused by the different test objects [25]. M. alternatus intestinal crude enzyme solution was used in the previous study but purified protease was used in this experiment. Degradation enzymes have been identified in the digestive track of Thermobia domestica, including cleavage double monooxygenase, which weaken cellulose fibers and make cellulose fibers easier to degrade [44]. By using the same expression vector to express two endogenous lignocellulase enzymes of Macrotermes barneyi, it was found that the synergistic factor was increased and the synergistic effect was better than when expressed separately [45]. The next step is to try to express lignocellulase with high activity and good properties in the same host.
By using real-time fluorescent quantitative PCR, we showed that the relative expression of MaCel1 in the early larvae of M. alternatus was higher than that in the late larvae. Our result is consistent with a previous study that showed lower cellulase activity in Ceratoides glabra with the increase of the larval age [20]. The cellulase activity of the A. glabripennis larvae was also measured under natural conditions and a similar pattern was found [21]. This phenomenon may be related to the feeding pattern of the M. alternatus larvae. The young stage is the peak feeding period, with a large amount of food digested and a greater demand for cellulase. In the advanced larval stage, the less it fed, the less it demanded cellulase. During the pupal stage of M. alternatus, the expression of MaCel1 was the lowest. At this stage, M. alternatus had stopped feeding, and the main way to obtain energy is to decompose and digest its own fat, thus the demand for cellulase was very low. After emergence of M. alternatus adults, they feed on fresh pine branches to supplement their nutrition, and the main component of pine branches is still cellulose, indicating that the MaCel1 should play an important role in the digestion process by M. alternatus adults. The endoglucanase activity in adults was also higher than that in the larvae from Cyrtotrachelus buqueti Guer [46]. The relative expression level of MaCel1 in adults was significantly higher than that in other stages due to multiple factors. Due to the complexity of the cellulase system of M. alternatus and the high expression level of other kinds of cellulase, MaCel1 only plays the auxiliary function [22]. On the other hand, it is also possible that the cellulase secreted by exogenous microorganisms plays a major role in the degradation of cellulose, as the cellulase activity of M. alternatus adults differs under different feeding conditions [47].
Research on the effects of RNAi on termites by targeting the conserved regions of the five endoglucanase genes of T. domestica showed that the injection of dsRNA and oral administration may cause significant silencing of the Coptotermes formosanus (CfEGs) gene, leading to death, decreased enzyme activity, and weight loss [48]. The cellulase activity in termites can be successfully reduced by using cellobiose imidazole and fluoromethyl cellobiose as cellulase inhibitors [49]. Thus, it is possible to control the population of M. alternatus by restricting its digestive function.
5. Conclusions
The results demonstrate the existence of the endogenous cellulase gene, MaCel1 in the digestive track of M. alternatus with 14 cellulase enzymes. The expression level of MaCel1 differed among developmental stages, being the highest in the adult stage. The β-D-Glucosidase activity of MaCel1 was higher than the β-Glucosinolate activity. The results of this study provide a theoretical basis for elucidating the digestive physiology of M. alternatus and developing new control strategies of M. alternatus, thereby controlling pine wilt disease.
Author Contributions
Conceived and designed the experiments: X.H. and F.Z.; performed the experiments: H.C., Y.L., and G.L.; analyzed the data: Y.L., H.C., and S.W.; contributed reagents/materials/analysis tools: R.W., Q.M., and X.C.; writing—review and editing: M.T.; wrote the paper: Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (grant numbers 31800548); the National Key Research and Development Program of China (grant number 2017YFD0600105); the Forestry Science Research Project of Fujian Forestry Department (grant number Minlinke [2019] 16); the Forest Science Peak Project of College of Forestry, Fujian Agriculture and Forestry University (grant numbers 71201800743, 71201800701); the Science and Technology innovation Special Fund project of Fujian Agriculture and Forestry University (grant numbers CXZX2019040S).
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Hemicellulase-associated enzyme family in the M. alternatus.
Table A1.
Hemicellulase-associated enzyme family in the M. alternatus.
| Family | Enzyme Activity or Function | Gene ID | EC# (Putative) |
|---|---|---|---|
| GH1 | Beta-glucosidase | c89763 | EC 3.2.1.21 |
| GH3 | Beta-glucosidase | c31589 | EC 3.2.1.21 |
| GH3 | Beta-glucosidase | c124724 | EC 3.2.1.21 |
| GH3 | Beta-glucosidase | c114480 | EC 3.2.1.21 |
| GH5 | Endo-β-1,4-glucanase | c39850 | EC 3.2.1.4 |
| GH5 | Endo-β-1,4-glucanase | c77902 | EC 3.2.1.4 |
| GH9 | Endo-β-1,4-glucanase | c81097 | EC 3.2.1.4 |
| GH9 | Endo-β-1,4-glucanase | c94844 | EC 3.2.1.4 |
| GH45 | Endo-β-1,4-glucanase | c71020 | EC 3.2.1.4 |
| GH45 | Endo-β-1,4-glucanase | c79607 | EC 3.2.1.4 |
| GH45 | Endo-β-1,4-glucanase | c86570 | EC 3.2.1.4 |
| GH48 | Exo-β-1,4-glucanase | c34374 | EC 3.2.1.91 |
| GH48 | Exo-β-1,4-glucanase | c83558 | EC 3.2.1.91 |
| GH48 | Exo-β-1,4-glucanase | c95129 | EC 3.2.1.91 |
References
- Apte, A.A.; Senger, R.S.; Fong, S.S. Designing novel cellulasen systems through agent-based modeling and global sensitivity analysis. Bioengineered 2014, 5, 243–253. [Google Scholar] [CrossRef]
- Peterson, B.F.; Stewart, H.L.; Scharf, M.E. Quantification of symbiotic contributions to lower termite lignocellulose digestion using antimicrobial treatments. Insect Biochem. Mol. Biol. 2015, 59, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Pothula, R.; Shirley, D.; Perera, O.P.; Klingeman, W.E.; Oppert, C.; Abdelgaffar, H.M.Y.; Johnson, B.R.; Jurat-Fuentes, J.L.; Cullen, D. The digestive system in zygentoma as an insect model for high cellulase activity. PLoS ONE 2019, 14, e0212505. [Google Scholar] [CrossRef] [PubMed]
- Shelomi, M.; Wipfler, B.; Zhou, X.; Pauchet, Y. Multifunctional cellulase enzymes are ancestral in Polyneoptera. Insect Mol. Biol. 2020, 29, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, L.R. The Physiological and symbiotic relationships between the intestinal protozoa of termites and their host, with special reference to Reticulitermes flavipes kollar. Biol. Bull. 1924, 46, 178–201. [Google Scholar] [CrossRef]
- Watanabe, H.; Tokuda, G. Animal cellulases. Cell. Mol. Life Sci. 2001, 58, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Li, H.J.; Young, S.E.; Poulsen, M.C.; Cameron, R. Symbiont-mediated digestion of plant biomass in fungus-farming insects. Annu. Rev. Entomol. 2020. [Google Scholar] [CrossRef]
- Watanabe, H.; Noda, H.; Tokuda, G.; Lo, N. A cellulase gene of termite origin. Nature 1998, 394, 330–331. [Google Scholar] [CrossRef]
- Su, L.J.; Wu, Z.W.; Gao, X.H.; Zhao, P.F.; Xiao, Y.X.; Chu, J.P.; Song, A.D. Identification of proteins in the gut of Tsaitermes ampliceps (Isoptera: Rhinotermitidae). Acta Entomol. Sin. 2020, 63, 825–834. [Google Scholar]
- Lo, N.; Tokuda, G.; Watanabe, H.; Rose, H.; Noda, H. Evidence from multiple gene sequences indicates that termites evolved from wood-feeding cockroaches. Curr. Biol. 2000, 10, 801–804. [Google Scholar] [CrossRef]
- Nakashima, K.; Watanabe, H.; Saitoh, H.; Tokuda, G.; Azuma, J.I. Dual cellulose-digesting system of the wood-feeding termite, Coptotermes formosanus Shiraki. Insect Biochem. Molec. 2002, 32, 777–784. [Google Scholar] [CrossRef]
- Scharf, M.E.; Dancia, W.S.; Pittendrigh, B.R.; Bennett, G.W. Caste- and development-associated gene expression in a lower termite. Genome Biol. 2003, 4, 183–210. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, G.; Lo, N.; Watanabe, H.; Arakawa, G.; Matsumoto, T.; Noda, H. Major alteration of the expression site of endogenous cellulases in members of an apical termite lineage. Mol. Ecol. 2004, 13, 3219–3228. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, G.; Lo, N.; Watanabe, H.; Slaytor, M.; Noda, H. Metazoan cellulase genes from termites: Intron/exon structures and sites of expression. BBA-Gene. Struct. Expr. 1999, 1447, 146–159. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, S.R.; Yoon, H.J.; Kim, I.; Lee, K.S.; Je, Y.H. cDNA cloning, expression, and enzymatic activity of a cellulase from the mulberry longicorn beetle, Apriona germari. Comp. Biochem. Physiol. 2004, 139, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, M.; Watanabe, H.; Lo, N.; Saito, H. Purification, characterization, cdna cloning and nucleotide sequencing of a cellulase from the yellow-spotted longicorn beetle, Psacothea hilaris. FEBS J. 2010, 270, 3455–3460. [Google Scholar] [CrossRef]
- Wei, Y.D.; Lee, K.S.; Gui, Z.Z.; Yoon, H.J.; Kim, I.; Zhang, G.Z.; Hyung, J.Y.; Iksoo, K.; Zhang, G.Z.; Guo, X.J.; et al. Molecular cloning, expression, and enzymatic activity of a novel endogenous cellulase from the mulberry longicorn beetle, Apriona germari. Comp. Biochem. Physiol. 2006, 145, 220–229. [Google Scholar]
- Wei, Y.D.; Lee, K.S.; Gui, Z.Z.; Yoon, H.J.; Kim, I.; Je, Y.H.; Lee, S.M.; Zhang, G.Z.; Guo, X.J.; Sohn, H.D.; et al. N-linked glycosylation of a beetle (Apriona germari) cellulase Ag-EGase II is necessary for enzymatic activity. Insect Biochem. Mol. Biol. 2006, 36, 435–441. [Google Scholar] [CrossRef]
- Wei, Y.D.; Lee, S.J.; Lee, K.S.; Gui, Z.Z.; Yoon, H.J.; Kim, I.; Je, Y.H.; Guo, X.J.; Sohn, H.D.; Jin, B.R. N-glycosylation is necessary for enzymatic activity of a beetle (Apriona germari) cellulase. Biochem. Biophys. Res. Commun. 2005, 329, 331–336. [Google Scholar] [CrossRef]
- Kim, N.; Choo, Y.M.; Lee, K.S.; Hong, S.J.; Seol, K.Y.; Je, Y.H.; Sohn, H.D.; Jin, B.R. Molecular cloning and characterization of a glycosyl hydrolase family 9 cellulase distributed throughout the digestive tract of the cricket Teleogryllus emma. Comp. Biochem. Physiol. 2008, 150, 368–376. [Google Scholar] [CrossRef]
- Calderón-Cortés, N.; Watanabe, H.; Cano-Camacho, H.; Zavala-Páramo, G.; Quesada, M. cDNA cloning, homology modelling and evolutionary insights into novel endogenous cellulases of the borer beetle Oncideres albomarginata Chamela (Cerambycidae). Insect Mol. Biol. 2010, 19, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, G. Plant cell wall degradation in insects: Recent progress on endogenous enzymes revealed by multi-omics technologies. In Advances in Insect Physiology; Academic Press: Cambridge, MA, USA, 2019; pp. 97–136. [Google Scholar]
- Xia, D.G.; Wei, Y.D.; Zhang, G.Z.; Zhao, Q.L.; Zhang, Y.H.; Xiang, Z.H.; Lu, C. cDNA cloning, expression, and enzymatic activity of a novel endogenous cellulase from the beetle Batocera horsfieldi. Gene 2013, 514, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Geib, S.; Jones, D.; Sellmer, J.; Morewood, D.; Hoover, K. Cellulose digestion in the larvae of the Asian longhorned beetle, (Anoplophora glabripennis). In Proceedings, 16th U.S. Department of Agriculture Interagency Research Forum on Gypsy Moth and Other Invasive Species; U.S. Department of Agriculture, Forest Service, Northeastern Research Station: Newtown Square, PA, USA, 2005. [Google Scholar]
- Li, X.J.; Luo, Y.Q.; Yan, X.F.; Tian, G.F.; Nian, Y.J.; Sun, H. Changes of cellulase activity in Anoplophora glabripennis larvae. For. Sci. 2011, 47, 204–207. [Google Scholar]
- Suo, F.M.; Lin, C.C.; Wang, H.J.; Ding, Z.W.; Xu, T.S. Study on the character of cellulase in Monochamus alternatus. For. Res. 2004, 17, 583–589. [Google Scholar]
- Suo, F.M.; Wang, H.J.; Ding, Z.W.; Xu, T.S. Study on the character of cellulase in Monochamus alternatus IV. Effect of feeding conditions on the Monochamus alternatus cellulase activity. For. Res. 2007, 20, 381–384. [Google Scholar]
- Wu, S.Q.; Zhu, X.L.; Liu, Z.X.; Shao, E.S.; Rebeca, C.L.; Guo, Y.J.; Xiong, Y.T.; Mou, Y.N.; Xu, R.X.; Hu, X.; et al. Identification of genes relevant to pesticides and biology from global transcriptome data of Monochamus alternatus Hope(Coleoptera: Cerambycidae) larvae. PLoS ONE 2016, 11, e0147855. [Google Scholar] [CrossRef]
- Wu, S.Q.; Yuan, W.M.; Tian, X.J.; Fan, B.; Fang, X.; Ye, J.R.; Ding, X.L. Specific and functional diversity of endophytic bacteria from pine wood nematode Bursaphelenchus xylophilus with different virulence. Int. J. Biol. Sci. 2013, 9, 34–44. [Google Scholar] [CrossRef]
- Kim, S.R.; Lee, W.K.; Lim, C.H.; Kim, M.; Kafatos, M.C.; Lee, S.H.; Lee, S.S. Hyperspectral analysis of pine wilt disease to determine an optimal detection index. Forests 2018, 9, 115. [Google Scholar] [CrossRef]
- Chen, H.; Hao, D.; Wei, Z.; Wang, L.; Lin, T. Bacterial communities associated with the pine wilt disease insect vector Monochamus alternatus (Coleoptera: Cerambycidae) during the larvae and pupae stages. Insects 2020, 11, 376. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Zha, Y.P.; Cai, S.S.; Hong, C.H.; Liang, P.; Chen, J.Y. Application of harmonic radar to analyze dispersal behavior of the Japanese pine sawyer beetle, Monochamus alternatus (Coleoptera: Cerambycidae). Entomol. Res. 2020, 50, 50–58. [Google Scholar] [CrossRef]
- Zhen, L.D.; Huang, X.F.; Yang, R.N.; Chen, J.Y.; Wang, M.Q. Functional analysis of two odorant-binding proteins, maltobp9 and maltobp10, in Monochamus alternatus hope. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, L.; Ning, J.; Wickham, J.D.; Tian, H.K.; Zhang, X.M.; Yang, M.L.; Wang, X.M.; Sun, J.H. miR-31-5p regulates cold acclimation of the wood-boring beetle Monochamus alternatus via ascaroside signaling. BMC Biology 2020, 18. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.J.; Wang, Y.F.; O’Donoghue, A.J.; Jiang, Z.Z.; Rebeca, C.L.; Liang, G.H.; Hu, X.; Wang, R.; Xu, L.; Guan, X.; et al. Engineering of multiple trypsin/chymotrypsin sites in Cry3A to enhance its activity against Monochamus alternatus Hope larvae. Pest Manag. Sci. 2020, 76, 3117–3126. [Google Scholar] [CrossRef] [PubMed]
- Gorman, M.J.; Dittmer, N.T.; Marshall, J.; Kanost, M.R. Characterization of the multicopper oxidase gene family in Anopheles gambiae. Insect Biochem. Mol. Biol. 2008, 38, 817–824. [Google Scholar] [CrossRef]
- Li, H.A.; Wang, C.Y.; Xiao, W.J.; Yang, Y.X.; Hu, P.; Dai, Y.J.; Jiang, Z.B. Dissecting the effect of polyethylene glycol on the enzymatic hydrolysis of diverse lignocellulose. Int. J. Biol. Macromol. 2019. [Google Scholar] [CrossRef]
- Wang, Z.H.; Hu, R.M.; Ye, X.Q.; Huang, J.H.; Chen, X.X.; Shi, M. Laccase 1 gene from Plutella xylostella (PxLac1) and its functions in humoral immune response. J. Insect Physiol. 2018, 107, 197–203. [Google Scholar] [CrossRef]
- Feng, B.; Guo, Q.S.; Mao, B.P.; Du, Y.J. Identification and selection of valid reference genes for assaying gene expression in the chemosensory tissues of Monochamus alternatus (Coleoptera: Cerambycidae) by RT-Qpcr. Acta Entomol. Sin. 2016, 59, 427–437. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Roman, L.T.; Natalie, D.F.; John, D.J.; Aviva, R.J.; Boris, K.; Eugene, V.K.; Dmitri, M.K.; Raja, M.; Sergei, L.M.; Anastasia, N.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 1–14. [Google Scholar]
- Hara, M.; Eto, H.; Kuboi, T. Tissue printing for myrosinase activity in roots of turnip and Japanese radish and horseradish: A technique for localizing myrosinases. Plant Sci. 2001, 160, 425–431. [Google Scholar] [CrossRef]
- Liu, J.; Song, K.; Teng, H.; Zhang, B.; Li, W.; Xue, H.; Yang, X. Endogenous cellulolytic enzyme systems in the longhorn beetle Mesosa myops (Insecta: Coleoptera) studied by transcriptomic analysis. Acta Biochim. Biophys. Sin. 2015, 47, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Sabbadin, F.; Hemsworth, G.R.; Ciano, L.; Henrissat, B.; Dupree, P.; Tryfona, T.; Marques, R.D.S.; Sweeney, S.T.; Besser, K.; Elias, L.; et al. An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Ning, N. Cloning and Expression of Lignocellulase Gene from Termite and Its Intestinal Microflora. Master’s Thesis, Shandong University, Jinan, China, 2017. [Google Scholar]
- Wang, M.J.; Wang, C.J.; Chen, X.Y.; Li, Y.Q.; Liang, J.J.; Luo, C.B. Determination of the optimum reaction conditions and screening of key genes for endoglucanase form Cyrtotrachelus buqueti Guer. J. Entomol. 2019, 62, 1150–1161. [Google Scholar]
- Li, X.J.; Yan, X.F.; Luo, Y.Q.; Tian, G.F.; Nian, Y.J.; Zhang, T.L. Cellulase activity and its relationship with host selection in the Anoplophora glabripennis, (Coleoptera:Cerambycidae). Acta Petrol. Sin. 2010, 10, 1179–1183. [Google Scholar]
- Wu, W.; Gu, D.; Yan, S.; Li, Z. RNA interference of endoglucanases in the formosan subterranean termite coptotermes Formosanus shiraki (Blattodea: Rhinotermitidae) by dsRNA injection or ingestion. J. Insect Physiol. 2019, 112, 15–22. [Google Scholar] [CrossRef]
- Zhou, X.G.; Wheeler, M.M.; Oi, F.M.; Scharf, M.E. Inhibition of termite cellulases by carbohydrate-based cellulase inhibitors: Evidence from in vitro biochemistry and in vivo feeding studies. Pestic. Biochem. Physiol. 2008, 90, 31–41. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).