Abstract
Common ash (Fraxinus excelsior L.) is a tree species of significant ecological and economic importance that has suffered a devastating decline since the 1990s in Europe. Native ash species are being threatened by the alien invasive fungus Hymenoscyphus fraxineus, which causes ash dieback. The main goal of the study was to develop markers for traits related to tolerance to ash dieback and to investigate whether genotypes selected for tolerance were genetically different from susceptible wild populations. We phenotyped 326 ash trees from Sweden for disease severity and genotyped them using 63 amplicon-derived single-nucleotide polymorphism (SNP) markers derived from genes in 40 scaffolds spanning 8 MB in total, which represents approximately 1% of the ash genome. We used a mixed linear model to test for an association between genotypic variation at these loci and disease severity of ash. In total, two SNPs were found to have significant associations. One non-synonymous SNP associated with the disease severity of ash was found in a gene predicted to encode a subtilisin-related peptidase S8/S53 domain. A second marginally significant marker was associated with an LRR gene. Our results demonstrate an inexpensive time-effective method for generating genomic data that could have potential for use in future tree breeding programs and provide information for marker-assisted selection. Our study also showed a low differentiation between genotypes selected for disease tolerance and the wild population of ash representing a range of susceptibilities to ash dieback, indicating opportunities for further selection without significantly losing genetic diversity in the ash population.
1. Introduction
Common ash (Fraxinus excelsior L.) is a broad-leaved tree species of major ecological and economical importance in European forests [1]. The species is predominantly distributed throughout northern and central Europe. Common ash is currently suffering from the ash dieback disease epidemic caused by the ascomycete fungus Hymenoscyphus fraxineus (T. Kowalski) Baral, Queloz and Hosoya [2,3], which is an alien invasive fungus pathogen in Europe that affects trees of all ages and has resulted in severe mortality throughout the natural distribution range of common ash [4,5,6]. Ash dieback has had a devastating impact on ash population since first noted in Sweden in 2001 [7,8]. This has resulted in the declaration of common ash as a threatened species in Sweden [9]. The loss of a high proportion of the ash trees will reduce the genetic diversity in the species, possibly making it more vulnerable to other disturbances and cause adverse ecological effects including reduced associated biodiversity [3,4,10,11,12]
Susceptibility to ash dieback disease, often scored as a high level of crown damage, has a strong negative effect on the growth and survival of ash trees [13,14]. Previous studies reported significant levels of phenotypic and genetic variability for susceptibility to ash dieback in common ash in natural forests and established field trials [14,15,16,17]. Although the observed differences in crown damage in natural settings could be due to disease escape, inoculation studies have shown that active defense is involved in the susceptibility phenotype of ash trees [18,19,20]. Susceptibility to ash dieback is a quantitative trait [21,22] and heritability for traits related to ash dieback severity (crown symptoms) is high (narrow sense heritability h2: 0.49–0.53) [13,14,23] (broad sense heritability H2: 0.42–0.54) [16,17], suggesting there is good scope for breeding less susceptible trees in future. Substantial genetic variation together with heritability estimates of susceptibility traits indicate a good potential of selection and breeding of ash genotypes tolerant to ash dieback [12].
Since it takes a long time to evaluate resistance properties against pathogens, partly due to the long generation times in forest trees, phenotyping selection is difficult, costly and time consuming. Molecular markers can help to identify superior genotypes for tree breeding and restoration. Marker-assisted selection may facilitate early selection of trees with favorable traits and could therefore accelerate the selection process [24]. Harper and co-workers [15] developed molecular markers to identify individuals with low susceptibility to ash dieback using an associative transcriptomics method based on gene expression variants. These markers were associated with tolerance to ash dieback and indicate a role of the MADS box transcription factor genes. However, only three markers [15] were able to significantly predict tolerance to the disease. These molecular markers were used to identify trees with a reduced susceptibility to ash dieback in Danish, British and Swedish ash populations [22,25] and had a moderate predictive capacity. These results are highly encouraging, but for efficient marker-assisted selection several more markers are needed in addition to the available markers. Stocks et al. [26] showed that single-nucleotide polymorphisms (SNPs) associated with fewer ash dieback symptoms allowed better predictions of genomic estimated breeding values (GEBVs), than randomly selected SNPs in a transpopulation genomic selection approach for ash dieback-tolerant trees, further underlining the need to characterize the genetics underlying tolerant phenotypes.
Next-generation sequencing (NGS) has revolutionized plant and animal research through the generation of molecular markers such as SNPs for high-throughput genotyping [27,28,29]. New NGS methods have allowed the construction of high-density linkage maps [30,31,32] and association genetics studies [33,34] in trees often use approaches for reduced representation sequencing. We have used a multiplex PCR approach for amplicon sequencing [29] with the aim to identify genetic markers for trait-related tolerance to ash dieback, which enables the enrichment of large numbers of amplicons in a single reaction. In multiplex sequencing, each sample is represented by a unique barcode sequence or tag added to the DNA products that are to be sequenced. The sample with the tag determines which sample the read originated from, enabling the assaying of multiple samples in a single sequencing run. After sequencing, the reads are sorted by the detection of the appropriate barcode.
The main objective of this study was to identify novel genetic markers for traits related to tolerance to ash dieback. We hypothesize that disease severity is associated with specific genetic variation in common ash. A secondary objective was to investigate if the population of ash genotypes selected for their tolerance phenotype are genetically differentiated from the susceptible wild ash population to ash dieback in Sweden.
2. Materials and Methods
2.1. Plant Materials and Phenotyping
In total, 111 tolerant and 215 susceptible ash genotypes from Sweden were included in this study. The material originated from several sources combining genetic variation from natural ash stands with material pre-selected for its tolerant phenotype. To represent the standing variation, 143 unrelated genotypes were assessed for disease severity and phenotypically selected in the counties Uppland and Öland in the year 2016. Secondly, 70 plus trees (trees with superior properties), originating from different regions of southern Sweden, still remaining in a seed orchard established before the ash dieback epidemic arrived were included [17]. Finally, we used 113 ash genotypes pre-selected phenotypically for their tolerance to ash dieback from southern Sweden and Gotland [25], of which 72 were tolerant with low disease severity scores (disease severity <10%) (Supplementary Table S1).
Disease severity assessments were conducted differently, using one of three methods (see Supplementary Table S1). For seed orchard trees, the disease severity assessment was based on a six-grade scale [17], ranging from 0 (none) to 6 (very serious damage) based on a visual scoring of crown damage to get a measure of health status. For the 113 ash trees from southern Sweden and Gotland described by Menkis et al. [25], the scoring system was based on a visual monitoring of health status where trees were classified as either tolerant (0%–10% crown damage) or susceptible (more than 10% crown damage). The remaining trees were phenotyped according to Kirisits and Freinschlag [35]. Briefly, the crown of each individual tree was divided into thirds. Thereafter, each third of the crown was assigned a score of ash dieback severity ranging from 0 (0% damage) to 6 (100% damage). The values of the three crown thirds were averaged to obtain an ash dieback severity rating in percent for each genotype [35]. The phenotypic data from trees in the seed orchard, Uppland and Öland, were divided into two categories with a score from 1—tolerant (disease severity score of 0–2.85), and 2—susceptible (greater than 2.85). In order to allow for marker-trait association, all scores were transformed into discrete unified disease scores corresponding to values 1 and 2 in subsequent analyses, where 1 was tolerant (disease severity <10%), and 2 was susceptible (>10%) trees (Supplementary Table S1).
2.2. DNA Extraction
Total genomic DNA from the leaf tissue of each genotype was isolated with CTAB (cetyl trimethylammonium bromide) buffer [36] with 2% (w/v) polyvinylpolypyrrolidone added. DNA samples were purified by the NucleoSpin® gDNA Clean-up kit to remove PCR inhibitors (MACHEREY-NAGEL, Düren, Germany). DNA concentration was measured using a NanoDrop Spectrometer ND 1000, SaveenWerner, Wilmington, U.S.A.
2.3. Selection of the Gene Models and Primer Design
In order to create gene model-derived amplicons for genotyping, the 1000 largest scaffolds in the BATG-0.5 release of the ash genome (http://www.ashgenome.org/, accessed on 13/02/2017) were identified. From each of the largest scaffolds, one or two gene models were selected based on the length of predicted CDS (coding sequence) of the gene models to design the primers. Fasta files were downloaded for the longest transcript CDS sequence from the BATG-0.5-CLCbioSSPACE genome assembly (http://www.ashgenome.org/, accessed on 13/02/2017). The selected target sequences were collected in a Fasta file and Batchprimer3 [37] was used to design primers with a melting temperature of 60–63 °C, product size of 95–105 bp and primer size of 18–24 bp. The returned amplicons with a product size of 97–100 bp primer pairs with the smallest Tm difference and smallest 3′ complementarity were selected. In total, 1000 amplicons were designed from the ash tree genomes database (Supplementary Table S2). We added forward (ctctctatgggcagtc) and reverse (ctcgtgtctccgact) heel sequences to each of the amplicons before ordering the primers. Additionally, 93 pairs of indexing primers with unique tags, to tag individual samples prior to pooling, were ordered (Supplementary Table S2).
2.4. Multiplex PCR and Library Preparation
The PCR conditions used to amplify the amplicons were modified from Nguyen et al. [29]. The PCR mixture (10 μL) consisted of a 1X Phusion HF PCR buffer (provides 1.5 mM MgCl2), 5 μM each of gene pool, 2 mM of each dNTP (Deoxynucleoside triphosphate), Phusion Hot Start II High-Fidelity DNA Polymerase and approximately 25 ng of genomic template DNA. PCR was done in a 2720 thermal cycler (AB Applied Biosystems) using the following temperature cycle: denaturation cycle at 98 °C for 1 min, followed by 10 cycles of 98 °C for 30 s, annealing at 61 °C for 45 s, extension at 72 °C for 30 s, followed by another 15 cycles of 98 °C for 30 s, annealing at 59 °C for 30 s, extension at 72 °C for sec and a final extension at 72 °C for 5 min. A mix of indexing primers of 2 μM was added to each PCR reaction mixture. The PCR conditions for the second round of amplification consisted of initial denaturation at 98 °C for 1 min, 3 cycles of 98 °C for 30 s, 50°C for 30 s, and 72 °C for 30 min, followed by 10 cycles of 98 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 min followed by a final extension at 72 °C for 10 min.
We used 20 gene pools with 50 amplicons in each pool. Amplification by PCR was performed using these 20 gene pools on 326 unrelated genotypes of ash in this study. These genotypes were divided into five batches to allow the tagging and tracing of each individual genotype. Briefly, we divided the genomic DNA of 326 genotypes into five batches with samples in each together with negative controls (water instead of DNA). Amplification by PCR was performed with each gene pool on each ash genotype. We assigned a pair of index primers to each genotype in the batch. The index primers were used with different samples in other batches. Therefore, the batches were run in separate library preps for the gene sequencing in order to avoid cross contamination. To balance the number of samples in the sequencing library with the aim to get approximately the same read depth of each sample, a pooling strategy was applied. In total 18 pools were prepared. These 18 pools were separated on 1.8% agarose TAE (Tris-acetate-EDTA) gel. Each library was excised (approximately 150 bp) from the gel and purified using a GeneJet kit (ThermoFisher Scientific, Carlsbad, California, USA) including an addition of both isopropanol and wash with a binding buffer step. The DNA quality of each library was quantified by a Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, California, USA).
The amplicon libraries were sequenced at the SNP&SEQ Platform (ScilifeLab, Uppsala). One sequencing library was prepared from each of the 18 Amplicon libraries using the ThruPLEX DNA-seq library preparation kit (Rubicon Genomics, ScilifeLab, Uppsala, Sweden). Sequencing was conducted using 150 cycles of paired-end sequencing of the 18 libraries in one run with the rapid mode of the HiSeq2500 system and v2 sequencing chemistry (Illumina Inc., ScilifeLab, Uppsala, Sweden). The raw sequences were submitted to the sequence Read Archive (SRA) portal (NCBI) under BioProject accession number PRJNA639842.
2.5. Bioinformatic Analysis, Demultiplexing and Variant Calling
The workflow is based on the Genome Analysis Toolkit (GATK) [38] and is illustrated in Supplementary Figure S1. The multiplexed samples from targeted amplicon sequencing were demultiplexed using an in-house developed script (http://github.com/troe27/hiplexdeplex, accessed on 21/6/2018) implemented in Phython 2.7 (https://www.python.org/, accessed on 21/6/2018) to sort reads based on their sequence tags. This script first combines forward and reverse reads and aligns 5′ and 3′ heels to the sequences and infers the tags of the read via their position relative to the heel sequences. The number of reads that fail to match 5′ or 3′ heel and number of reads that match the heel but do not have the tags; chimeric reads were discarded. Reads were organized into new files with heel and tag sequences removed. The data were processed using setting h5score 0.9; h3score 0.9 (to match to the heel at 5′ and 3′ found in the read, there has to be at least 90% similarity in order to be considered as a valid heel); paired params 7:10:5 (Kmer size:minimum length of high scoring segment pair:minimum overlap) and filtering of 1:50:150:20:15 (no filtering of reads:minimum length:maximum length:maximum phred-score cut-off: minimum phred-score cut off).
The initial quality control of the sequencing data was made by the sequencing service provider SNP&SEQ Platform (SciLifeLab, Uppsala, Sweden). First, demultiplexed Illumina sequence reads were aligned to the reference sequence (BATG-0.5-CLCbioSSPACE) (http://www.ashgenome.org/, accessed on 28/6/2018) with Burrows–Wheeler Aligner (BWA mem) version 0.7.5a [39]. In this step, SAM files were generated, which were converted to BAM files using SAMtools [40]. Reads were then processed (PCR duplicates removed, read groups added to alignment file) with Picard tools (broadinstitute.github.io/picard/, accessed on 3/8/2018). SAMtools mpileup version 0.1.19 [40] was used to generate variants and Bcftools version 1.2 [40] was used for variant calling from the output of the SAMtools mpileup version 0.1.19 [40]. All vcf files were merged with the help of Bcftools [40]. We used VCFtools v0.1.12b [41] to filter variants. The dataset was filtered for SNPs with a minor allele frequency (MAF) of 0.05 and a minimum coverage of 70%; i.e., the proportion of individuals for which the base at the position could be called.
The extent of LD (Linkage disequilibrium) was estimated by calculating the squared allele frequency correlation coefficient (r2) between all pairs of markers with only the p-values < 0.05 for each pair of markers considered as significant using the software package TASSEL v.5.2.5 [42].
2.6. Population Structure Analysis
To minimize the impact of linkage between SNPs in the analysis, we selected a single SNP representing each scaffold. The population structure was determined using a Principal component analysis (PCA) obtained from Tassel software version 5.2.5 [42] and the Bayesian clustering algorithm implemented in STRUCTURE version 2.3.4 [43] to estimate the number of hypothetical subpopulations (K) and to estimate the membership probability of each genotype to the subpopulations. The optimal K value was determined based on the ΔK from the Structure Harvester v0.6.94 program [44]. The admixture model was run with correlated allele frequencies with no prior information using 100,000 burn-in time and 100,000 Markov chain Monte Carlo (MCMC) iterations. Cluster values (k) from 1 to 10 were chosen and 5 independent runs for each k were chosen to obtain consistent results.
2.7. Association Analysis
We preliminary ran the general linear model (GLM) and the mixed linear model (MLM) implemented in the program TASSEL, version 5.2.5 [42], with 63 SNPs to find association between SNP markers and disease severity for ash genotypes. We then filtered scaffolds with multiple SNPs to one SNP per scaffold by retaining the SNP with lower p-values derived from the TASSEL output and ran an association analysis with one SNP per scaffold. An association analysis using the GLM model was made with the SNPs as a fixed effect and a matrix of population structure as a covariate (GLM+PCA model); in order to reduce false associations, a permutation test with 10,000 replicates was implemented in TASSEL, version 5.2.5.
The MLM model takes into consideration both population structure and relatedness (PCA+K models). The kinship matrix (K) which takes in account the relatedness among genotypes as random effects was estimated using the pairwise kinship matrix as a covariate in the mixed model. The MLM was used with the centered IBS (identity by state) matrix as kinship matrices calculated in Tassel V 5.2.50 [42]. The PCA matrix of the population structure was accounted for, incorporating the first three PCA as covariates in the model [42]. For MLM, we analyzed the 249 genotypes for 40 SNP markers associated with a disease severity with a p-value below 0.05. Finally, we estimated the proportion of significantly associated markers by applying a false discovery rate (FDR) procedure according to Benjamini–Hochberg-adjusted p-value (p < 0.05) (https://tools.carbocation.com/FDR, accessed on 13/2/2020). The markers considered to be significantly associated with the trait are represented in the Manhattan plot. Quantile–quantile (QQ) plots of the expected and observed p-values was plotted to evaluate the adequacy of controlling type I errors.
The localization of SNPs in coding regions was based on the annotation of gene models as provided by the Ash Genome Database (http://www.ashgenome.org/, accessed on 25/10/2017). The open reading frame (ORF) for the gene models, each containing SNPs, were identified using Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/orf_find.html, accessed on 14/11/2019) and compared against the NCBI protein database using BLASTp (basic local alignment search tool for protein), accessed on 4/11/2019 and the protein with the highest E-value was downloaded and aligned with the reading frame for SNP location using Clustal W accessed on 5/11/2019 [45] implemented in BioEdit v. 7.1. accessed on 5/11/2019 [46]. This approach enabled us to locate SNPs by looking at the coding region. For those SNPs in the coding region, the resulting amino acid sequences of variants were translated to determine whether SNP variants were synonymous or non-synonymous. We used the conserved domain database (CDD) accessed on 1/10/2019 [47] to identify the conserved domain of the protein.
2.8. Prediction of Functional Effect of Non-Synonymous SNP and Secondary Structure
We predicted the functional effect of non-synonymous SNPs by Proven accessed on 19/11/2019 [48] to predict if amino acid substitutions caused by these SNPs were deleterious or neutral in nature. A secondary structure prediction was performed by PSIPRED (PSI BLAST-based secondary structure prediction) accessed on 19/10/2019 [49] and I-TASSER accessed on 30/10/2019 [50]. We also used I-TASSER to predict solvent accessibility.
3. Results
3.1. Generation of SNP Markers
Out of 1000 randomly selected amplicons, 655 amplicons (66%) were amplified and produced reads. Amplicons with more than 100 reads in total were retained, leaving 567 amplicons to be mapped to the ash genome. The Illumina HiSeq2500 (SNP&SEQ Platform, ScilifeLab, Uppsala, Sweden) sequencing of 326 individuals generated on an average 0.19 M reads per genotype of which on average 73,834 reads (38.8%) were kept after demultiplexing (Supplementary Table S3). The overall mapping rate was 99.81%–97.83% and the variant calling analysis identified 156,735 putative SNPs in 655 scaffolds in the ash genome. The average size of the SNP holding scaffold was 145 kb. These SNPs were filtered at a call rate of 70% among the sample trees and with a MAF of 0.05. A total of 63 SNPs were identified from 40 scaffolds with an average size of 197.5 kb. The SNP data were filtered to one SNP per scaffold. Forty SNPs and 249 genotypes were retained for a marker-trait association analysis. The pattern of physical LD was examined over the scaffolds harboring the 40 SNPs, covering a total of 8 Mb and the length of the included scaffolds ranged from 503 to 1.35 kb. As expected, none of the marker pairs showed complete linkage. The average LD for statistically significant marker pair values in different scaffolds was r2 = 0.033 (p < 0.05), and LD estimates were statistically significant for 3.85% of the scaffold marker pairs (p < 0.05) (data not shown).
3.2. Low Levels of Differentiation between Material Selected for Disease Tolerance Phenotype and Susceptible Wild Population
To examine the potential effects derived from population stratification, we analyzed the population structure using the 40 SNPs both in TASSEL and STRUCTURE v2.3.4. No population structure was observed in the PCA analysis generated in TASSEL. The first two principal components explained 5.8 and 5.5% of the genetic variance in the population. STRUCTURE has also provided some evidence for K = 2 (data not shown) according to the maximum ΔK value (35.4), indicating little population structure in the studied ash population. A low genetic differentiation was detected between tolerant and susceptible genotypes, and genotypes selected for tolerance phenotypes and the rest of the materials (Table 1). A low FST was also observed between the Uppland and Öland population.

Table 1.
Pairwise FST comparison of ash population.
Tolerant: total tolerant ash genotypes; susceptible: total susceptible ash genotypes; selected for tolerance: selected for tolerance phenotypes only, e.g., disease severity; other materials: selected for other traits as wood quality traits (Seed orchard) and unrelated genotypes with disease severity data collected from Uppland and Öland in the year 2016; Uppland: ash genotypes from Uppland; Öland: genotypes from Öland.
3.3. Marker-Traits Association Identifies Two Scaffolds Associated with the Tolerance Phenotype
To investigate if any association could be detected between the SNPs and the tolerance to ash dieback, we performed marker-traits analyses in TASSEL using an MLM+PCA+K model. The model detected a total of two statistically significant associations (p-value < 0.05) one of which remained significant after correction for multiple testing (FDR < 0.05, Table 2, Figure 1). As a comparison, a GLM+PCA model was also run. This model detected four statistically significant associations between four SNP markers, all of which were identical to the associations with the MLM+PCA+K model and the disease severity of ash (p-value < 0.05, Supplementary Table S4). However, the MLM+PCA+K model showed the smallest departure from the expected p-value (−log10) in the QQ plots, and was therefore least prone to producing false positives. Therefore, the MLM+PCA+K model and its results were considered as the most reliable in the association analysis (Supplementary Figure S2).

Table 2.
Single-nucleotide polymorphism (SNP) locus annotations and significance values for disease severity in ash at p-value < 0.05 and false discovery rate (FDR) < 0.05 using mixed linear model (MLM).
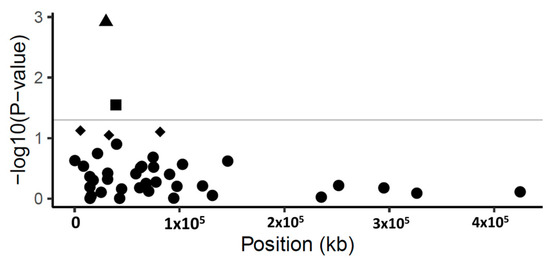
Figure 1.
Manhattan plot of p-values (−log10 scale) for SNP associations to disease severity in common ash. The p-values are plotted against position of the SNP in the scaffolds. Grey line indicates the threshold value (−log10 of p-value of 1.3) for declaring a significant association. Triangle symbol indicates the statistically significant SNP (p-value < 0.001, FDR = 0.04) in scaffold 5992; square symbols show marginally significant SNPs (p-value < 0.05). Diamond-shaped symbol indicates non-significant SNPs (p-value > 0.05) and circles are the non-associated SNPs with disease severity.
The marker-trait association generated a statistically significant association with one scaffold. This scaffold (scaffold 5992) comprised five gene models (Supplementary Figure S3). The SNP SCONTIG5992_29927 on scaffold 5992 was significantly associated to the disease severity of ash (p-value < 0.001, FDR = 0.04), explaining 5.4% of the phenotypic variance (Table 2). This SNP, SCONTIG5992_29927, is located at 29,927 bp in a gene model (FRAEX38873_v2_000299890.1), predicted to encode a Peptidase S8 subtilisin-related Peptidase S8/S53 domain. The substitution encoded by the SNP SCONTIG5992_29927 is located in the coding region changing the amino acid at position 658 in the predicted protein from a tyrosine to an aspartic acid. Another marginally significant SNP (p < 0.05 but FDR > 0.05) SCONTIG5992_29954 was detected on scaffold 5992. This SNP was located 27 bp downstream of SCONTIG5992_29927 in the same gene model (Supplementary Figure S3). This SNP substitutes the amino acid at position 667 in the predicted protein from an alanine to an isoleucine. Both substitutions are predicted to be buried in the mature protein and located outside the active and catalytic site according to PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/, accessed on 19/10/2019) and I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/, accessed on 30/10/2019) analyses. The 5.87 kb scaffold 5992 contains four other gene models, of which three are annotated as a pectin methylesterase (FRAEX38873_v2_000299870.1), proteasome subunit alpha type 5 (FRAEX38873_v2_000299880.1), and fyve finger-containing phosphoinositide (FRAEX38873_v2_000299900.1) (Supplementary Figure S3).
One marginally significant association, i.e., p < 0.05 and FDR > 0.05, was also detected in the marker-trait association, namely SCONTIG6368_39377, in scaffold 6368 contributing to 3.0% of PVE (phenotypic variance explained). This harbors four gene models (Supplementary Figure S3) and the SNP SCONTIG6368_39377 is located within the gene model FRAEX38873_v2_000311990.1 at position 39,377 bp. This gene model encodes a Leucine-rich repeat protein (Table 2, Figure 1); based on the available predicted protein sequence, this SNP is also non-synonymous and would lead to the substitution of an arginine to a glycine (Table 2, Figure 1).
4. Discussion
The selection for and deployment of disease-tolerant tree genotypes are keys to the restoration of sites destroyed by forest disease epidemics and help prevent the functional extirpation of threatened tree species [51] and associated biodiversity. It seems possible to achieve this with the ongoing ash dieback epidemic by identifying ash with improved tolerance using both phenotypic and genotypic selection methods [17,25]. However, phenotypic selection methods, although being simple and straightforward, are relatively slow as they generally involve scoring crown damage in a large number of trees or following trees over a long time to identify putatively resistant individuals [17,23,25]. Therefore, genotypic selection based on, e.g., molecular markers, is desirable. The main objective of the current study was to identify novel genetic markers for tolerance to ash dieback. Furthermore, we wanted to investigate if the collected population of tolerant ash phenotypes was genetically different from the standing natural variation. To address these objectives, we developed and analyzed a set of 1000 new SNP markers using a multiplex PCR approach for amplicon sequencing [29] that after filtering were concentrated to 40 markers in independent scaffolds.
We used 249 phenotyped and genotyped ash trees from Sweden for marker-trait associations with disease severity in ash. One SNP, significantly associated with the disease severity of ash, was found in a gene predicted to encode a subtilisin-related peptidase S8/S53 domain-containing protein. Furthermore, one other scaffold contained marginally significant associations with disease severity. The two significant, or marginally significant, SNPs explained 5.4 and 3.0%, respectively, of the phenotypic variation. These values are well in line with what has been predicted for ash and for forest trees in general where most traits, including disease resistance and tolerance traits, appear to be under the control of many genes with a small-to-modest effect [22,26,52,53]. This means that single markers will not have a very high predictive capacity, but as many loci controlling disease tolerance and resistance are likely to be additive, combining multiple markers, the predictive power in genotypic selection increases [15,21,22]. However, before molecular markers are put to practical use they need to be validated in independent populations [24,51]. An understanding of the causative variation’s influence on the trait may be helpful in designing appropriate validation experiments [54,55].
The SNP SCONTIG5992_29927, which was significantly associated with the disease severity scores, was positioned in a subtilisin-like protease. Several subtilisin-like serine proteases have been linked to plant–microbe interactions and immunity [56,57,58,59]. The functions of subtilisins in plant–pathogen interactions are diverse and poorly understood, but a virus-induced gene silencing of the cotton subtilisin gene GbSBT1 reduced the resistance related to Verticillium dahliae Klebahn in previously resistant cotton (Gossypium hirsutum L.) accessions, and the heterologous expression of the gene enhanced the resistance of Arabidopsis thaliana (L.) Heynh. to Fusarium oxysporum Schltdl., and to V. dahliae infections [60]. Thus, at least some subtilisins can confer direct resistance to pathogens. Whether the SNP SCONTIG5992_29927 in the FRAEX38873_v2_000299890.1 subtilisin are actual causal variants found within the gene influencing the level of disease severity of ash dieback remains to be validated, but it is a promising candidate for marker-assisted selection that should be studied further.
The ash dieback disease epidemic, since its arrival over two decades ago, has caused large losses of ash trees throughout Europe, and maximum mortality rates of over 70% have been estimated in woodland studies [5]. The disease is likely to affect the level of tolerance and genetic diversity among survivors in the species [13,14,23,61]. Genetic diversity is important to maintain high levels of overall population fitness, and to provide sufficient variation to respond to pests and pathogens and adapt to climatic changes [22,51,62,63]. Concerns have been raised that the ongoing ash dieback disease epidemic will reduce the effective population size and genetic diversity in common ash making the regeneration of ash populations difficult, at least in Lithuania which has one of the longest histories with this forest disease epidemic [12]. In the current study, we found little population differentiation and low differentiation between genotypes selected for a tolerance phenotype and standing natural populations of ash in Sweden, a result which is well in agreement with Stock et al. [26], indicating that there are still opportunities for further selection without significantly losing existing diversity in ash.
5. Conclusions
The highly multiplexed PCR amplification method paired with association studies with phenotypic traits proved to be a viable approach for generating candidate markers for tolerance in forest trees. By covering a larger proportion of the genome it should be possible to identify more interesting candidate markers, which could be used for marker-assisted selection for future tree breeding programs. Our study, using 40 SNPs and 249 genotypes, indicates that selection for tolerant phenotypes can be done while maintaining existing diversity in ash. The marker-trait association studies yielded one strong marker candidate for the selection of tolerant ash in the non-synonymous SNP in a subtilisin gene. After validation, this marker could be used for marker-assisted selection together with other molecular markers in tree improvement and breeding programs. Further possible markers are also indicated from our study.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/10/1083/s1. Table S1: A list of genotypes sampled from various regions of Sweden, Table S2: List of gene models used to design primers. Figure S1: The illustration of workflow of demultiplexingand Genome Analysis toolkit (GATK) pipeline, Table S3: A list of total reads kept per genotypes, Table S4: Significantly associated SNP markers, Figure S2: Comparison of qqplotin GLM and MLM, Figure S3: Schematic illustration of the distribution of gene models in the Scaffold 5992 and 6368 and position of SNPs in the FRAEX38873_v2_000299890.1 and FRAEX38873_v2_000311990.1
Author Contributions
Conceived the study: J.S., M.C., M.E., L.-G.S.; Planned the study: J.S., M.C., M.E., L.-G.S., A.M., K.L., R.C.; Performed experiments: R.C., M.S.Å., K.I., A.M., M.E.; Analyzed data: R.C., T.R., M.B.D., K.L., M.E.; Drafted the Manuscript—R.C., K.L., M.E., J.S.; Commented on MS—J.S., M.E., M.C., L.-G.S., A.M., K.I., M.B.D., K.L., M.S.Å., T.R. All authors have read and agreed to the published version of the manuscript.
Funding
The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS), grant numbers 2013-965 and 2019-00597 provided financial support to the study.
Acknowledgments
The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS), grant numbers 2013-965 and 2019-00597 provided financial support to the study. The authors would like to thank Åke Olson and Maria Jonsson for assistance in the ash sampling from Öland.
Conflicts of Interest
The authors declare no competing interests.
References
- FRAXIGEN. Ash species in Europe: Biological characteristics and practical guidelines for sustainable use. Folia Oecologica 2005, 33, 137. [Google Scholar]
- Baral, H.O.; Queloz, V.; Hosoya, T. Hymenoscyphus fraxineus, the correct scientific name for the fungus causing ash dieback in Europe. IMA Fungus 2014, 5, 79–80. [Google Scholar] [CrossRef] [PubMed]
- Enderle, R.; Stenlid, J.; Vasaitis, R. An overview of ash (Fraxinus spp.) and the ash dieback disease in Europe. CAB Rev. 2019, 14, 1–12. [Google Scholar] [CrossRef]
- Pautasso, M.; Aas, G.; Queloz, V.; Holdenrieder, O. European ash (Fraxinus excelsior) dieback–A conservation biology challenge. Biol. Conserv. 2013, 158, 37–49. [Google Scholar] [CrossRef]
- Coker, T.L.; Rozsypálek, J.; Edwards, A.; Harwood, T.P.; Butfoy, L.; Buggs, R.J. Estimating mortality rates of European ash (Fraxinus excelsior) under the ash dieback (Hymenoscyphus fraxineus) epidemic. Plants People Planet 2019, 1, 48–58. [Google Scholar] [CrossRef]
- Pautasso, M.; Schlegel, M.; Holdenrieder, O. Forest Health in a Changing World. Microb. Ecol. 2015, 69, 826–842. [Google Scholar] [CrossRef]
- Barklund, P. Askdöd grasserar över Syd-och Mellansverige. SkogsEko 2005, 3, 11–13. [Google Scholar]
- Cleary, M.; Nguyen, D.; Stener, L.; Stenlid, J.; Skovsgaard, J. Ash and ash dieback in Sweden: A review of disease history, current status, pathogen and host dynamics, host tolerance and management options in forests and landscapes. Dieback Eur. Ash (Fraxinus spp.) Conseq. Guidel. Sustain. Manag. 2017, 195–208. [Google Scholar]
- Stenlid, J.; Oliva, J.; Boberg, J.B.; Hopkins, A.J. Emerging diseases in European forest ecosystems and responses in society. Forests 2011, 2, 486–504. [Google Scholar] [CrossRef]
- Hultberg, T.; Sandström, J.; Felton, A.; Öhman, K.; Rönnberg, J.; Witzell, J.; Cleary, M. Ash dieback risks an extinction cascade. Biol. Conserv. 2020, 244, 108516. [Google Scholar] [CrossRef]
- Jönsson, M.T.; Thor, G. Estimating coextinction risks from epidemic tree death: Affiliate lichen communities among diseased host tree populations of Fraxinus excelsior. PLoS ONE 2012, 7, e45701. [Google Scholar] [CrossRef] [PubMed]
- Pliura, A.; Bakys, R.; Suchockas, V.; Marčiulyniene, D.; Gustiene, V.; Verbyla, V.; Lygis, V. Ash dieback in Lithuania: Disease history, research on impact and genetic variation in disease resistance, tree breeding and options for forest management. Dieback Eur. Ash (Fraxinus spp.) Conseq. Guidel. Sustain. Manag. 2017, 150–165. [Google Scholar]
- Kjær, E.D.; McKinney, L.V.; Nielsen, L.R.; Hansen, L.N.; Hansen, J.K. Adaptive potential of ash (Fraxinus excelsior) populations against the novel emerging pathogen Hymenoscyphus pseudoalbidus. Evol. Appl. 2012, 5, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Lobo, A.; Hansen, J.K.; McKinney, L.V.; Nielsen, L.R.; Kjær, E.D. Genetic variation in dieback resistance: Growth and survival of Fraxinus excelsior under the influence of Hymenoscyphus pseudoalbidus. Scand. J. Forest Res. 2014, 29, 519–526. [Google Scholar] [CrossRef]
- Harper, A.L.; McKinney, L.V.; Nielsen, L.R.; Havlickova, L.; Li, Y.; Trick, M.; Fraser, F.; Wang, L.; Fellgett, A.; Sollars, E.S.; et al. Molecular markers for tolerance of European ash (Fraxinus excelsior) to dieback disease identified using Associative Transcriptomics. Sci. Rep. 2016, 6, 19335. [Google Scholar] [CrossRef]
- McKinney, L.V.; Nielsen, L.R.; Hansen, J.K.; Kjær, E.D. Presence of natural genetic resistance in Fraxinus excelsior (Oleraceae) to Chalara fraxinea (Ascomycota): An emerging infectious disease. Heredity 2011, 106, 788. [Google Scholar] [CrossRef]
- Stener, L.G. Clonal differences in susceptibility to the dieback of Fraxinus excelsior in southern Sweden. Scand. J. For. Res. 2013, 28, 205–216. [Google Scholar] [CrossRef]
- Kjær, E.D.; McKinney, L.V.; Hansen, L.N.; Olrik, D.C.; Lobo, A.; Thomsen, I.M.; Hansen, J.K.; Nielsen, L.R. Genetics of ash dieback resistance in a restoration context–experiences from Denmark. Dieback Eur. Ash 2017, 106–114. [Google Scholar]
- Lobo, A.; McKinney, L.V.; Hansen, J.K.; Kjær, E.D.; Nielsen, L.R. Genetic variation in dieback resistance in Fraxinus excelsior confirmed by progeny inoculation assay. For. Pathol. 2015, 45, 379–387. [Google Scholar] [CrossRef]
- McKinney, L.V.; Thomsen, I.M.; Kjær, E.D.; Nielsen, L.R. Genetic resistance to Hymenoscyphus pseudoalbidus limits fungal growth and symptom occurrence in Fraxinus excelsior. For. Pathol. 2012, 42, 69–74. [Google Scholar] [CrossRef]
- McKinney, L.V.; Nielsen, L.R.; Collinge, D.B.; Thomsen, I.M.; Hansen, J.K.; Kjær, E.D. The ash dieback crisis: Genetic variation in resistance can prove a long-term solution. Plant Pathol. 2014, 63, 485–499. [Google Scholar] [CrossRef]
- Sollars, E.S.; Harper, A.L.; Kelly, L.J.; Sambles, C.M.; Ramirez-Gonzalez, R.H.; Swarbreck, D.; Kaithakottil, G.; Cooper, E.D.; Uauy, C.; Havlickova, L.; et al. Genome sequence and genetic diversity of European ash trees. Nature 2017, 541, 212. [Google Scholar] [CrossRef] [PubMed]
- Pliura, A.; Lygis, V.; Suchockas, V.; Bartkevicius, E. Performance of twenty four European Fraxinus excelsior populations in three Lithuanian progeny trials with a special emphasis on resistance to Chalara fraxinea. Balt. For. 2011, 17, 17–34. [Google Scholar]
- Neale, D.B.; Savolainen, O. Association genetics of complex traits in conifers. Trends Plant Sci. 2004, 9, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Menkis, A.; Bakys, R.; Stein Åslund, M.; Davydenko, K.; Elfstrand, M.; Stenlid, J.; Vasaitis, R. Identifying Fraxinus excelsior tolerant to ash dieback: Visual field monitoring versus a molecular marker. For. Pathol. 2019, e12572. [Google Scholar] [CrossRef]
- Stocks, J.J.; Metheringham, C.L.; Plumb, W.J.; Lee, S.J.; Kelly, L.J.; Nichols, R.A.; Buggs, R.J.A. Genomic basis of European ash tree resistance to ash dieback fungus. Nat. Ecol. Evol. 2019, 3, 1686–1696. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Dumont, T.; Pope, B.J.; Hammet, F.; Southey, M.C.; Park, D.J. A high-plex PCR approach for massively parallel sequencing. Biotechniques 2013, 55, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Marchese, A.; Marra, F.P.; Caruso, T.; Mhelembe, K.; Costa, F.; Fretto, S.; Sargent, D.J. The first high-density sequence characterized SNP-based linkage map of olive (‘Olea europaea’L. subsp. ‘europaea’) developed using genotyping by sequencing. Aust. J. Crop Sci. 2016, 10, 857. [Google Scholar] [CrossRef]
- Wu, D.; Koch, J.; Coggeshall, M.; Carlson, J. The first genetic linkage map for Fraxinus pennsylvanica and syntenic relationships with four related species. Plant Mol. Biol. 2019, 99, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Zhigunov, A.V.; Ulianich, P.S.; Lebedeva, M.V.; Chang, P.L.; Nuzhdin, S.V.; Potokina, E.K. Development of F1 hybrid population and the high-density linkage map for European aspen (Populus tremula L.) using RADseq technology. BMC Plant Biol. 2017, 17, 180. [Google Scholar] [CrossRef] [PubMed]
- Grattapaglia, D.; de Alencar, S.; Pappas, G. Genome-wide genotyping and SNP discovery by ultra-deep Restriction-Associated DNA (RAD) tag sequencing of pooled samples of E. grandis and E. globulus. BMC Proc. BioMed Cent. 2011, 5, 45. [Google Scholar] [CrossRef]
- Konar, A.; Choudhury, O.; Bullis, R.; Fiedler, L.; Kruser, J.M.; Stephens, M.T.; Gailing, O.; Schlarbaum, S.; Coggeshall, M.V.; Staton, M.E.; et al. High-quality genetic mapping with ddRADseq in the non-model tree Quercus rubra. BMC Genom. 2017, 18, 417. [Google Scholar] [CrossRef] [PubMed]
- Kirisits, T.; Freinschlag, C. Ash dieback caused by Hymenoscyphus pseudoalbidus in a seed plantation of Fraxinus excelsior in Austria. J. Agric. Ext. Rural. Dev. 2012, 4, 184–191. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Report. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.-C.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008, 9, 253. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Li, H.; Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Eprint Arxiv 2013, arXiv:1303.13033997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Earl, D.A. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; pp. 95–98. [Google Scholar]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014, 43, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Buchan, D.W.; Jones, D.T. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019, 47, 402–407. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, 174–181. [Google Scholar] [CrossRef]
- Sniezko, R.A.; Koch, J. Breeding trees resistant to insects and diseases: Putting theory into application. Biol. Invasions 2017, 19, 3377–3400. [Google Scholar] [CrossRef]
- Namkoong, G. Introduction to Quantitative Genetics in Forestry; United States Forest Service: Washington, DC, USA, 1979. [Google Scholar]
- Hall, D.; Hallingbäck, H.R.; Wu, H.X. Estimation of number and size of QTL effects in forest tree traits. Tree Genet. Genomes 2016, 12, 110. [Google Scholar] [CrossRef]
- Nemesio-Gorriz, M.; Hammerbacher, A.; Ihrmark, K.; Källman, T.; Olson, Å.; Lascoux, M.; Stenlid, J.; Gershenzon, J.; Elfstrand, M. Different alleles of a gene encoding leucoanthocyanidin reductase (PaLAR3) influence resistance against the fungus Heterobasidion parviporum in Picea abies. Plant Physiol. 2016, 171, 2671–2681. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Deng, D.; Wang, Z.; Duan, C.; Wu, X.; Wang, X.; Zong, X.; Zhu, Z. A novel er1 allele and the development and validation of its functional marker for breeding pea (Pisum sativum L.) resistance to powdery mildew. Theor. Appl. Genet. 2016, 129, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Antão, C.M.; Malcata, F.X. Plant serine proteases: Biochemical, physiological and molecular features. Plant Physiol. Biochem. 2005, 43, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Monteiro, F.; Sebastiana, M. Subtilisin-like proteases in plant–pathogen recognition and immune priming: A perspective. Front. Plant Sci. 2014, 5, 739. [Google Scholar] [CrossRef] [PubMed]
- Laplaze, L.; Ribeiro, A.; Franche, C.; Duhoux, E.; Auguy, F.; Bogusz, D.; Pawlowski, K. Characterization of a Casuarina glauca nodule-specific subtilisin-like protease gene, a homolog of Alnus glutinosa ag12. Mol. Plant Microbe Interact. 2000, 13, 113–117. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tornero, P.; Conejero, V.; Vera, P. Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J. Biol. Chem. 1997, 272, 14412–14419. [Google Scholar] [CrossRef]
- Duan, X.; Zhang, Z.; Wang, J.; Zuo, K. Characterization of a novel cotton subtilase gene GbSBT1 in response to extracellular stimulations and its role in Verticillium resistance. PLoS ONE 2016, 11, e0153988. [Google Scholar] [CrossRef]
- Semizer-Cuming, D.; Finkeldey, R.; Nielsen, L.R.; Kjær, E.D. Negative correlation between ash dieback susceptibility and reproductive success: Good news for European ash forests. Ann. For. Sci. 2019, 76, 16. [Google Scholar] [CrossRef]
- Aitken, S.N.; Yeaman, S.; Holliday, J.A.; Wang, T.; Curtis-McLane, S. Adaptation, migration or extirpation: Climate change outcomes for tree populations. Evol. Appl. 2008, 1, 95–111. [Google Scholar] [CrossRef]
- Namkoong, G. Maintaining genetic diversity in breeding for resistance in forest trees. Annu. Rev. Phytopathol. 1991, 29, 325–342. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).