Characterization and Development of Genomic SSRs in Pecan (Carya illinoinensis)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
2.2. gSSR Mining and Primer Design
2.3. Amplification and Validation of gSSRs
2.4. Genetic Diversity Analysis of Pecan and Transferability of gSSRs to Other Species
3. Results
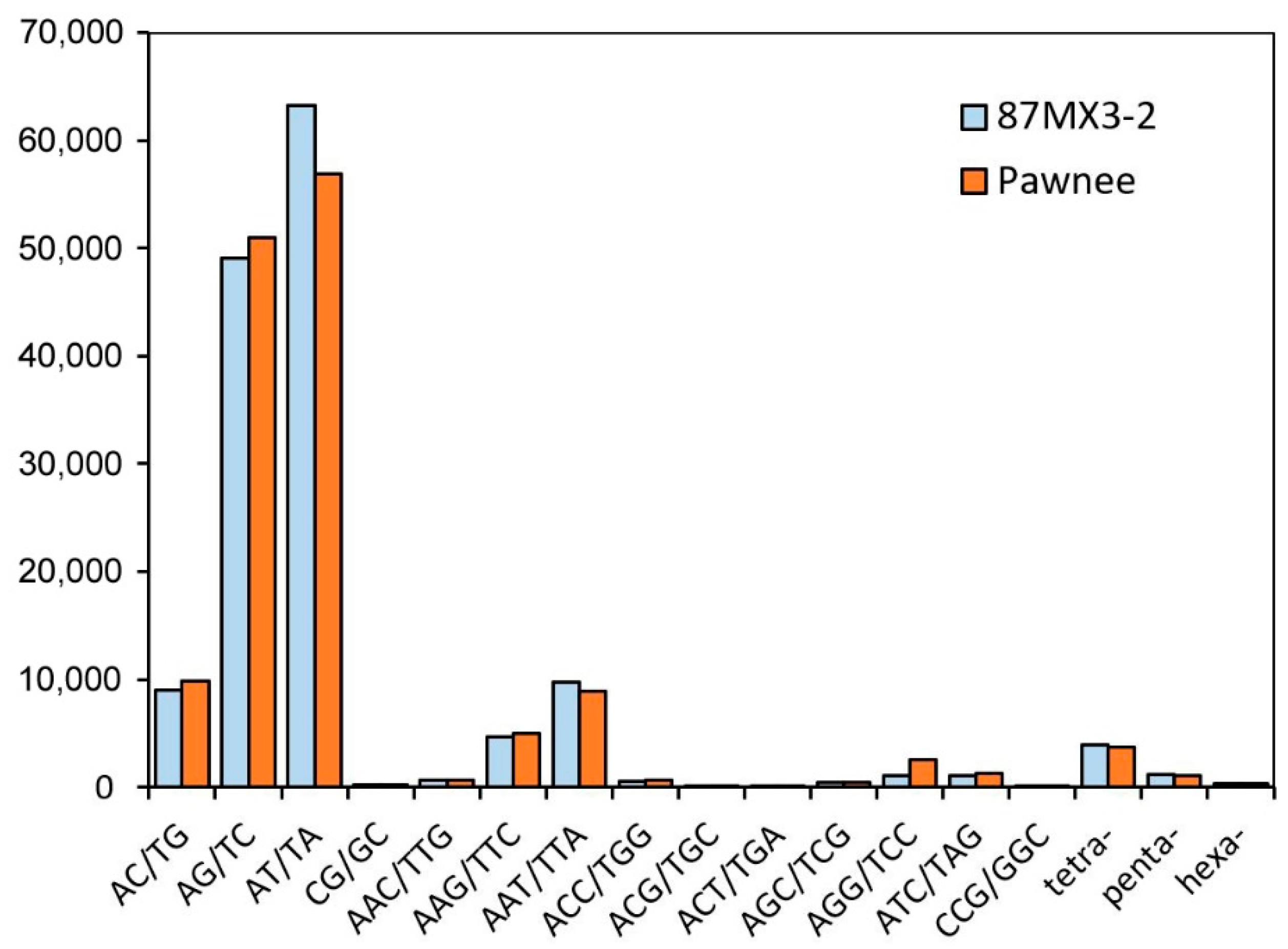
3.1. Characteristics of gSSRs
3.2. Primer Design and Validation
3.3. Genetic Diversity Analysis and Cross-Species Transferability of gSSRs
3.4. Population Structure and Cluster Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Grauke, L.J.; Wood, B.W.; Harris, M.K. Crop Vulnerability: Carya. J. Am. Soc. Hortic. Sci. 2016, 51, 653–663. [Google Scholar] [CrossRef]
- Gong, Y.; Pegg, R.B. Separation of ellagitannin-rich phenolics from U.S. pecans and Chinese hickory nuts using fused-core HPLC columns and their characterization. J. Agric. Food Chem. 2017, 65, 5810–5820. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yao, X.; Ren, H.; Chang, J.; Wang, K. RNA-Seq reveals flavonoid biosynthesis-related genes in pecan (Carya illinoinensis) kernels. J. Agric. Food Chem. 2019, 67, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xiao, L.; Zhang, Z.; Zhang, R.; Wang, Z.; Huang, C.; Huang, R.; Luan, Y.; Fan, T.; Wang, J.; et al. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. GigaScience 2019, 8, giz036. [Google Scholar] [CrossRef]
- Hilbig, J.; Policarpi, P.D.B.; Grinevicius, V.M.A.d.S.; Mota, N.S.R.S.; Toaldo, I.M.; Luiz, M.T.B.; Pedrosa, R.C.; Block, J.M. Aqueous extract from pecan nut [Carya illinoinensis (Wangenh) C. Koch] shell show activity against breast cancer cell line MCF-7 and Ehrlich ascites tumor in Balb-C mice. J. Ethnopharmacol. 2018, 211, 256–266. [Google Scholar] [CrossRef]
- Prado, A.C.P.d.; Manion, B.A.; Seetharaman, K.; Deschamps, F.C.; Barrera Arellano, D.; Block, J.M. Relationship between antioxidant properties and chemical composition of the oil and the shell of pecan nuts [Carya illinoinensis (Wangenh) C. Koch]. Ind. Crops Prod. 2013, 45, 64–73. [Google Scholar] [CrossRef]
- Qin, L.; Qian, H.; He, Y. Microbial lipid production from enzymatic hydrolysate of pecan nutshell pretreated by combined pretreatment. Appl. Biochem. Biotechnol. 2017, 183, 1336–1350. [Google Scholar] [CrossRef]
- Villarreal-Lozoya, J.E.; Lombardini, L.; Cisneros-Zevallos, L. Phytochemical constituents and antioxidant capacity of different pecan [Carya illinoinensis (Wangenh.) K. Koch] cultivars. Food Chem. 2007, 102, 1241–1249. [Google Scholar] [CrossRef]
- Zhang, C.; Yao, X.; Ren, H.; Wang, K.; Chang, J. Isolation and characterization of three chalcone synthase genes in pecan (Carya illinoinensis). Biomolecules 2019, 9, 236. [Google Scholar] [CrossRef]
- Vendrame, W.A.; Kochert, G.; Wetzstein, H.Y. AFLP analysis of variation in pecan somatic embryos. Plant Cell Rep. 1999, 18, 853–857. [Google Scholar] [CrossRef]
- Beedanagari, S.R.; Dove, S.K.; Wood, B.W.; Conner, P.J. A first linkage map of pecan cultivars based on RAPD and AFLP markers. Theor. Appl. Genet. 2005, 110, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.D.; Wang, T.; Zhai, M.; Li, Y.R.; Guo, Z.R. Genetic diversity and identification of Chinese-grown pecan using ISSR and SSR markers. Molecules 2011, 16, 10078–10092. [Google Scholar] [CrossRef] [PubMed]
- Grauke, L.J.; Iqbal, M.J.; Reddy, A.S.; Thompson, T.E. Developing microsatellite DNA markers in pecan. J. Am. Soc. Hortic. Sci. 2003, 128, 374–380. [Google Scholar] [CrossRef]
- Bentley, N.; Grauke, L.J.; Klein, P. Genotyping by sequencing (GBS) and SNP marker analysis of diverse accessions of pecan (Carya illinoinensis). Tree Genet. Genom. 2019, 15, 8. [Google Scholar] [CrossRef]
- Bharti, R.; Kumar, S.; Parekh, M.J. Development of genomic simple sequence repeat (gSSR) markers in cumin and their application in diversity analyses and cross-transferability. Ind. Crops Prod. 2018, 111, 158–164. [Google Scholar] [CrossRef]
- Vieira, M.L.; Santini, L.; Diniz, A.L.; Munhoz Cde, F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Zhong, Y.; Yang, A.; Li, Z.; Zhang, H.; Liu, L.; Wu, Z.; Li, Y.; Liu, T.; Xu, M.; Yu, F. Genetic diversity and population genetic structure of Cinnamomum camphora in south China revealed by EST-SSR markers. Forests 2019, 10, 1019. [Google Scholar] [CrossRef]
- Grauke, L.J.; Mendoza-Herrera, M.A.; Binzel, M.L. Plastid microsatellite markers in Carya. Acta Hortic. 2010, 859, 237–246. [Google Scholar] [CrossRef]
- Grauke, L.J.; Mendoza-Herrera, M.A.; Miller, A.J.; Wood, B.W. Geographic patterns of genetic variation in native pecans. Tree Genet. Genom. 2011, 7, 917. [Google Scholar] [CrossRef]
- Li, J.; Zeng, Y.; Shen, D.; Xia, G.; Huang, Y.; Huang, Y.; Chang, J.; Huang, J.; Wang, Z. Development of SSR markers in hickory (Carya cathayensis Sarg.) and their transferability to other species of Carya. Curr. Genom. 2014, 15, 357–379. [Google Scholar] [CrossRef]
- Conner, P.J. Roadblocks and hindrances to the development of molecular tools in pecan (Carya illinoinensis): A breeder’s perspective. Acta Hortic. 2015, 1070, 83–87. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Du, Q.; Chen, J.; Tian, J.; Yang, X.; Zhang, D. Allelic variation in PtoPsbW associated with photosynthesis, growth, and wood properties in Populus tomentosa. Mol. Genet. Genom. 2016, 292, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhou, C.; Song, Z.; Weng, Q.; Li, M.; Ji, H.; Mo, X.; Huang, H.; Lu, W.; Luo, J. The first identification of genomic loci in plants associated with resistance to galling insects: A case study inEucalyptusL’Hér. (Myrtaceae). Sci. Rep. 2018, 8, 2319. [Google Scholar] [CrossRef] [PubMed]
- Baison, J.; Vidalis, A.; Zhou, L.; Chen, Z.Q.; Li, Z.; Sillanpää, M.J.; Bernhardsson, C.; Scofield, D.; Forsberg, N.; Grahn, T.; et al. Genome-wide association study identified novel candidate loci affecting wood formation in Norway spruce. Plant J. 2019, 100, 83–100. [Google Scholar] [CrossRef]
- Cao, K.; Zhou, Z.; Wang, Q.; Guo, J.; Zhao, P.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, X.; et al. Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 2016, 7, 13246. [Google Scholar] [CrossRef]
- Müller, B.S.F.; de Almeida Filho, J.E.; Lima, B.M.; Garcia, C.C.; Missiaggia, A.; Aguiar, A.M.; Takahashi, E.; Kirst, M.; Gezan, S.A.; Silva-Junior, O.B.; et al. Independent and Joint-GWAS for growth traits in Eucalyptus by assembling genome-wide data for 3373 individuals across four breeding populations. New Phytol. 2019, 221, 818–833. [Google Scholar] [CrossRef]
- Xu, L.Y.; Wang, L.Y.; Wei, K.; Tan, L.Q.; Su, J.J.; Cheng, H. High-density SNP linkage map construction and QTL mapping for flavonoid-related traits in a tea plant (Camellia sinensis) using 2b-RAD sequencing. BMC Genom. 2018, 19, 955. [Google Scholar] [CrossRef]
- Jenkins, J.; Wilson, B.; Grimwood, J.; Schmutz, J.; Grauke, L.J. Twoards a reference pecan genome sequence. Acta Hortic. 2015, 1070, 101–108. [Google Scholar] [CrossRef]
- Jia, Z.; Wang, G.; Xuan, J.; Zhang, J.; Zhai, M.; Jia, X.; Guo, Z.; Li, M. Comparative transcriptome analysis of pecan female and male inflorescences. Russ. J. Plant Physiol. 2018, 65, 186–196. [Google Scholar] [CrossRef]
- Liu, S.; An, Y.; Li, F.; Li, S.; Liu, L.; Zhou, Q.; Zhao, S.; Wei, C. Genome-wide identification of simple sequence repeats and development of polymorphic SSR markers for genetic studies in tea plant (Camellia sinensis). Mol. Breed. 2018, 38, 59. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, L.; Peng, Z.; Sun, H.; Yue, X.; Lou, Y.; Dong, L.; Wang, L.; Gao, Z. Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci. Rep. 2015, 5, 8018. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gu, Y.Q.; Hu, Y.; You, F.M.; Dandekar, A.M.; Leslie, C.A.; Aradhya, M.; Dvorak, J.; Luo, M.C. Characterizing the walnut genome through analyses of BAC end sequences. Plant Mol. Biol. 2012, 78, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.C.; Zhang, Y.Z.; Ya, H.Y.; Gao, Q.B. Characterization of SSR genomic abundance and identification of SSR markers for population genetics in Chinese jujube (Ziziphus jujuba Mill.). PeerJ 2016, 4, e1735. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhao, J.; Liu, M.; Liu, P.; Dai, L.; Zhao, Z. Genome-wide characterization of simple sequence repeat (SSR) loci in Chinese jujube and jujube SSR primer transferability. PLoS ONE 2015, 10, e0127812. [Google Scholar] [CrossRef]
- Tribhuvan, K.U.; SV, A.M.; Sharma, P.; Das, A.; Kumar, K.; Tyagi, A.; Solanke, A.U.; Sharma, R.; Jadhav, P.V. Identification of genomic SSRs in cluster bean (Cyamopsis tetragonoloba) and demonstration of their utility in genetic diversity analysis. Ind. Crops Prod. 2019, 133, 221–231. [Google Scholar] [CrossRef]
- Huang, C.; Yin, Q.; Khadka, D.; Meng, K.; Fan, Q.; Chen, S.; Liao, W. Identification and development of microsatellite (SSRs) makers of Exbucklandia (HAMAMELIDACEAE) by high-throughput sequencing. Mol. Biol. Rep. 2019, 46, 3381–3386. [Google Scholar] [CrossRef]
- Zaloğlu, S.; Kafkas, S.; Doğan, Y.; Güney, M. Development and characterization of SSR markers from pistachio (Pistacia vera L.) and their transferability to eight Pistacia species. Sci. Hortic. 2015, 189, 94–103. [Google Scholar] [CrossRef]
- Xu, X.; Zhou, C.; Zhang, Y.; Zhang, W.; Gan, X.; Zhang, H.; Guo, Y.; Gan, S. A novel set of 223 EST-SSR markers in Casuarina L. ex Adans.: Polymorphisms, cross-species transferability, and utility for commercial clone genotyping. Tree Genet. Genomes 2018, 14, 30. [Google Scholar] [CrossRef]
- Yan, Z.; Wu, F.; Luo, K.; Zhao, Y.; Yan, Q.; Zhang, Y.; Wang, Y.; Zhang, J. Cross-species transferability of EST-SSR markers developed from the transcriptome of Melilotus and their application to population genetics research. Sci. Rep. 2017, 7, 17959. [Google Scholar] [CrossRef]
- Zhang, J.B.; Li, R.Q.; Xiang, X.G.; Manchester, S.R.; Lin, L.; Wang, W.; Wen, J.; Chen, Z.D. Integrated fossil and molecular data reveal the biogeographic diversification of the eastern asian-eastern north american disjunct hickory genus (Carya Nutt.). PLoS ONE 2013, 8, e70449. [Google Scholar] [CrossRef]
- Conner, P.J.; Wood, B.W. Identification of pecan cultivars and their genetic relatedness as determined by randomly amplified polymorphic DNA analysis. J. Am. Soc. Hortic. Sci. 2001, 126, 474–480. [Google Scholar] [CrossRef]
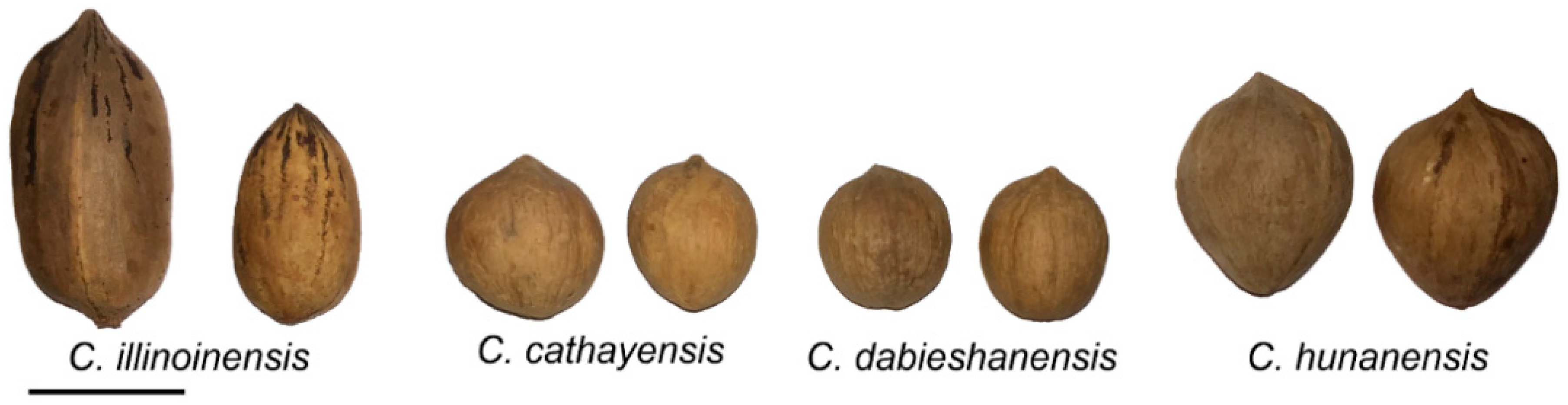


| No. | Name | Species 1 | Type 2 | Origin 3 | No. | Name | Species | Type | Origin | No. | Name | Species | Type | Origin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Osage | C. il | C | USA | 28 | J1 | C. il | ES | Jinhua, ZJ | 55 | JLSX | C. il | SP | Jiande, ZJ |
| 2 | Pawnee | C. il | C | USA | 29 | J2 | C. il | ES | Jinhua, ZJ | 56 | 0071A | C. il | SP | Jiande, ZJ |
| 3 | McMillan | C. il | C | USA | 30 | 1101 | C. il | ES | Nanjing, JS | 57 | 0072A | C. il | SP | Jiande, ZJ |
| 4 | Lakota | C. il | C | USA | 31 | 1103 | C. il | ES | Nanjing, JS | 58 | 0073A | C. il | SP | Jiande, ZJ |
| 5 | Carter | C. il | C | USA | 32 | 1104 | C. il | ES | Nanjing, JS | 59 | SDHD1 | C. il | SP | Jiande, ZJ |
| 6 | Colby | C. il | C | USA | 33 | 1106 | C. il | ES | Nanjing, JS | 60 | SDHDX1 | C. il | SP | Jiande, ZJ |
| 7 | Stuart | C. il | C | USA | 34 | 1201 | C. il | ES | Nanjing, JS | 61 | JP-18 | C. hu | SP | Jinping, GZ |
| 8 | Greenriver | C. il | C | USA | 35 | 1202 | C. il | ES | Nanjing, JS | 62 | JP-17 | C. hu | SP | Jinping, GZ |
| 9 | Waco | C. il | C | USA | 36 | JL72 | C. il | SP | Jiande, ZJ | 63 | JP-19 | C. hu | SP | Jinping, GZ |
| 10 | Major | C. il | C | USA | 37 | JL72D | C. il | SP | Jiande, ZJ | 64 | JP-20 | C. hu | SP | Jinping, GZ |
| 11 | Oconee | C. il | C | USA | 38 | JL1 | C. il | SP | Jiande, ZJ | 65 | JP-18 | C. hu | SP | Jinping, GZ |
| 12 | Syrup Mill | C. il | C | USA | 39 | JL2 | C. il | SP | Jiande, ZJ | 66 | TTZ-15 | C. da | SP | Luan, AH |
| 13 | Navaho | C. il | C | USA | 40 | JL3 | C. il | SP | Jiande, ZJ | 67 | TTZ-21 | C. da | SP | Luan, AH |
| 14 | Gloria Grande | C. il | C | USA | 41 | JL00011 | C. il | SP | Jiande, ZJ | 68 | TTZ-20 | C. da | SP | Luan, AH |
| 15 | Forkert | C. il | C | USA | 42 | SD1-2 | C. il | SP | Jiande, ZJ | 69 | TTZ-22 | C. da | SP | Luan, AH |
| 16 | Choctaw | C. il | C | USA | 43 | SJ2 | C. il | SP | Jiande, ZJ | 70 | TTZ-11 | C. da | SP | Luan, AH |
| 17 | Creek | C. il | C | USA | 44 | SJ1 | C. il | SP | Jiande, ZJ | 71 | GL2 | C. ca | S | Jinhua, ZJ |
| 18 | Mohawk | C. il | C | USA | 45 | YZXT | C. il | SP | Jiande, ZJ | 72 | XK24 | C. ca | S | Jinhua, ZJ |
| 19 | Elliott | C. il | C | USA | 46 | YZXD | C. il | SP | Jiande, ZJ | 73 | DY1 | C. ca | S | Jinhua, ZJ |
| 20 | Mahan | C. il | C | USA | 47 | YZXG1 | C. il | SP | Jiande, ZJ | 74 | DY6 | C. ca | S | Jinhua, ZJ |
| 21 | YLC21 | C. il | C | Jiande, ZJ | 48 | YZXG2 | C. il | SP | Jiande, ZJ | 75 | GL8 | C. ca | S | Jinhua, ZJ |
| 22 | XH1 | C. il | ES | Hangzhou, ZJ | 49 | JL4 | C. il | SP | Jiande, ZJ | 76 | XK88 | C. ca | S | Jinhua, ZJ |
| 23 | XH4 | C. il | ES | Hangzhou, ZJ | 50 | CHK1 | C. il | SP | Hangzhou, ZJ | 77 | DY5 | C. ca | S | Jinhua, ZJ |
| 24 | XH5 | C. il | ES | Hangzhou, ZJ | 51 | CHK2 | C. il | SP | Hangzhou, ZJ | 78 | GL7 | C. ca | S | Jinhua, ZJ |
| 25 | XH6 | C. il | ES | Hangzhou, ZJ | 52 | CHK3 | C. il | SP | Hangzhou, ZJ | 79 | XK89 | C. ca | S | Jinhua, ZJ |
| 26 | XH14 | C. il | ES | Hangzhou, ZJ | 53 | CHK4 | C. il | SP | Hangzhou, ZJ | 80 | XK40 | C. ca | S | Jinhua, ZJ |
| 27 | DFH | C. il | ES | Jinhua, ZJ | 54 | JLCD | C. il | SP | Jiande, ZJ |
| Features | Values | |
|---|---|---|
| ‘87MX3-2’ | ‘Pawnee’ | |
| Total number of sequences examined | 124,560 | 43,183 |
| Total size of examined sequences (bp) | 531,741,495 | 651,240,950 |
| Total number of identified SSRs | 145,714 | 143,041 |
| Number of SSR containing sequences | 48,009 | 4955 |
| Number of sequences containing more than 1 SSR | 27,045 | 1961 |
| Number of SSRs present in compound formation | 21,060 | 17,816 |
| NO. 1 | Loci | Fluorescence Dye | Na 2 | Ho 3 | He 4 | PIC 5 | Size (bp) |
|---|---|---|---|---|---|---|---|
| 1 | Ciz003 | ROX | 10 | 0.695 | 0.741 | 0.735 | 137–178 |
| 2 | Ciz009 | ROX | 5 | 0.525 | 0.517 | 0.513 | 157–163 |
| 3 | Ciz011 | ROX | 2 | 0.068 | 0.097 | 0.096 | 202–204 |
| 4 | Ciz022 | FAM | 5 | 0.650 | 0.630 | 0.625 | 223–237 |
| 5 | Ciz031 | FAM | 18 | 0.542 | 0.901 | 0.893 | 237–257 |
| 6 | Ciz036 | TRAMA | 1 | 0.000 | 0.000 | 0.000 | 264 |
| 7 | Ciz038 | HEX | 3 | 0.300 | 0.551 | 0.546 | 213–216 |
| 8 | Ciz039 | ROX | 4 | 0.200 | 0.390 | 0.387 | 198–202 |
| 9 | Ciz040 | FAM | 5 | 0.367 | 0.475 | 0.471 | 219–227 |
| 10 | Ciz043 | TRAMA | 14 | 0.712 | 0.800 | 0.793 | 260–278 |
| 11 | Ciz045 | FAM | 6 | 0.224 | 0.549 | 0.544 | 182–253 |
| 12 | Ciz046 | FAM | 12 | 0.237 | 0.755 | 0.748 | 250–262 |
| 13 | Ciz047 | FAM | 5 | 1.000 | 0.646 | 0.641 | 236–241 |
| 14 | Ciz050 | FAM | 3 | 0.070 | 0.102 | 0.101 | 243–261 |
| 15 | Ciz052 | ROX | 9 | 0.517 | 0.601 | 0.596 | 203–223 |
| 16 | Ciz055 | TRAMA | 16 | 0.690 | 0.899 | 0.891 | 249–268 |
| 17 | Ciz058 | FAM | 13 | 0.550 | 0.807 | 0.801 | 199–247 |
| 18 | Ciz059 | TRAMA | 7 | 0.633 | 0.708 | 0.702 | 232–265 |
| 19 | Ciz070 | TRAMA | 12 | 0.695 | 0.824 | 0.817 | 265–280 |
| 20 | Ciz071 | TRAMA | 5 | 0.817 | 0.751 | 0.744 | 269–273 |
| 21 | Ciz073 | TRAMA | 5 | 0.390 | 0.512 | 0.507 | 273–285 |
| 22 | Ciz074 | ROX | 3 | 0.339 | 0.333 | 0.330 | 180–186 |
| 23 | PM-CA10 | HEX | 2 | 0.051 | 0.050 | 0.050 | 92–113 |
| 24 | PM-CIN13 | HEX | 6 | 0.441 | 0.510 | 0.506 | 102–116 |
| 25 | PM-CIN20 | ROX | 2 | 0.017 | 0.017 | 0.016 | 139–142 |
| 26 | PM-CIN22 | HEX | 5 | 0.117 | 0.590 | 0.585 | 96–103 |
| 27 | PM-CIN27 | HEX | 12 | 0.800 | 0.775 | 0.768 | 62–92 |
| 28 | PM-CIN4 | HEX | 16 | 0.783 | 0.796 | 0.789 | 90–143 |
| 29 | PM-GA38 | HEX | 9 | 0.627 | 0.602 | 0.596 | 81–100 |
| 30 | PM-GA41 | HEX | 6 | 0.105 | 0.637 | 0.631 | 69–90 |
| - | Mean | - | 7.367 | 0.439 | 0.552 | 0.547 | - |
| - | St. Dev | - | 4.752 | 0.283 | 0.266 | 0.264 | - |
| Locus | C. cathayensis (n = 10) | C. dabieshanensis (n = 5) | C. hunanensis (n = 5) | |||
|---|---|---|---|---|---|---|
| Na | Size (bp) | Na | Size (bp) | Na | Size (bp) | |
| Ciz003 | - | - | - | - | - | - |
| Ciz009 | 1 | 167 | 1 | 167 | 2 | 167–176 |
| Ciz011 | 1 | 203 | 1 | 203 | 1 | 203 |
| Ciz022 | 2 | 228–230 | 1 | 228 | 1 | 230 |
| Ciz031 | 2 | 249–255 | 3 | 252–255 | 1 | 253 |
| Ciz036 | 1 | 264 | 1 | 264 | 1 | 264 |
| Ciz038 | 2 | 216–218 | 1 | 218 | - | - |
| Ciz039 | 1 | 199 | 3 | 195–202 | 2 | 196–197 |
| Ciz040 | 1 | 222 | 1 | 222 | 2 | 216–222 |
| Ciz043 | 1 | 260 | 1 | 260 | 2 | 260–261 |
| Ciz045 | 6 | 205–227 | 4 | 205–224 | 1 | 224 |
| Ciz046 | - | - | - | - | - | - |
| Ciz047 | 1 | 238 | 2 | 238–241 | 2 | 238–241 |
| Ciz050 | 4 | 239–254 | 2 | 230–240 | 2 | 222–240 |
| Ciz052 | 3 | 251–255 | 1 | 254 | 1 | 254 |
| Ciz055 | 1 | 263 | 1 | 263 | 3 | 260–264 |
| Ciz058 | - | - | 3 | 233–241 | - | - |
| Ciz059 | 2 | 266–267 | 2 | 264–267 | 1 | 264 |
| Ciz070 | - | - | - | - | - | - |
| Ciz071 | 2 | 255–266 | 2 | 255–266 | 2 | 255–266 |
| Ciz073 | 4 | 266–270 | 1 | 270 | 2 | 267–270 |
| Ciz074 | 3 | 175–180 | 3 | 177–183 | - | - |
| PM-CA10 | 1 | 134 | 3 | 116–136 | - | - |
| PM-CIN13 | 3 | 104–111 | - | - | - | - |
| PM-CIN20 | 1 | 139 | 1 | 139 | 1 | 139 |
| PM-CIN22 | 1 | 95 | 1 | 95 | 1 | 94 |
| PM-CIN27 | 2 | 62–66 | - | - | 1 | 68 |
| PM-CIN4 | 3 | 113–118 | 2 | 113–114 | 2 | 105–118 |
| PM-GA38 | 2 | 80–90 | 2 | 80–82 | 2 | 82–92 |
| PM-GA41 | 2 | 95–101 | 1 | 101 | 1 | 99 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Yao, X.; Ren, H.; Chang, J.; Wu, J.; Shao, W.; Fang, Q. Characterization and Development of Genomic SSRs in Pecan (Carya illinoinensis). Forests 2020, 11, 61. https://doi.org/10.3390/f11010061
Zhang C, Yao X, Ren H, Chang J, Wu J, Shao W, Fang Q. Characterization and Development of Genomic SSRs in Pecan (Carya illinoinensis). Forests. 2020; 11(1):61. https://doi.org/10.3390/f11010061
Chicago/Turabian StyleZhang, Chengcai, Xiaohua Yao, Huadong Ren, Jun Chang, Jun Wu, Weizhong Shao, and Qing Fang. 2020. "Characterization and Development of Genomic SSRs in Pecan (Carya illinoinensis)" Forests 11, no. 1: 61. https://doi.org/10.3390/f11010061
APA StyleZhang, C., Yao, X., Ren, H., Chang, J., Wu, J., Shao, W., & Fang, Q. (2020). Characterization and Development of Genomic SSRs in Pecan (Carya illinoinensis). Forests, 11(1), 61. https://doi.org/10.3390/f11010061





