Genetic Diversity and Population Structure of Natural Pinus koraiensis Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Sampling
2.2. DNA Extraction and Microsatellite Genotyping
2.3. Data Analysis
2.3.1. Genetic Diversity Indices
2.3.2. Population Structure Analysis
3. Results
3.1. Genetic Diversity
3.2. Genetic Differentiation
3.3. Population Structure
3.4. Correlations Between Genetic Diversity Parameters and Geoclimatic Variables
4. Discussion
4.1. Genetic Diversity
4.2. Genetic Differentiation and Population Structure
4.3. Implications for Conservation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ma, J.L.; Zhang, L.W.; Cheng, D.; Li, J.W. Geographic distribution of Pinus koraiensis in the world. J. Northeast. For. Univ. 1992, 20, 40–47. (In Chinese) [Google Scholar]
- Potenko, V.V.; Velikov, A.V. Genetic diversity and differentiation of natural populations of Pinus koraiensis (Sieb. et Zucc.) in Russia. Silvae Genet. 1998, 47, 202–208. [Google Scholar]
- Feng, F.J.; Sui, X.; Chen, M.M.; Zhao, D.; Han, S.J.; Li, M.H. Mode of pollen spread in clonal seed orchard of Pinus koraiensis. J. Biophys. Chem. 2010, 1, 33–39. [Google Scholar] [CrossRef][Green Version]
- Feng, F.J.; Han, S.J.; Wang, H.M. Genetic diversity and genetic differentiation of natural Pinus koraiensis population. J. For. Res. 2006, 17, 21–24. [Google Scholar] [CrossRef]
- Aizawa, M.; Kim, Z.S.; Yoshimaru, H. Phylogeography of the Korean pine (Pinus koraiensis) in northeast Asia: Inferences from organelle gene sequences. J. Plant Res. 2012, 125, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.P.; Zhou, L.; Zhou, W.M.; Ding, H.; Wang, Q.W.; Wang, Y.; Wu, X.Q.; Dai, L.M. Forest management in Northeast China: History, problems, and challenges. Environ. Manag. 2011, 48, 1122–1135. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Yang, J.; Yu, D.; Dai, L.; Contrereas, M. Responses of regeneration and species coexistence to single-tree selective logging for a temperate mixed forest in eastern Eurasia. Ann. For. Sci. 2016, 73, 449–460. [Google Scholar]
- Sun, Y.; Zhu, J.; Sun, O.J.; Yan, Q. Photosynthetic and growth responses of Pinus koraiensis seedlings to canopy openness: Implications for the restoration of mixed-broadleaved Korean pine forests. Environ. Exp. Bot. 2016, 129, 118–126. [Google Scholar] [CrossRef]
- Chen, X.W.; Zhou, G.S.; Zhang, X.S. Spatial characteristics and change for tree species along the North East China Transect (NECT). Plant Ecol. 2003, 164, 65–74. [Google Scholar] [CrossRef]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef]
- Gapare, W.J. Merging applied gene conservation activities with advanced generation breeding initiatives: A case study of Pinus radiata D. Don. New For. 2014, 45, 311–331. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Shi, X.M.; Wen, Q.; Cao, M.; Guo, X.; Xu, L.A. Genetic Diversity and Structure of Natural Quercus variabilis Population in China as Revealed by Microsatellites Markers. Forests 2017, 8, 495. [Google Scholar] [CrossRef]
- Potter, K.M.; Jetton, R.M.; Bower, A.; Jacobs, D.F.; Man, G.; Hipkins, V.D.; Westwood, M. Banking on the future: Progress, challenges and opportunities for the genetic conservation of forest trees. New For. 2017, 48, 153–180. [Google Scholar] [CrossRef]
- Reed, D.H.; Frankham, R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Eltaher, S.; Sallam, A.; Belamkar, V.; Emara, H.A.; Nower, A.A.; Salem, K.F.M. Genetic diversity and population structure of F3:6 Nebraska winter wheat genotypes using genotyping-by-sequencing. Front. Genet. 2018, 9, 76. [Google Scholar] [CrossRef]
- Sutherland, W.J.; Albon, S.D.; Allison, H.; Armstrong-Brown, S.; Bailey, M.J.; Brereton, T.; Boyd, I.L.; Carey, P.; Edwards, J.; Gill, M.; et al. The identification of priority policy options for UK nature conservation. J. Appl. Ecol. 2010, 47, 955–965. [Google Scholar] [CrossRef]
- Guo, H.Y.; Wang, Z.L.; Huang, Z.; Chen, Z.; Yang, H.B.; Kang, X.Y. Genetic Diversity and Population Structure of Alnus cremastogyne as Revealed by Microsatellite Markers. Forests 2019, 10, 278. [Google Scholar] [CrossRef]
- Thakur, S.; Choudhary, S.; Singh, A.; Ahmad, K.; Sharma, G.; Majeed, A.; Bhardwaj, P. Genetic diversity and population structure of Melia azedarach in North-Western Plains of India. Trees 2016, 30, 1483–1494. [Google Scholar] [CrossRef]
- Kim, Z.S.; Hwang, J.W.; Lee, S.W.; Yang, C.; Gorovoy, P.G. Genetic variation of Korean pine (Pinus koraiensis Sieb. et Zucc.) at allozyme and RAPD markers in Korea, China and Russia. Silvae Genet. 2005, 54, 235–246. [Google Scholar] [CrossRef]
- Feng, F.J.; Chen, M.M.; Zhang, D.D.; Sui, X.; Han, S.J. Application of SRAP in the genetic diversity of Pinus koraiensis of different provenances. Afr. J. Biotechnol. 2009, 8, 1000–1008. [Google Scholar]
- Yu, J.H.; Chen, C.M.; Tang, Z.H.; Yuan, S.S.; Wang, C.J.; Zu, Y.G. Isolation and characterization of 13 novel polymorphic microsatellite markers for Pinus koraiensis (Pinaceae). Am. J. Bot. 2012, 99, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Zhen, Z.; Zhang, H.G.; Tang, J.H. EST-SSR marker development and transcriptome sequencing analysis of different tissues of Korean pine (Pinus koraiensis Sieb. et Zucc.). Biotechnol. Biotechnol. Equip. 2017, 31, 679–689. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Goudet, J. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Trouet, V.; Van Oldenborgh, G.J. KNMI climate explorer: A web-based research tool for high-resolution paleoclimatology. Tree Ring Res. 2013, 69, 3–13. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2010, 14, 2611–2620. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes-application to human mitochondrial-DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [PubMed]
- Iwaizumi, M.G.; Miyata, S.; Hirao, T.; Tamura, M.; Watanabe, A. Historical seed use and transfer affects geographic specificity in genetic diversity and structure of old planted Pinus thunbergii populations. For. Ecol. Manag. 2018, 408, 211–219. [Google Scholar] [CrossRef]
- Toth, E.G.; Vendramin, G.G.; Bagnoli, F.; Cseke, K.; Hoehn, M. High genetic diversity and distinct origin of recently fragmented Scots pine (Pinus sylvestris L.) populations along the Carpathians and the Pannonian Basin. Tree Genet. Genomes 2017, 13, 47. [Google Scholar] [CrossRef]
- Mandak, B.; Hadincova, V.; Mahelka, V.; Wildova, R. European Invasion of North American Pinus strobus at Large and Fine Scales: High Genetic Diversity and Fine-Scale Genetic Clustering over Time in the Adventive Range. PLoS ONE 2013, 8, e68514. [Google Scholar] [CrossRef]
- Xu, Y.L.; Cai, N.H.; Woeste, K.; Kang, X.Y.; He, C.Z.; Li, G.Q.; Chen, S.; Duan, A.A. Genetic diversity and population structure of Pinus yunnanensis by simple sequence repeat markers. For. Sci. 2016, 62, 38–47. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Linhart, Y.B.; Mitton, J.B. Relationships between life-history characteristics and electrophoretically detectable genetic-variation in plants. Annu. Rev. Ecol. Syst. 1979, 10, 173–200. [Google Scholar] [CrossRef]
- Hellmann, J.J.; Pineda-Krch, M. Constraints and reinforcement on adaptation under climate change: Selection of genetically correlated traits. Biol. Conserv. 2007, 137, 599–609. [Google Scholar] [CrossRef]
- Gaudeul, M.; Taberlet, P.; Till-Bottraud, I. Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers. Mol. Ecol. 2000, 9, 1625–1637. [Google Scholar] [CrossRef]
- Karmin, M.; Saag, L.; Vicente, M.; Sayres, M.A.W.; Jarve, M.; Talas, U.G.; Rootsi, S.; Ilumaee, A.M.; Maegi, R.; Mitt, M.; et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 2015, 25, 459–466. [Google Scholar] [CrossRef]
- Peery, M.Z.; Kirby, R.; Reid, B.N.; Stoelting, R.; Doucet-Beer, E.; Robinson, S.; Vasquez-Carrillo, C.; Pauli, J.N.; Palsboll, P.J. Reliability of genetic bottleneck tests for detecting recent population declines. Mol. Ecol. 2012, 21, 3403–3418. [Google Scholar] [CrossRef]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Belletti, P.; Ferrazzini, D.; Piotti, A.; Monteleone, I.; Ducci, F. Genetic variation and divergence in Scots pine (Pinus sylvestris L.) within its natural range in Italy. Eur. J. For. Res. 2012, 131, 1127–1138. [Google Scholar] [CrossRef]
- Iwaizumi, M.G.; Tsuda, Y.; Ohtani, M.; Tsumura, Y.; Takahashi, M. Recent distribution changes affect geographic clines in genetic diversity and structure of Pinus densiflora natural populations in Japan. For. Ecol. Manag. 2013, 304, 407–416. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Petit, R.J.; Duminil, J.; Fineschi, S.; Hampe, A.; Salvini, D.; Vendramin, G.G. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005, 14, 689–701. [Google Scholar] [CrossRef]
- Rubio-Moraga, A.; Candel-Perez, D.; Lucas-Borja, M.E.; Tiscar, P.A.; Vinegla, B.; Linares, J.C.; Gómez-Gómez, L.; Ahrazem, O. Genetic diversity of Pinus nigra Arn. populations in southern Spain and northern Morocco revealed by inter-simple sequence repeat profiles. Int. J. Mol. Sci. 2012, 13, 5645–5658. [Google Scholar] [CrossRef]
- Wang, B.; Mao, J.F.; Zhao, W.; Wang, X.R. Impact of geography and climate on the genetic differentiation of the subtropical pine Pinus yunnanensis. PLoS ONE 2013, 8, e67345. [Google Scholar] [CrossRef]
- Young, A.; Merriam, H. Effects of forest fragmentation on the spatial genetic structure of Acer saccharum Marsh.(sugar maple) populations. Heredity 1994, 72, 201. [Google Scholar] [CrossRef]
- Aldrich, P.R.; Hamrick, J.L.; Chavarriaga, P.; Kochert, G. Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Mol. Ecol. 1998, 7, 933–944. [Google Scholar] [CrossRef]
- Forest, F.; Grenyer, R.; Rouget, M.; Davies, T.J.; Cowling, R.M.; Faith, D.P.; Balmford, A.; Manning, J.C.; Proches, S.; van der Bank, M.; et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 2007, 445, 757. [Google Scholar] [CrossRef]
- Stojnić, S.; Avramidou, E.V.; Fussi, B.; Westergren, M.; Orlović, S.; Matović, B.; Trudić, B.; Kraigher, H.; Aravanopoulos, F.A.; Konnert, M. Assessment of Genetic Diversity and Population Genetic Structure of Norway Spruce (Picea abies (L.) Karsten) at Its Southern Lineage in Europe. Implications for Conservation of Forest Genetic Resources. Forests 2019, 10, 258. [Google Scholar] [CrossRef]
- Carabeo, M.; Simeone, M.C.; Cherubini, M.; Mattia, C.; Chiocchini, F.; Bertini, L.; Caruso, C.; La Mantia, T.; Villani, F.; Mattioni, C. Estimating the genetic diversity and structure of Quercus trojana Webb populations in Italy by SSRs: Implications for management and conservation. Can. J. For. Res. 2017, 47, 331–339. [Google Scholar] [CrossRef]
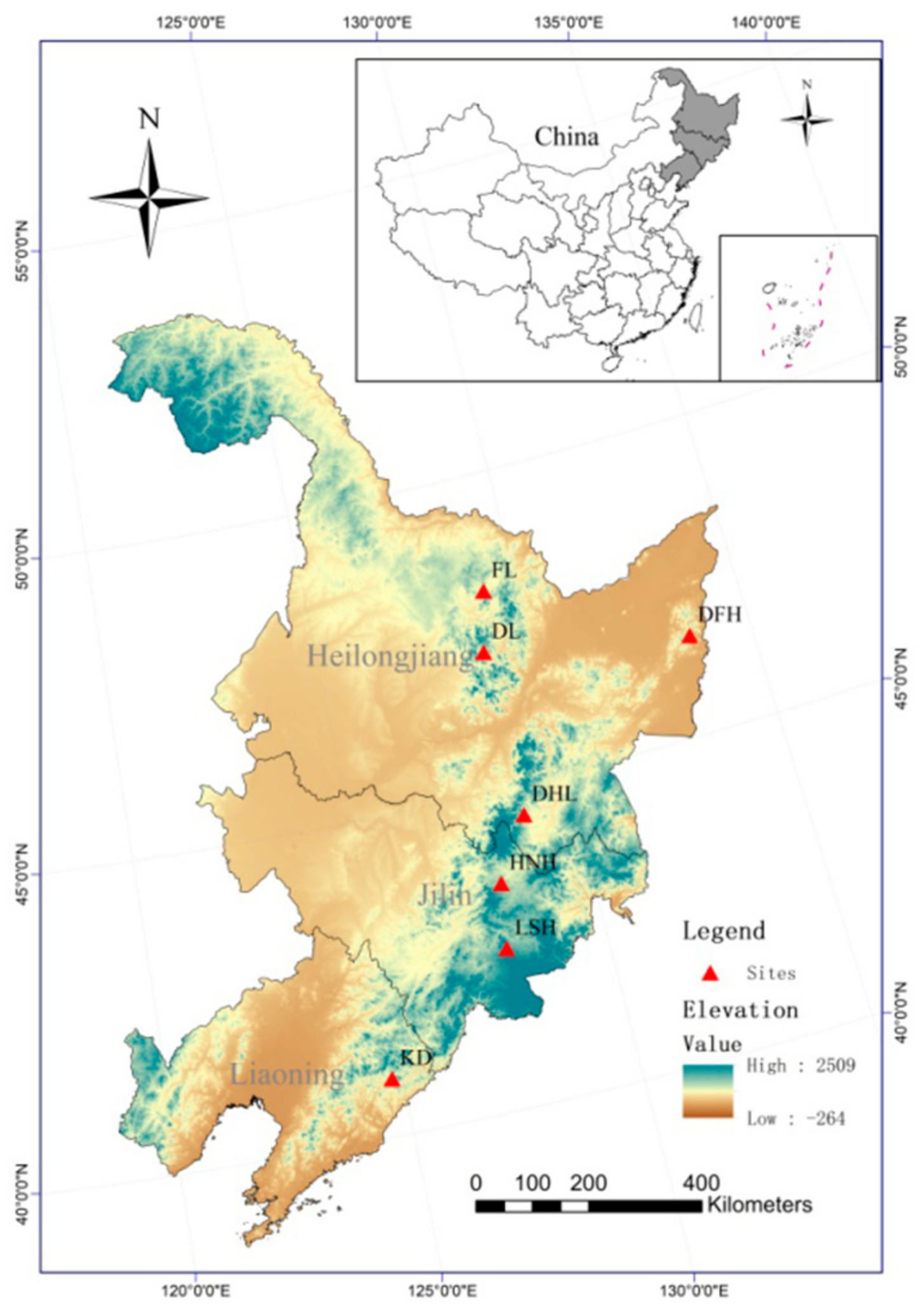
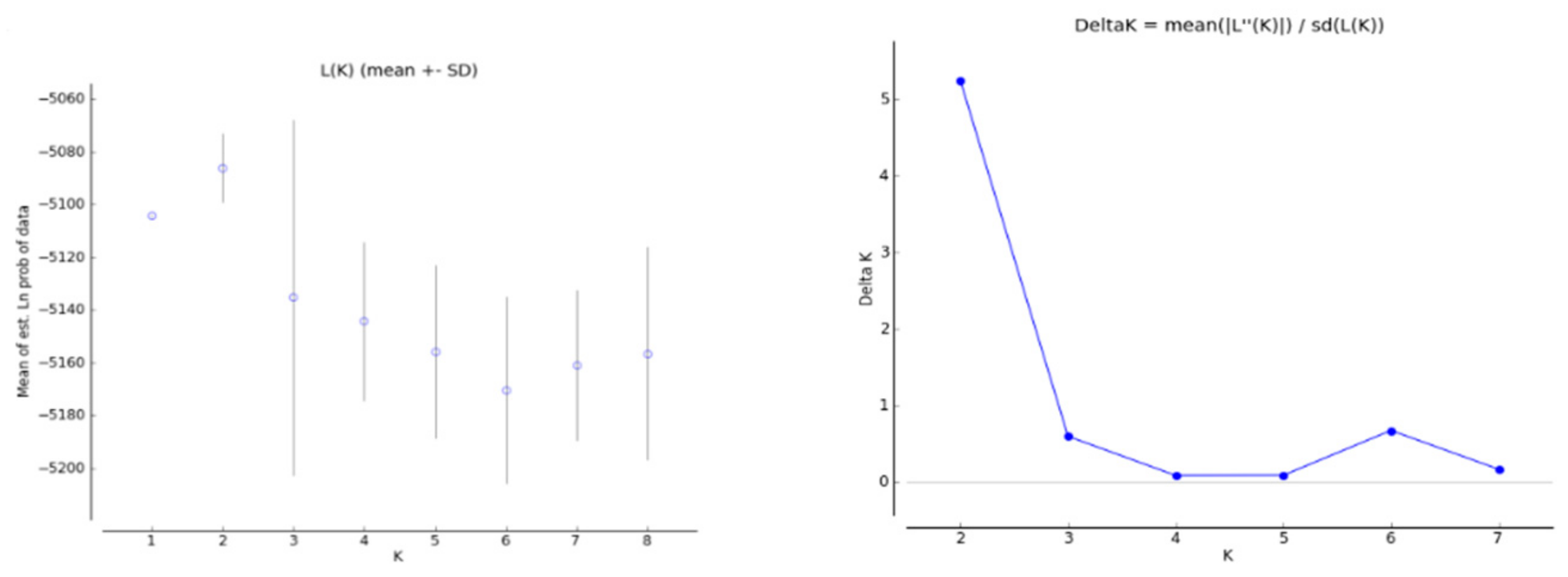
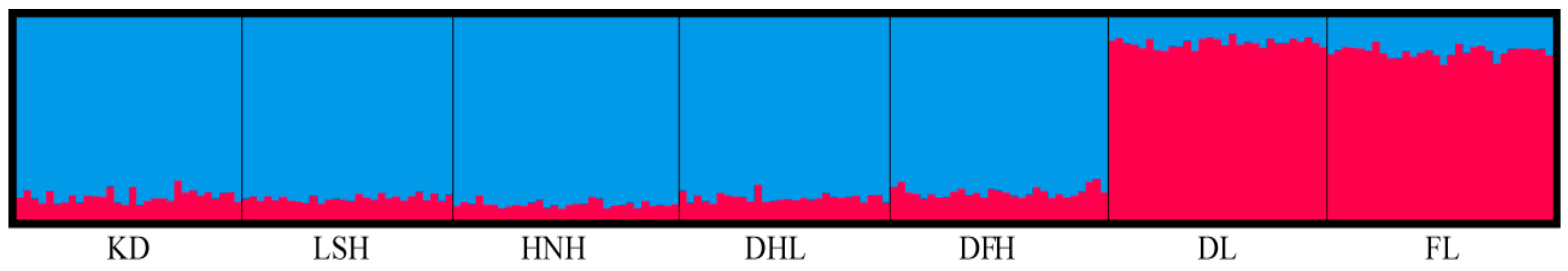
| Population | ID | N | Location (Province) | Latitude (°) | Longitude (°) | Annual Mean Temperature (°C) | Annual Mean Precipitation (mm) |
|---|---|---|---|---|---|---|---|
| Kuandian | KD | 30 | Liaoning | 40.91 | 124.78 | 6.29 | 973.04 |
| Lushuihe | LSH | 28 | Jilin | 42.53 | 127.80 | 4.12 | 783.68 |
| Huangnihe | HNH | 30 | Jilin | 43.55 | 128.01 | 3.76 | 635.42 |
| Dahailin | DHL | 28 | Heilongjiang | 44.52 | 128.86 | 1.14 | 619.72 |
| Dongfanghong | DFH | 29 | Heilongjiang | 46.58 | 133.58 | 3.17 | 648.99 |
| Dailin | DL | 29 | Heilongjiang | 47.18 | 128.85 | 0.30 | 574.59 |
| Fenglin | FL | 30 | Heilongjiang | 48.13 | 129.19 | 0.04 | 596.15 |
| All populations | 204 |
| Locus | Dye | Primer Sequences (5′–3′) | Repeat Motif | Range (bp) | Ta (°C) |
|---|---|---|---|---|---|
| P5 | FAM | F: ATTCCTACTTTTCCCGTTT R: ACAGAGACCCCGTTTACAT | (CA)11 | 108–118 | 55 |
| P6 | FAM | F: TCAAATTACCAGACAATAA R: GAATTCGCCAATGAAATCA | (TA)3(GT)15 | 107–128 | 55 |
| P29 | HEX | F: TGTCAACTTTGAACCCTGAA R: AGGCCAATCCTCATACATTT | (CA)9 | 134–148 | 55 |
| P45 | HEX | F: CTTACATTTTGCTGCTTTTC R: TTGTCAGTTTTAGGTTGGAT | (TG)16(AG)17 | 165–203 | 55 |
| P51 | HEX | F: CCTAAGAGCAATGTAAAATG R: AGCTTGACAACGACTAACT | (AG)15 | 188–225 | 55 |
| P52 | FAM | F: CCATCCTTCAAATTTTCCT R: GCCATTCTTTCTACCACTT | (AG)26 | 113–145 | 56 |
| P62 | FAM | F: CAAGGAGGAAAACAATAAGG R: CTACAACAGAAACTAGCCAGA | (CT)10 | 127–133 | 56 |
| P63 | HEX | F: CTCCTTCTTCATCCATCCATT R: TGAGGTGAGCCTGCATATAGT | (CT)19 | 218–252 | 55 |
| P79 | HEX | F: CCACCGCCAAGTCCATTA R: GCTTTGTTAGCCGTCCAG | (CAA)7 | 183–201 | 55 |
| Locus | Na | Ne | I | Ho | He | uHe | Ar | FIS |
|---|---|---|---|---|---|---|---|---|
| P5 | 2.3 | 1.6 | 0.565 | 0.425 | 0.358 | 0.364 | 2.5 | −0.186 * |
| P6 | 7.1 | 2.3 | 1.214 | 0.688 | 0.561 | 0.571 | 7.4 | −0.227 * |
| P29 | 3.1 | 1.6 | 0.649 | 0.416 | 0.351 | 0.357 | 3.3 | −0.186 * |
| P45 | 11.6 | 6.1 | 2.043 | 0.922 | 0.836 | 0.851 | 11.9 | −0.102 * |
| P51 | 11.7 | 6.6 | 2.122 | 0.922 | 0.846 | 0.861 | 11.7 | −0.089 * |
| P52 | 8.3 | 3.7 | 1.560 | 0.868 | 0.714 | 0.726 | 8.3 | −0.216 * |
| P62 | 3.9 | 2.4 | 0.987 | 0.833 | 0.574 | 0.584 | 4.5 | −0.451 |
| P63 | 9.1 | 4.8 | 1.779 | 0.921 | 0.787 | 0.800 | 9.5 | −0.171 * |
| P79 | 2.7 | 1.9 | 0.699 | 0.676 | 0.466 | 0.474 | 2.6 | −0.450 |
| Mean | 6.7 | 3.4 | 1.291 | 0.741 | 0.610 | 0.621 | 6.8 | −0.231 * |
| Population | Na | Ne | I | Ho | He | uHe | Ar | FIS |
|---|---|---|---|---|---|---|---|---|
| KD | 5.9 | 3.3 | 1.204 | 0.700 | 0.581 | 0.591 | 5.8 | −0.188 ** |
| LSH | 7.1 | 3.4 | 1.303 | 0.718 | 0.605 | 0.616 | 7.1 | −0.170 ** |
| HNH | 7.1 | 3.4 | 1.332 | 0.730 | 0.618 | 0.629 | 7.0 | −0.164 ** |
| DHL | 6.7 | 3.8 | 1.339 | 0.718 | 0.616 | 0.627 | 7.0 | −0.148 ** |
| DFH | 6.7 | 3.1 | 1.214 | 0.743 | 0.586 | 0.596 | 6.6 | −0.252 ** |
| DL | 6.6 | 3.5 | 1.324 | 0.808 | 0.636 | 0.647 | 6.5 | −0.254 ** |
| FL | 6.6 | 3.6 | 1.321 | 0.770 | 0.629 | 0.640 | 6.5 | −0.208 ** |
| Mean | 6.7 | 3.4 | 1.291 | 0.741 | 0.610 | 0.621 | 6.6 | −0.198 * |
| KD | LSH | HNH | DHL | DFH | DL | FL | |
|---|---|---|---|---|---|---|---|
| 0.000 | KD | ||||||
| 0.009 | 0.000 | LSH | |||||
| 0.007 | 0.008 | 0.000 | HNH | ||||
| 0.010 | 0.007 | 0.008 | 0.000 | DHL | |||
| 0.008 | 0.008 | 0.007 | 0.009 | 0.000 | DFH | ||
| 0.016 * | 0.016 * | 0.017 * | 0.021 * | 0.019 * | 0.000 | DL | |
| 0.015 * | 0.014 * | 0.014 * | 0.017 * | 0.014 * | 0.007 * | 0.000 | FL |
| Analysis | Source of Variation | df | Sum of Squares | Variance Component | Percentage of Variation |
|---|---|---|---|---|---|
| Non-hierarchical AMOVA | Among populations | 6 | 45.619 | 0.108 | 2.35% * |
| Within populations | 197 | 879.895 | 4.466 | 97.65% * | |
| Total | 203 | 925.515 | 4.574 | 100.00% | |
| Hierarchical AMOVA | Between groups | 1 | 19.414 | 0.169 | 3.62% * |
| Among populations | 5 | 26.206 | 0.027 | 0.57% | |
| Within populations | 197 | 879.895 | 4.466 | 95.81% * | |
| Total | 203 | 925.515 | 4.662 | 100.00% |
| Parameter | Latitude | Longitude | Tmean | Tmax | Tmin | Prec | PCA1 |
|---|---|---|---|---|---|---|---|
| Na | −0.202 | 0.018 | 0.202 | −0.092 | 0.202 | 0.128 | 0.202 |
| Ne | 0.464 | 0.214 | −0.679 | −0.929 ** | −0.643 | −0.750 | −0.643 |
| I | 0.321 | 0.214 | −0.500 | −0.893 ** | −0.393 | −0.679 | −0.536 |
| Ho | 0.901 ** | 0.631 | −0.811 * | −0.432 | −0.757 * | −0.811 * | −0.847 * |
| He | 0.714 | 0.286 | −0.786 * | −0.786 * | −0.714 | −0.929 * | −0.821 * |
| uHe | 0.714 | 0.286 | −0.786 * | −0.786 * | −0.714 | −0.929 * | −0.821 * |
| Ar | −0.250 | −0.071 | 0.214 | −0.179 | 0.179 | 0.107 | 0.179 |
| FIS | −0.536 | −0.321 | 0.357 | −0.179 | 0.393 | 0.286 | 0.429 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, Y.W.; Lewis, B.J.; Zhou, W.M.; Mao, C.R.; Wang, Y.; Zhou, L.; Yu, D.P.; Dai, L.M.; Qi, L. Genetic Diversity and Population Structure of Natural Pinus koraiensis Populations. Forests 2020, 11, 39. https://doi.org/10.3390/f11010039
Tong YW, Lewis BJ, Zhou WM, Mao CR, Wang Y, Zhou L, Yu DP, Dai LM, Qi L. Genetic Diversity and Population Structure of Natural Pinus koraiensis Populations. Forests. 2020; 11(1):39. https://doi.org/10.3390/f11010039
Chicago/Turabian StyleTong, Yue W., Bernard J. Lewis, Wang M. Zhou, Cheng R. Mao, Yan Wang, Li Zhou, Da P. Yu, Li M. Dai, and Lin Qi. 2020. "Genetic Diversity and Population Structure of Natural Pinus koraiensis Populations" Forests 11, no. 1: 39. https://doi.org/10.3390/f11010039
APA StyleTong, Y. W., Lewis, B. J., Zhou, W. M., Mao, C. R., Wang, Y., Zhou, L., Yu, D. P., Dai, L. M., & Qi, L. (2020). Genetic Diversity and Population Structure of Natural Pinus koraiensis Populations. Forests, 11(1), 39. https://doi.org/10.3390/f11010039







