Modeling of Aboveground Biomass with Landsat 8 OLI and Machine Learning in Temperate Forests
Abstract
:1. Introduction
2. Materials and Methods
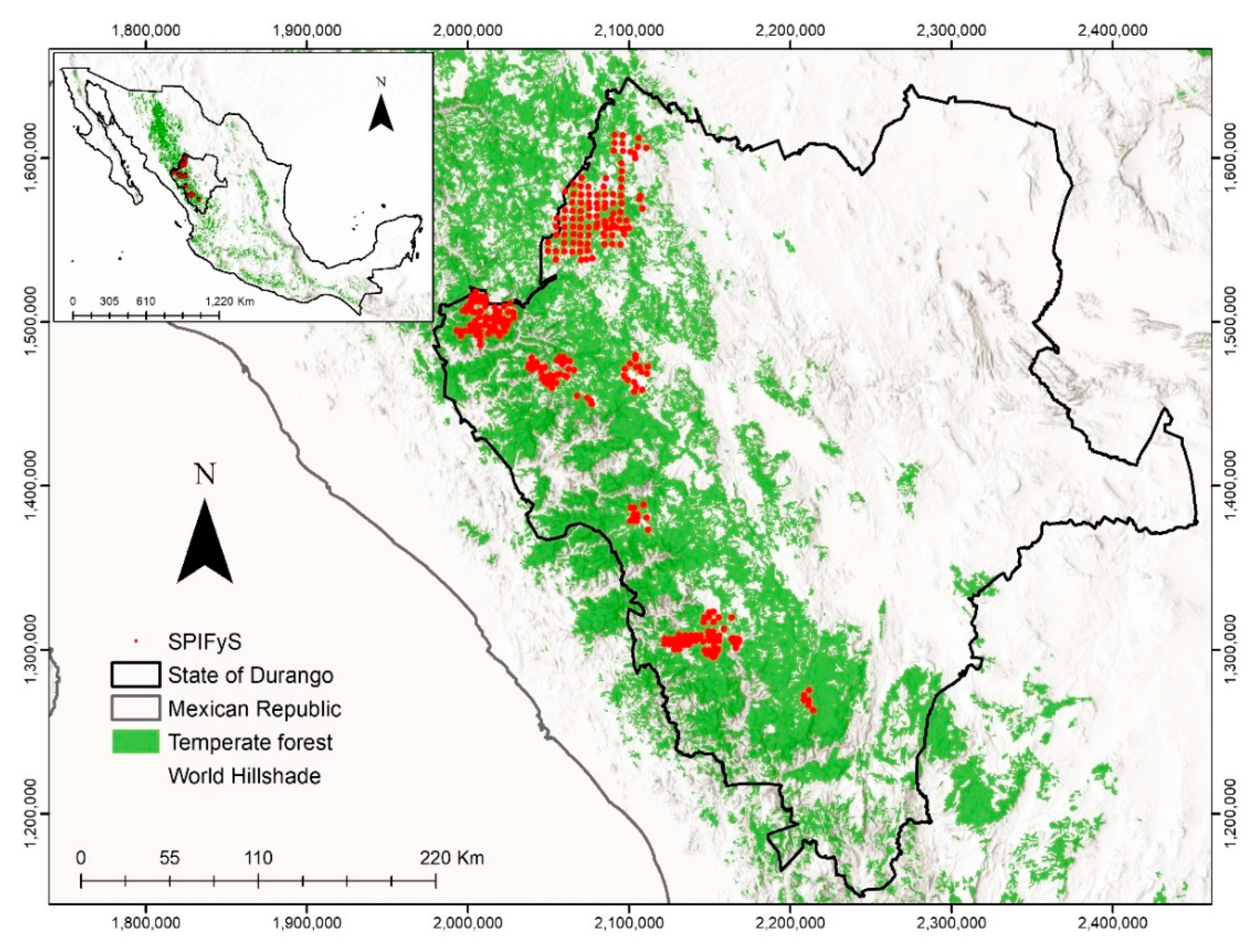
2.1. Study Area
2.2. Field Data
2.3. Spectral Data from the Landsat 8 Operational Land Imager (OLI)
2.4. Vegetation Indices
- NIR: Near Infrared.
- R: Red.
- L: Is the soil brightness correction factor and its value is 0.5.
2.5. Texture Indices
2.6. Topographic and Climatic Variables
2.7. Statistical Analysis
2.7.1. Support Vector Regression (SVR)
2.7.2. Random Forest (RF)
- = observed AGB.
- = predicted AGB.
- = average AGB.
- = number of observations
- = number of model parameters.
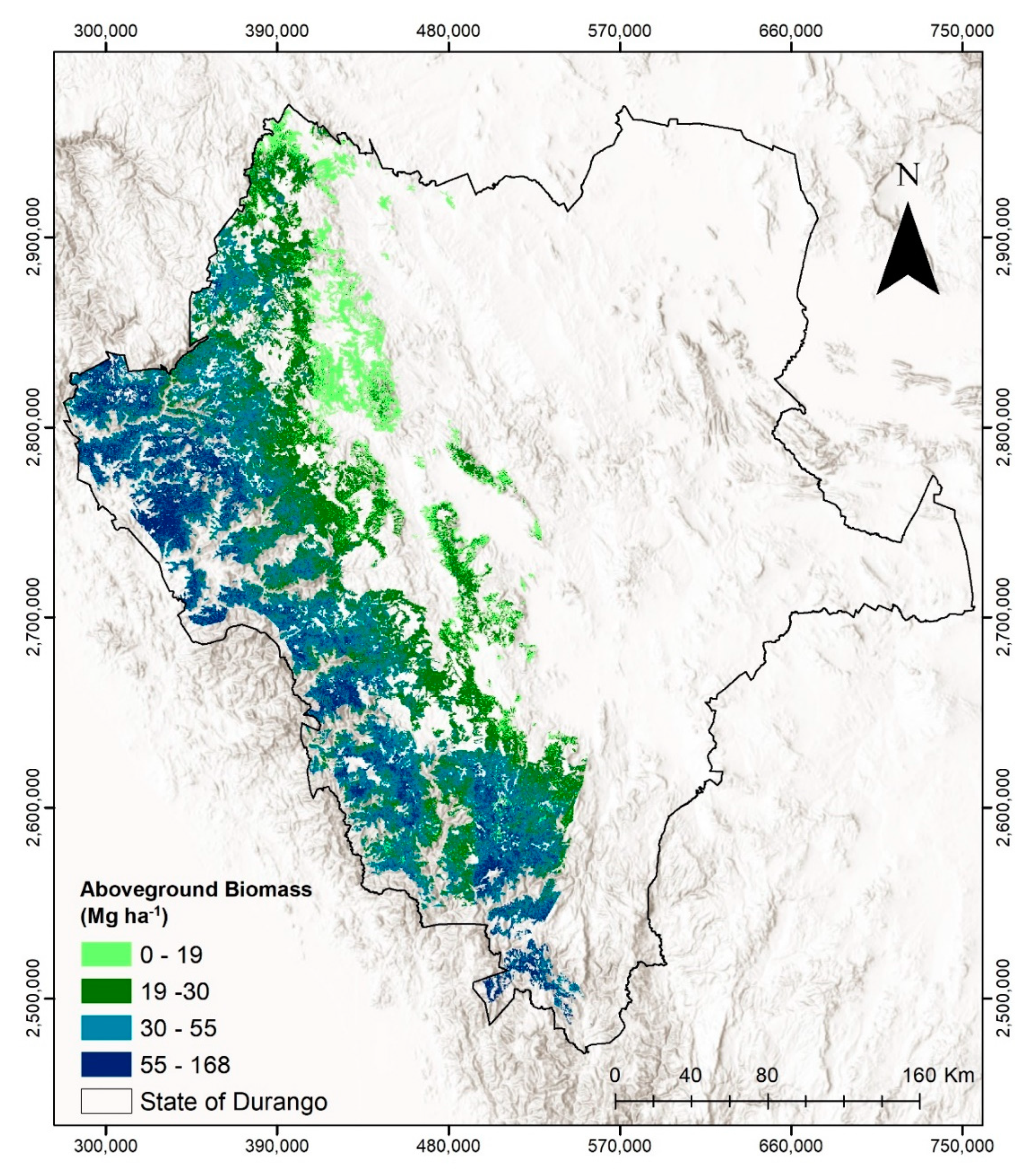
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hu, T.; Su, Y.; Xue, B.; Liu, J.; Zhao, X.; Fang, J.; Guo, Q. Mapping global forest aboveground biomass with spaceborne LiDAR, optical imagery, and forest inventory data. Remote Sens. 2016, 8, 565. [Google Scholar] [CrossRef] [Green Version]
- Galbraith, D.; Levy, P.E.; Sitch, S.; Huntingford, C.; Cox, P.; Williams, M.; Meir, P. Multiple mechanisms of amazonian forest biomass losses in three dynamic global vegetation models under climate change. New Phytol. 2010, 187, 647–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Veiga, P.; Quegan, S.; Carreiras, J.; Persson, H.J.; Fransson, J.E.; Hoscilo, A.; Ziółkowski, D.; Stereńczak, K.; Lohberger, S.; Stängel, M.; et al. Forest biomass retrieval approaches from earth observation in different biomes. Int. J. Appl. Earth Obs. Geoinf. 2019, 77, 53–68. [Google Scholar] [CrossRef]
- De Jong, W.; van Ommen, J.R. Biomass as a Sustainable Energy Source for the Future: Fundamentals of Conversion Processes; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Morris, J. Recycle, bury, or burn wood waste biomass?: Lca answer depends on carbon accounting, emissions controls, displaced fuels, and impact costs. J. Ind. Ecol. 2017, 21, 844. [Google Scholar] [CrossRef]
- Foody, G.M. Remote sensing of tropical forest environments: Towards the monitoring of environmental resources for sustainable development. Int. J. Remote Sens. 2003, 24, 4035–4046. [Google Scholar] [CrossRef]
- Bunker, D.E.; DeClerck, F.; Bradford, J.C.; Colwell, R.K.; Perfecto, I.; Phillips, O.L.; Sankaran, M.; Naeem, S. Species loss and aboveground carbon storage in a tropical forest. Science 2005, 310, 1029–1031. [Google Scholar] [CrossRef] [Green Version]
- Picard, N.; Saint-André, L.; Henry, M. Manual de Construcción de Ecuaciones Alométricas para Estimar el Volumen y la Biomasa de los Árboles: Del Trabajo de Campo a la Predicción; FAO: Rome, Italy, 2012. [Google Scholar]
- Njana, M.A.; Meilby, H.; Eid, T.; Zahabu, E.; Malimbwi, R.E. Importance of tree basic density in biomass estimation and associated uncertainties: A case of three mangrove species in Tanzania. Ann. For. Sci. 2016, 73, 1073–1087. [Google Scholar] [CrossRef]
- Zianis, D.; Mencuccini, M. On simplifying allometric analyses of forest biomass. For. Ecol. Manag. 2004, 187, 311–332. [Google Scholar] [CrossRef]
- Walker, W.; Baccini, A.; Nepstad, M.; Horning, N.; Knight, D.; Braun, E.; Bausch, A. Guia de Campo para la Estimacion de Biomasa y Carbono Forestal (Field Guide to Estimate Forest Biomass and Carbon), version 1.0; Woods Hole Research Center: Falmouth, MA, USA, 2011; p. 53. [Google Scholar]
- Asner, G.P.; Hughes, R.F.; Varga, T.A.; Knapp, D.E.; Kennedy-Bowdoin, T. Environmental and biotic controls over aboveground biomass throughout a tropical rain forest. Ecosystems 2009, 12, 261–278. [Google Scholar] [CrossRef]
- Huang, W.; Swatantran, A.; Johnson, K.; Duncanson, L.; Tang, H.; Dunne, J.O.N.; Hurtt, G.; Dubayah, R. Local discrepancies in continental scale biomass maps: A case study over forested and non-forested landscapes in maryland, USA. Carbon Balance Manag. 2015, 10, 19. [Google Scholar] [CrossRef] [Green Version]
- Guitet, S.; Hérault, B.; Molto, Q.; Brunaux, O.; Couteron, P. Spatial structure of above-ground biomass limits accuracy of carbon mapping in rainforest but large scale forest inventories can help to overcome. PLoS ONE 2015, 10, e0138456. [Google Scholar] [CrossRef] [PubMed]
- López-Serrano, P.M.; López Sánchez, C.A.; Solís-Moreno, R.; Corral-Rivas, J.J. Geospatial estimation of above ground forest biomass in the Sierra Madre Occidental in the state of Durango, Mexico. Forests 2016, 7, 70. [Google Scholar] [CrossRef] [Green Version]
- López-Sánchez, C.A.; García-Ramírez, P.; Resl, R.; Hernández-Díaz, J.C.; Lopez-Serrano, P.M.; Wehenkel, C. Modelling dasometric attributes of mixed and uneven-aged forests using Landsat-8 spectral data in the sierra madre occidental, mexico. iForest-Biogeosci. For. 2017, 10, 288. [Google Scholar] [CrossRef] [Green Version]
- Liang, S. Recent developments in estimating land surface biogeophysical variables from optical remote sensing. Prog. Phys. Geogr. 2007, 31, 501–516. [Google Scholar] [CrossRef] [Green Version]
- McRoberts, R.E.; Tomppo, E.O. Remote sensing support for national forest inventories. Remote Sens. Environ. 2007, 110, 412–419. [Google Scholar] [CrossRef]
- Powell, S.L.; Cohen, W.B.; Healey, S.P.; Kennedy, R.E.; Moisen, G.G.; Pierce, K.B.; Ohmann, J.L. Quantification of Live Aboveground Forest Biomass Dynamics with Landsat Time-series and Field Inventory Data: A Comparison of Empirical Modeling Approaches. Remote Sens. Environ. 2010, 114, 1053–1068. [Google Scholar] [CrossRef]
- Cartus, O.; Kellndorfer, J.; Walker, W.; Franco, C.; Bishop, J.; Santos, L.; Fuentes, J. A national detailed map of forest aboveground carbon stocks in Mexico. Remote Sens. 2014, 6, 5559–5588. [Google Scholar] [CrossRef] [Green Version]
- López-Serrano, P.M.; López-Sánchez, C.A.; Álvarez-González, J.G.; García-Gutiérrez, J. A comparison of machine learning techniques applied to Landsat-5 tm spectral data for biomass estimation. Can. J. Remote Sens. 2016, 42, 690–705. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2014, 9, 63–105. [Google Scholar] [CrossRef]
- López-Serrano, P.M.; Corral-Rivas, J.J.; Díaz-Varela, R.A.; Álvarez-González, J.G.; López-Sánchez, C.A. Evaluation of radiometric and atmospheric correction algorithms for aboveground forest biomass estimation using Landsat 5 tm data. Remote Sens. 2016, 8, 369. [Google Scholar] [CrossRef] [Green Version]
- Bannari, A.; Morin, D.; Bonn, F.; Huete, A.R. A Review of Vegetation Indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- Barbosa, J.M.; Broadbent, E.N.; Bitencourt, M.D. Remote sensing of aboveground biomass in tropical secondary forests: A review. Int. J. For. Res. 2014, 2014, 715796. [Google Scholar] [CrossRef]
- Mohd Zaki, N.A.; Latif, Z.A. Carbon Sinks and Tropical Forest Biomass Estimation: A Review on Role of Remote Sensing in Aboveground-Biomass Modelling. Geocarto Int. 2016, 32, 701–716. [Google Scholar] [CrossRef]
- Beaudoin, A.; Bernier, P.Y.; Guindon, L.; Villemaire, P.; Guo, X.J.; Stinson, G.; Bergeron, T.; Magnussen, S.; Hall, R.J. Mapping attributes of Canada’s forests at moderate resolution through kNN and MODIS imagery. Can. J. For. Res. 2014, 44, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Fagua, J.C.; Cabrera, E.; Gonzalez, V.H. The effect of highly variable topography on the spatial distribution of aniba perutilis (lauraceae) in the colombian andes. Rev. de Biol. Trop. 2013, 61, 301–309. [Google Scholar] [CrossRef]
- Rana, P.; Korhonen, L.; Gautam, B.; Tokola, T. Effect of field plot location on estimating tropical forest above-ground biomass in Nepal using airborne laser scanning data. ISPRS J. Photogramm. Remote Sens. 2014, 94, 55–62. [Google Scholar] [CrossRef]
- Van der Laan, C.; Verweij, P.A.; Quiñones, M.J.; Faaij, A.P. Analysis of biophysical and anthropogenic variables and their relation to the regional spatial variation of aboveground biomass illustrated for North and East Kalimantan, Borneo. Carbon Balance Manag. 2014, 9. [Google Scholar] [CrossRef] [Green Version]
- Cutler, M.E.J.; Boyd, D.S.; Foody, G.M.; Vetrivela, A. Estimating tropical forest biomass with a combination of SAR image texture and Landsat TM data: An assessment of predictions between regions. ISPRS J. Photogramm. Remote Sens. 2012, 70, 66–67. [Google Scholar] [CrossRef] [Green Version]
- Brosofske, K.D.; Froese, R.E.; Falkowski, M.J.; Banksota, A. A review of methods for mapping and prediction of inventory attributes for operational forest management. For. Sci. 2014, 60, 1–24. [Google Scholar] [CrossRef]
- Zhang, X.; Ni-Meister, W. Remote sensing of forest biomass. In Biophysical Applications of Satellite Remote Sensing; Springer: New York, NY, USA, 2014. [Google Scholar]
- Tonolli, S.; Dalponte, M.; Neteler, M.; Rodeghiero, M.; Vescovo, L.; Gianelle, A.D. Fusion of airborne LiDAR and satellite multispectral data for the estimation of timber volume in the Southern Alps. Rem. Sens. Environ. 2011, 115, 2486–2498. [Google Scholar] [CrossRef]
- Rodriguez-Veiga, P.; Saatchi, S.; Tansey, K.; Balzter, H. Magnitude, spatial distribution and uncertainty of forest biomass stocks in Mexico. Rem. Sens. Environ. 2016, 183, 265–281. [Google Scholar] [CrossRef] [Green Version]
- Straub, C.; Weinacker, H.; Koch, B. A comparison of different methods for forest resource estimation using information from airborne laser scanning and CIR orthophotos. Eur. J. For. Res. 2010, 129, 1069–1080. [Google Scholar] [CrossRef]
- Karjalainen, M.; Kankare, V.; Vastaranta, M.; Holopainen, M.; Hyyppa, J. Prediction of plot-level forest variables using TerraSAR-X stereo SAR data. Remote Sens. Environ. 2012, 117, 338–347. [Google Scholar] [CrossRef]
- Chen, G.; Hay, G.J.; St-Onge, B. A GEOBIA framework to estimate forest parameters from LiDAR transects, Quickbird imagery and machine learning: A case study in Quebec, Canada. Int. J. Appl. Earth Obs. Geoinf. 2012, 15, 28–37. [Google Scholar] [CrossRef]
- Marabel, M.; Alvarez-Taboada, F. Spectroscopic determination of aboveground biomass in grasslands using spectral transformations, support vector machine and partial least squares regression. Sensors 2013, 13, 10027–10051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, T.M.; Wang, Y.; Hodges, D.G.; Guess, F.M. Decision Tree Applications for Forestry and Forest Products Manufacturers. In Proceedings of the 2008 Southern Forest Economics Workers Annual Meeting; Siry, J., Bettinger, P., Harris, T., Tye, T., Baldwin, S., Merry, K., Eds.; Center for Forest Business Publ. No. 30; University of Georgia: Athens, GA, USA, 2009; pp. 104–115. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Biau, G.; Scornet, E. A random forest guided tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, J.; Gonzalez-Ferreiro, E.; Mateos-Garcia, D.; Riquelme-Santos, J.C.; Miranda, D. A Comparative Study between Two Regression Methods on LiDAR Data: A Case Study. In Proceedings of the 6th International Conference on Hybrid Artificial Intelligent Systems, Wroclaw, Poland, 23–25 May 2011; Part II. pp. 311–318. [Google Scholar]
- Chen, S.-T. Mining informative hydrologic data by using support vector machines and elucidating mined data according to information entropy. Entropy 2015, 17, 1023–1041. [Google Scholar] [CrossRef] [Green Version]
- Drucker, H.; Burges, C.J.; Kaufman, L.; Smola, A.J.; Vapnik, V. Support vector regression machines. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 1997; pp. 155–161. [Google Scholar]
- Cherkassky, V.; Ma, Y. Practical Selection of SVM Parameters and Noise Estimation for SVM Regression. Neural Netw. 2004, 17, 113–126. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhou, X.; Zhu, X.; Dong, Z.; Guo, W. Estimation of biomass in wheat using random forest regression algorithm and remote sensing data. Crop J. 2016, 4, 212–219. [Google Scholar] [CrossRef] [Green Version]
- Shataee, S. Forest Attributes Estimation Using Aerial Laser Scanner and TM Data. For. Syst. 2013, 22, 484–496. [Google Scholar]
- Gagliasso, D.; Hummel, S.; Temesgen, H. A comparison of selected parametric and non-parametric imputation methods for estimating forest biomass and basal area. Open J. For. 2014, 4, 42–48. [Google Scholar] [CrossRef] [Green Version]
- García-Gutiérrez, J.; Martinez-Alvarez, F.; Troncoso, A.; Riquelme, J.C. A comparison of machine learning regression techniques for LiDAR-derived estimation of forest variables. Neurocomputing 2015, 167, 24–31. [Google Scholar] [CrossRef]
- Wehenkel, C.; Corral-Rivas, J.J.; Hernández-Díaz, J.C.; Gadow, K. Estimating Balanced Structure Areas in multi-species forests on the Sierra Madre Occidental, Mexico. Ann. For. Sci. 2011, 68, 385–394. [Google Scholar] [CrossRef] [Green Version]
- Corral-Rivas, J.; Vargas, B.; Wehenkel, C.; Aguirre, O.; Álvarez, J.; Rojo, A. Guía para el establecimiento de Sitios de Inventario Periódico Forestal y de Suelos del Estado de Durango; Facultad de Ciencias Forestales, Universidad Juárez del Estado de Durango: Durango, Mexico, 2009. [Google Scholar]
- Corral-Rivas, J.; Diéguez-Aranda, U.; Corral-Rivas, S.; Castedo-Dorado, F. A merchantable volume system for major pine species in El Salto, Durango (Mexico). For. Ecol. Manag. 2007, 238, 118–129. [Google Scholar] [CrossRef]
- Vargas-Larreta, B.; López-Sánchez, C.A.; Corral-Rivas, J.J.; López-Martínez, J.O.; Aguirre-Calderón, C.G.; Álvarez-González, J.G. Allometric equations for estimating biomass and carbon stocks in the temperate forests of north-western mexico. Forests 2017, 8, 269. [Google Scholar] [CrossRef] [Green Version]
- Chavez, P.S. An improved dark-object subtraction technique for atmospheric scattering correction of multispectral data. Remote Sens. Environ. 1988, 24, 459–479. [Google Scholar] [CrossRef]
- Goslee, S. Package “Landsat”. R Package Documentation. Available online: https://cran.r-project.org/web/packages/landsat/landsat.pdf (accessed on 16 December 2019).
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Zhou, J.; Guo, R.Y.; Sun, M.; Di, T.T.; Wang, S.; Zhai, J.; Zhao, Z. The effects of glcm parameters on lai estimation using texture values from quickbird satellite imagery. Sci. Rep. 2017, 7, 7366. [Google Scholar] [CrossRef] [PubMed]
- PCI Geomatics; PCI Geomatics Inc.: Toronto, ON, Canada, 2013.
- INEGI. Continuo de Elevaciones Mexicano 3.0; Instituto Nacional de Estadística y Geografía: Aguascalientes, Mexico, 2013.
- Fleming, C.; Giles, J.; Marsh, S. Elevation Models for Geoscience; Geological Society of London: London, UK, 2010. [Google Scholar]
- Horning, N. Remote Sensing for Ecology and Conservation: A Handbook of Techniques; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- INIFAP. Red nacional de estaciones agroclimáticas automatizadas (RNEAA). Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias. Available online: https://clima.inifap.gob.mx/lnmysr (accessed on 16 December 2019).
- Oliver, M.A.; Webster, R. Kriging: A method of interpolation for geographical information systems. Int. J. Geogr. Inf. Syst. 1990, 4, 313–332. [Google Scholar] [CrossRef]
- Clark, P.J.; Evans, F.C. Distance to nearest neighbor as a measure of spatial relationships in populations. Ecology 1954, 35, 445–453. [Google Scholar] [CrossRef]
- Horn, B.K. Hill shading and the reflectance map. Proc. IEEE 1981, 69, 14–47. [Google Scholar] [CrossRef] [Green Version]
- Zevenbergen, L.W.; Thorne, C.R. Quantitative analysis of land surface topography. Earth Surf. Process. Landf. 1987, 12, 47–56. [Google Scholar] [CrossRef]
- Moore, I.D.; Grayson, R.; Ladson, A. Digital terrain modelling: A review of hydrological, geomorphological, and biological applications. Hydrol. Process. 1991, 5, 3–30. [Google Scholar] [CrossRef]
- McCune, B.; Keon, D. Equations for potential annual direct incident radiation and heat load index. J. Veg. Sci. 2002, 13, 603–606. [Google Scholar] [CrossRef]
- Stage, A.R. An expression for the effect of aspect, slope, and habitat type on tree growth. For. Sci. 1976, 22, 457–460. [Google Scholar]
- Awad, M.; Khanna, R. Efficient Learning Machines: Theories, Concepts, and Applications for Engineers and System Designers; Apress: New York, NY, USA, 2015. [Google Scholar]
- Yu, P.-S.; Chen, S.-T.; Chang, I.-F. Support vector regression for real-time flood stage forecasting. J. Hydrol. 2006, 328, 704–716. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. Package ‘e1071’. 2017. Available online: https://cran.r-project.org/web/packages/e1071/e1071.pdf (accessed on 26 January 2018).
- RColorBrewer, S.; Liaw, A.; Wiener, M.; MLiaw, A. Package ‘randomForest’. 2018. Available online: https://cran.r-project.org/web/packages/randomForest/randomForest.pdf (accessed on 16 December 2019).
- Möller, A.; Tutz, G.; Gertheiss, J. Random forests for functional covariates. J. Chemom. 2016, 30, 715–725. [Google Scholar] [CrossRef]
- Ni, X.; Cao, C.; Zhou, Y.; Ding, L.; Choi, S.; Shi, Y.; Park, T.; Fu, X.; Hu, H.; Wang, X. Estimation of forest biomass patterns across northeast china based on allometric scale relationship. Forests 2017, 8, 288. [Google Scholar] [CrossRef] [Green Version]
- Shen, W.; Li, M.; Huang, C.; Wei, A. Quantifying live aboveground biomass and forest disturbance of mountainous natural and plantation forests in northern guangdong, china, based on multi-temporal Landsat, Palsar and field plot data. Remote Sens. 2016, 8, 595. [Google Scholar] [CrossRef] [Green Version]
- Baccini, A.; Laporte, N.; Goetz, S.J.; Sun, M.; Dong, H. A First Map of Tropical Africa’s Above-ground Biomass Derived from Satellite Imagery. Environ. Res. Lett. 2008, 3, 045011. [Google Scholar] [CrossRef] [Green Version]
- Vafaei, S.; Soosani, J.; Adeli, K.; Fadaei, H.; Naghavi, H.; Pham, T.D.; Bui, D.T. Improving accuracy estimation of Forest Aboveground Biomass based on incorporation of ALOS-2 PALSAR-2 and Sentinel-2A imagery and machine learning: A case study of the Hyrcanian forest area (Iran). Remote Sens. 2018, 10, 172. [Google Scholar] [CrossRef] [Green Version]
- Gleason, C.J.; Im, J. Forest biomass estimation from airborne LiDAR data using machine learning approaches. Remote Sens. Environ. 2012, 125, 80–91. [Google Scholar] [CrossRef]
- Li, M.; Im, J.; Quackenbush, L.J.; Liu, T. Forest Biomass and Carbon Stock Quantification Using Airborne LiDAR Data: A Case Study Over Huntington Wildlife Forest in the Adirondack Park. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2014, 7, 3143–3156. [Google Scholar] [CrossRef]
- Gallaun, H.; Zanchi, G.; Nabuurs, G.-J.; Hengeveld, G.; Schardt, M.; Verkerk, P.J. Eu-wide maps of growing stock and above-ground biomass in forests based on remote sensing and field measurements. For. Ecol. Manag. 2010, 260, 252–261. [Google Scholar] [CrossRef]
- Jachowski, N.R.; Quak, M.S.; Friess, D.A.; Duangnamon, D.; Webb, E.L.; Ziegler, A.D. Mangrove biomass estimation in Southwest Thailand using machine learning. Appl. Geogr. 2013, 45, 311–321. [Google Scholar] [CrossRef]
- Wu, C.; Huanhuan, S.; Aihua, S.; Jinsong, D.; Muye, G.; Jinxia, Z.; Hongwei, X.; Ke, W.J. Comparison of machine-learning methods for above-ground biomass estimation based on Landsat imagery. J. Appl. Remote Sens. 2016, 10, 035010. [Google Scholar] [CrossRef]
- Freitas, S.R.; Mello, M.C.S.; Cruz, C.B.M. Relationships between forest structure and vegetation indices in Atlantic Rainforest. For. Ecol. Manag. 2005, 218, 353–362. [Google Scholar] [CrossRef]
- Pizaña, J.M.G.; Hernández, J.M.N.; Romero, N.C. Remote sensing-based biomass estimation. In Environmental Applications of Remote Sensing; IntechOpen: London, UK, 2016. [Google Scholar]
- Trotter, C.M.; Dymond, J.R.; Goulding, C.J. Estimation of timber volume in a coniferous plantation forest using Landsat TM. Int. J. Remote Sens. 1997, 18, 2209–2223. [Google Scholar] [CrossRef]
- Huete, A.; Liu, H.; van Leeuwen, W.J. The Use of Vegetation Indices in Forested Regions: Issues of Linearity and Saturation. In Proceedings of the 1997 IEEE International Geoscience and Remote Sensing Symposium Proceedings. Remote Sensing—A Scientific Vision for Sustainable Development, Singapore, 3–8 August 1997; pp. 1966–1968. [Google Scholar]
- Kelsey, K.C.; Neff, J.C. Estimates of Aboveground Biomass from Texture Analysis of Landsat Imagery. Remote Sens. 2014, 6, 6407–6422. [Google Scholar] [CrossRef] [Green Version]
- Wood, E.M.; Pidgeon, A.M.; Radeloff, V.C.; Keuler, N.S. Image texture as a remotely sensed measure of vegetation structure. Remote Sens. Environ. 2012, 121, 516–526. [Google Scholar] [CrossRef]
- Nichol, J.E.; Sarker, M.L.R. Improved biomass estimation using the texture parameters of two high-resolution optical sensors. IEEE Trans. Geosci. Remote Sens. 2011, 49, 930–948. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Wu, B.; Wang, Y. Estimating spatiotemporal patterns of aboveground biomass using Landsat TM and MODIS images in the Mu Us Sandy Land, China. Agric. For. Meteorol. 2015, 200, 119–128. [Google Scholar] [CrossRef]
- Propastin, P. Modifying geographically weighted regression for estimating aboveground biomass in tropical rainforests by multispectral remote sensing data. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 82–90. [Google Scholar] [CrossRef]
- Baccini, A. Forest biomass estimation over regional scales using multisource data. Geophys. Res. Lett. 2004, 31. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Huang, H.; Wang, S. Estimation of tropical forest biomass using Landsat TM imagery and permanent plot data in Xishuangbanna, China. Int. J. Remote Sens. 2011, 32, 5741–5756. [Google Scholar] [CrossRef]




| Variable | Mean | Standard Deviation | Minimum Value | Maximum Value |
|---|---|---|---|---|
| Number of trees per ha | 164 | 74.48 | 34 | 566 |
| Basal area (m2·ha−1) | 5.61 | 2.10 | 0.84 | 14.45 |
| Volume (m3·ha−1) | 51.88 | 28.52 | 3.26 | 187.26 |
| Aboveground biomass (Mg·ha−1) | 28.83 | 16.76 | 1.72 | 101.71 |
| Band | Name | Wavelength (μm) | Spatial Resolution (m) |
|---|---|---|---|
| BAND 02 | Blue | 0.45–0.51 | 30 × 30 |
| BAND 03 | Green | 0.53–0.59 | 30 × 30 |
| BAND 04 | Red | 0.64–0.67 | 30 × 30 |
| BAND 05 | Near Infrared (NIR) | 0.85–0.88 | 30 × 30 |
| BAND 06 | Shortwave infrared (SWIR 1) | 1.57–1.65 | 30 × 30 |
| BAND 07 | Shortwave infrared (SWIR 2) | 2.11–2.29 | 30 × 30 |
| BAND 09 | Cirrus | 1.36–1.38 | 30 × 30 |
| Texture Variables | Formula |
|---|---|
| Mean | |
| Standard deviation | |
| Homogeneity | |
| Contrast | |
| Dissimilarity | |
| Entropy | |
| Second moment | |
| Correlation |
| Physical Variable | Equation | Reference |
|---|---|---|
| Topographic | ||
| Slope | [66] | |
| Elevation | [67] | |
| Aspect | [68] | |
| Curvature | [69] | |
| Plan curvature | ||
| Profile curvature | ||
| Wetness Index | [70] | |
| Heat load index | [71] | |
| Aspect/Slope ratio | [72] | |
| Climatic | ||
| Maximum Temperature (TM) | (°C) | [65] |
| Mean Annual Temperature (Tmed) | (°C) | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Serrano, P.M.; Cárdenas Domínguez, J.L.; Corral-Rivas, J.J.; Jiménez, E.; López-Sánchez, C.A.; Vega-Nieva, D.J. Modeling of Aboveground Biomass with Landsat 8 OLI and Machine Learning in Temperate Forests. Forests 2020, 11, 11. https://doi.org/10.3390/f11010011
López-Serrano PM, Cárdenas Domínguez JL, Corral-Rivas JJ, Jiménez E, López-Sánchez CA, Vega-Nieva DJ. Modeling of Aboveground Biomass with Landsat 8 OLI and Machine Learning in Temperate Forests. Forests. 2020; 11(1):11. https://doi.org/10.3390/f11010011
Chicago/Turabian StyleLópez-Serrano, Pablito M., José Luis Cárdenas Domínguez, José Javier Corral-Rivas, Enrique Jiménez, Carlos A. López-Sánchez, and Daniel José Vega-Nieva. 2020. "Modeling of Aboveground Biomass with Landsat 8 OLI and Machine Learning in Temperate Forests" Forests 11, no. 1: 11. https://doi.org/10.3390/f11010011






