Effects of Landscape Fragmentation on Genetic Diversity of Male-Biased Dioecious Plant Pistacia chinensis Bunge Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
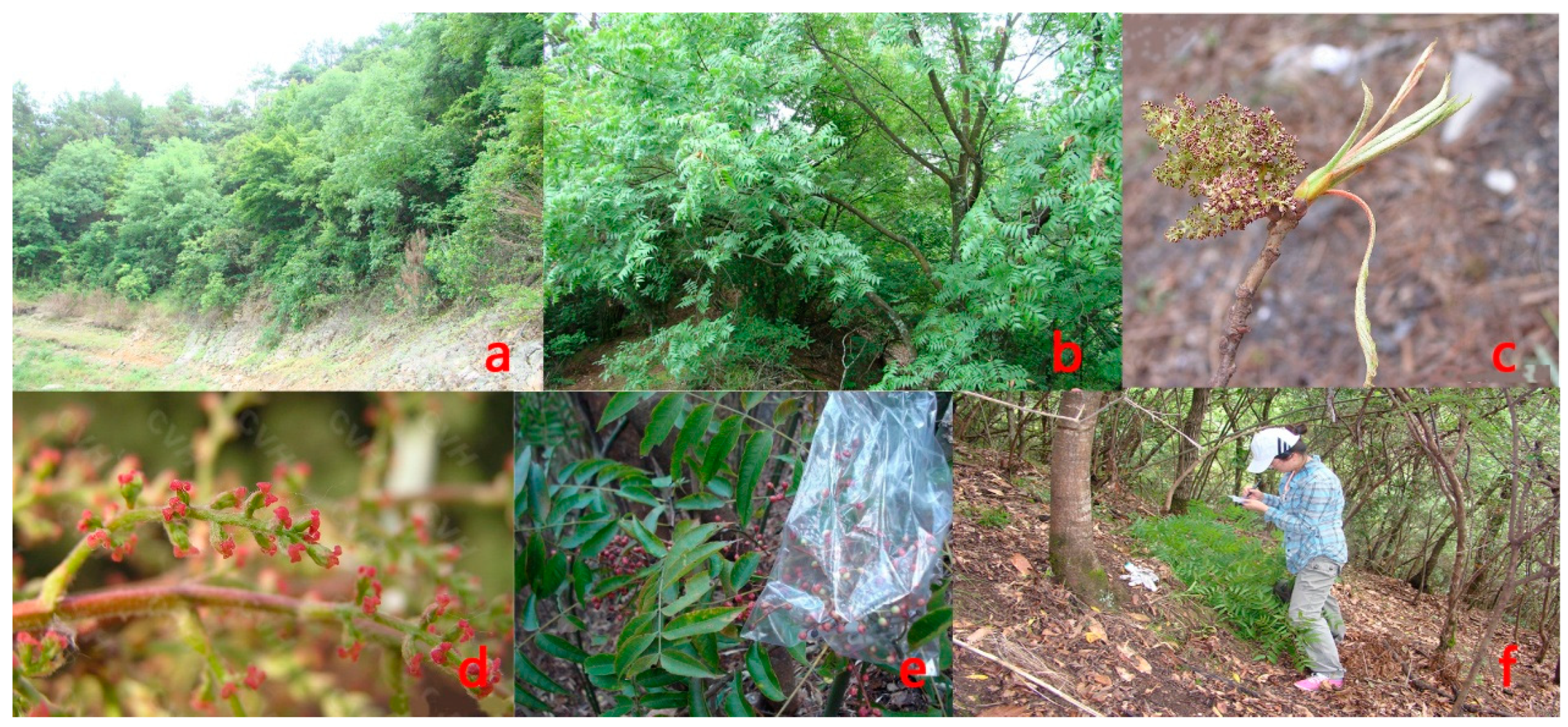
2.2. Study Species
2.3. Field Sampling and Data Collection
2.4. DNA Extraction and Microsatellite Genotyping
2.5. Statistical Analysis
3. Results
3.1. Microsatellite Loci
3.2. The Total Genetic Diversity and Differentiation of Three Population Classes of P. chinensis
3.3. The Genetic Diversity and Differentiation of Six Pre- and Post-Fragmentation Populations
3.4. The Genetic Diversity and Differentiation of Six Male and Female Populations
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aguilar, R.; Quesada, M.; Ashworth, L.; Herrerias-Diego, Y.; Lobo, J. Genetic consequences of habitat fragmentation in plant populations: Susceptible signals in plant traits and methodological approaches. Mol. Ecol. 2008, 17, 5177–5188. [Google Scholar] [CrossRef]
- Holderegger, R.; Buehler, D.; Gugerli, F.; Manel, S. Landscape genetics of plants. Trends Plant Sci. 2010, 15, 675–683. [Google Scholar] [CrossRef]
- Sork, V.L.; Waits, L. Contributions of landscape genetics-approaches, insights, and future potential. Mol. Ecol. 2010, 19, 3489–3495. [Google Scholar] [CrossRef]
- Bacles, C.F.E.; Lowe, A.J.; Ennos, R.A. Genetic effects of chronic habitat fragmentation on tree species: The case of Sorbus aucuparia in a deforested Scottish landscape. Mol. Ecol. 2004, 13, 573–584. [Google Scholar] [CrossRef]
- Dubreuil, M.; Riba, M.; Gonzalez-Martinez, S.C.; Vendramin, G.G.; Sebastiani, F.; Mayol, M. Genetic effects of chronic habitat fragmentation revisited: Strong genetic structure in a temperate tree, Taxus baccata (Taxaceae), with great dispersal capability. Am. J. Bot. 2010, 97, 303–310. [Google Scholar] [CrossRef]
- Vranckx, G.; Jacquemyn, H.; Muys, B.; Honnay, O. Meta-analysis of susceptibility of woody plants to loss of genetic diversity through habitat fragmentation. Conserv. Biol. 2012, 26, 228–237. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, M.M.; Shen, D.W.; Chen, X.Y. Habitat loss other than fragmentation per se decreased nuclear and chloroplast genetic diversity in a monoecious tree. PLoS ONE 2012, 7, e39146. [Google Scholar] [CrossRef]
- Lauterbach, D.; Ristow, M.; Gemeinholzer, B. Population genetics and fitness in fragmented populations of the dioecious and endangered Silene otites (Caryophyllaceae). Plant Syst. Evol. 2011, 298, 155–164. [Google Scholar] [CrossRef]
- Yu, L.; Lu, J.B. Does landscape fragmentation influence sex ratio of dioecious plants? A case study of Pistacia chinensis in the Thousand-Island Lake region of China. PLoS ONE 2011, 6, e22903. [Google Scholar] [CrossRef]
- Öster, M.; Eriksson, O. Sex ratio mediated pollen limitation in the dioecious herb Antennaria dioica. Ecoscience 2007, 14, 387–398. [Google Scholar] [CrossRef]
- Stehlik, I.; Friedman, J.; Barrett, S.C. Environmental influence on primary sex ratio in a dioecious plant. Proc. Natl. Acad. Sci. USA 2008, 105, 10847–10852. [Google Scholar] [CrossRef]
- Hilfiker, K.; Gugerli, F.; Schütz, J.-P.; Rotach, P.; Holderegger, R. Low RAPD variation and female-biased sex ratio indicate genetic drift in small populations of the dioecious conifer Taxus baccata in Switzerland. Conserv. Genet. 2004, 5, 357–365. [Google Scholar] [CrossRef]
- Vandepitte, K.; Roldán-Ruiz, I.; Leus, L.; Jacquemyn, H.; Honnay, O. Canopy closure shapes clonal diversity and fine-scale genetic structure in the dioecious understorey perennial Mercurialis perennis. J. Ecol. 2009, 97, 404–414. [Google Scholar] [CrossRef]
- Nosrati, H.; Husainpourfeizi, M.A.; Khorasani, M.; Razban-Haghighi, A.; Nikniazi, M. Sex ratio and genetic diversity in the dioecious Pistacia atlantica (Anacardiaceae). J. Agrobiol. 2012, 29, 41–46. [Google Scholar] [CrossRef]
- Vandepitte, K.; Honnay, O.; De Meyer, T.; Jacquemyn, H.; Roldán-Ruiz, I. Patterns of sex ratio variation and genetic diversity in the dioecious forest perennial Mercurialis perennis. Plant Ecol. 2009, 206, 105–114. [Google Scholar] [CrossRef]
- Bashalkhanov, S.; Eckert, A.J.; Rajora, O.P. Genetic signatures of natural selection in response to air pollution in red spruce (Picea rubens, Pinaceae). Mol. Ecol. 2013, 22, 5877–5889. [Google Scholar] [CrossRef]
- Sun, Q.; Lu, J.B.; Wu, J.G.; Zhang, F.F. Effects of island area on plant species distribution and conservation implications in the Thousand Island Lake region. Biodivers. Sci. 2008, 16, 1–7. [Google Scholar]
- Lu, J.; Jiang, L.; Yu, L.; Sun, Q. Local factors determine plant community structure on closely neighbored islands. PLoS ONE 2011, 6, e19762. [Google Scholar] [CrossRef]
- Ding, L.Z.; Lu, J.B.; Xu, G.F.; Wu, J.G. Effects of ecological protection and development on landscape pattern in the Thousand-Island Lake region, Zhejiang Province. Biodivers. Sci. 2004, 12, 473–480. [Google Scholar]
- Smith, J.; Borgis, S.; Seifert, V. Studies in urban ecology: The first wave of biological invasion by Pistacia chinensis in Armidale, New South Wales, Australia. Aust. Geogr. Stud. 2000, 38, 263–274. [Google Scholar] [CrossRef]
- Niu, Z.T.; Li, T.; Guan, G.Z.; Zhang, Y.J. Resource survey, cultivation techniques and multipurpose utilization of Pistacia chinensis Bunge. Nonwood For. Res. 2005, 23, 68–71. [Google Scholar]
- Qin, F.; Guo, T.B.; Liu, Z.G.; Song, M.H. Literature review of researches on Pistacia chinensis Bunge. Nonwood For. Res. 2007, 25, 90–96. [Google Scholar]
- Tang, M.; Zhang, P.; Zhang, L.; Li, M.; Wu, L. A Potential Bioenergy Tree: Pistacia chinensis Bunge. Energy Procedia 2012, 16, 737–746. [Google Scholar] [CrossRef][Green Version]
- Liu, Q.S.; Li, J.X. Study on growth principle of Pistacia chinensis. J. Henan For. Sci. Technol. 1999, 19, 3–6. [Google Scholar]
- Wu, Z.Z.; Zhang, Z.X.; Wang, Z.J.; Li, J.X.; Wang, X.Y. SSR analysis on genetic diversity of natural populations of Pistacia chinensis Bunge. Chin. J. Appl. Environ. Biol. 2010, 16, 803–806. [Google Scholar]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.; Wills, D.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T. Hp-rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT, A Program to Estimate and Test Gene Diversities and Fixation Indices (Version 2.9. 3). 2001. Available online: http://www.unil.ch/izea/softwares/fstat.htlm (accessed on 1 January 2001).
- Cushman, S.A.; Landguth, E.L. Spurious correlations and inference in landscape genetics. Mol. Ecol. 2010, 19, 3592–3602. [Google Scholar] [CrossRef] [PubMed]
- Haag, T.; Santos, A.S.; Sana, D.A.; Morato, R.G.; Cullen, L.; Crawshaw, P.G.; De Angelo, C.; Di Bitetti, M.S.; Salzano, F.M.; Eizirik, E. The effect of habitat fragmentation on the genetic structure of a top predator: Loss of diversity and high differentiation among remnant populations of Atlantic Forest jaguars (Panthera onca). Mol. Ecol. 2010, 19, 4906–4921. [Google Scholar] [CrossRef] [PubMed]
- Jacquemyn, H.; Roldán-Ruiz, I.; Honnay, O. Evidence for demographic bottlenecks and limited gene flow leading to low genetic diversity in a rare thistle. Conserv. Genet. 2010, 11, 1979–1987. [Google Scholar] [CrossRef]
- Karlin, E.F.; Hotchkiss, S.C.; Boles, S.B.; Stenoien, H.K.; Hassel, K.; Flatberg, K.I.; Shaw, A.J. High genetic diversity in a remote island population system: Sans sex. New Phytol. 2012, 193, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Comes, H.P.; Mao, Y.R.; Qi, X.S.; Qiu, Y.X. Genetic effects of recent habitat fragmentation in the Thousand-Island Lake region of southeast China on the distylous herb Hedyotis chrysotricha (Rubiaceae). Am. J. Bot. 2012, 99, 1715–1725. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.H.; Miao, J.; Song, H.W.; Yuan, G.J. The change rule of pollen flying time and space in Pistacia chinensis seed orchard. Jiangsu Agric. Sci. 2012, 40, 190–191. [Google Scholar]
- Sezen, U.U.; Chazdon, R.L.; Holsinger, K.E. Genetic consequences of tropical second-growth forest regeneration. Science 2005, 307, 891. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.D.; Epperson, B.K.; Fortin, M.J.; Holderegger, R.; James, P.M.; Rosenberg, M.S.; Scribner, K.T.; Spear, S. Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol. Ecol. 2010, 19, 3565–3575. [Google Scholar] [CrossRef]
- Gaino, A.P.S.C.; Silva, A.M.; Moraes, M.A.; Alves, P.F.; Moraes, M.L.T.; Freitas, M.L.M.; Sebbenn, A.M. Understanding the effects of isolation on seed and pollen flow, spatial genetic structure and effective population size of the dioecious tropical tree species Myracrodruon urundeuva. Conserv. Genet. 2010, 11, 1631–1643. [Google Scholar] [CrossRef]
- Bacles, C.F.; Jump, A.S. Taking a tree’s perspective on forest fragmentation genetics. Trends Plant Sci. 2011, 16, 13–18. [Google Scholar] [CrossRef]
- Hou, Y.; Lou, A. Population genetic diversity and structure of a naturally isolated plant species, Rhodiola dumulosa (Crassulaceae). PLoS ONE 2011, 6, e24497. [Google Scholar] [CrossRef]
- Quinteros-Casaverde, N.; Flores-Negrón, C.F.; Williams, D.A. Low genetic diversity and fragmentation effects in a wind-pollinated tree, Polylepis multijuga Plige (Rosaceae) in the high Andes. Conserv. Genet. 2012, 13, 593–603. [Google Scholar] [CrossRef]
- Wei, X.; Jiang, M. Contrasting relationships between species diversity and genetic diversity in natural and disturbed forest tree communities. New Phytol. 2012, 193, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Yang, F.; Yi, C.Y.; Li, C.Y. Research advances in sex-specific responses of dioecious plants to environmental stresses. Chin. J. Appl. Ecol. 2007, 18, 2626–2631. [Google Scholar]
- O’Connell, L.M.; Mosseler, A.; Rajora, O.P. Extensive long-distance pollen dispersal in a fragmented landscape maintains genetic diversity in white spruce. J. Hered. 2007, 98, 640–645. [Google Scholar] [CrossRef] [PubMed]
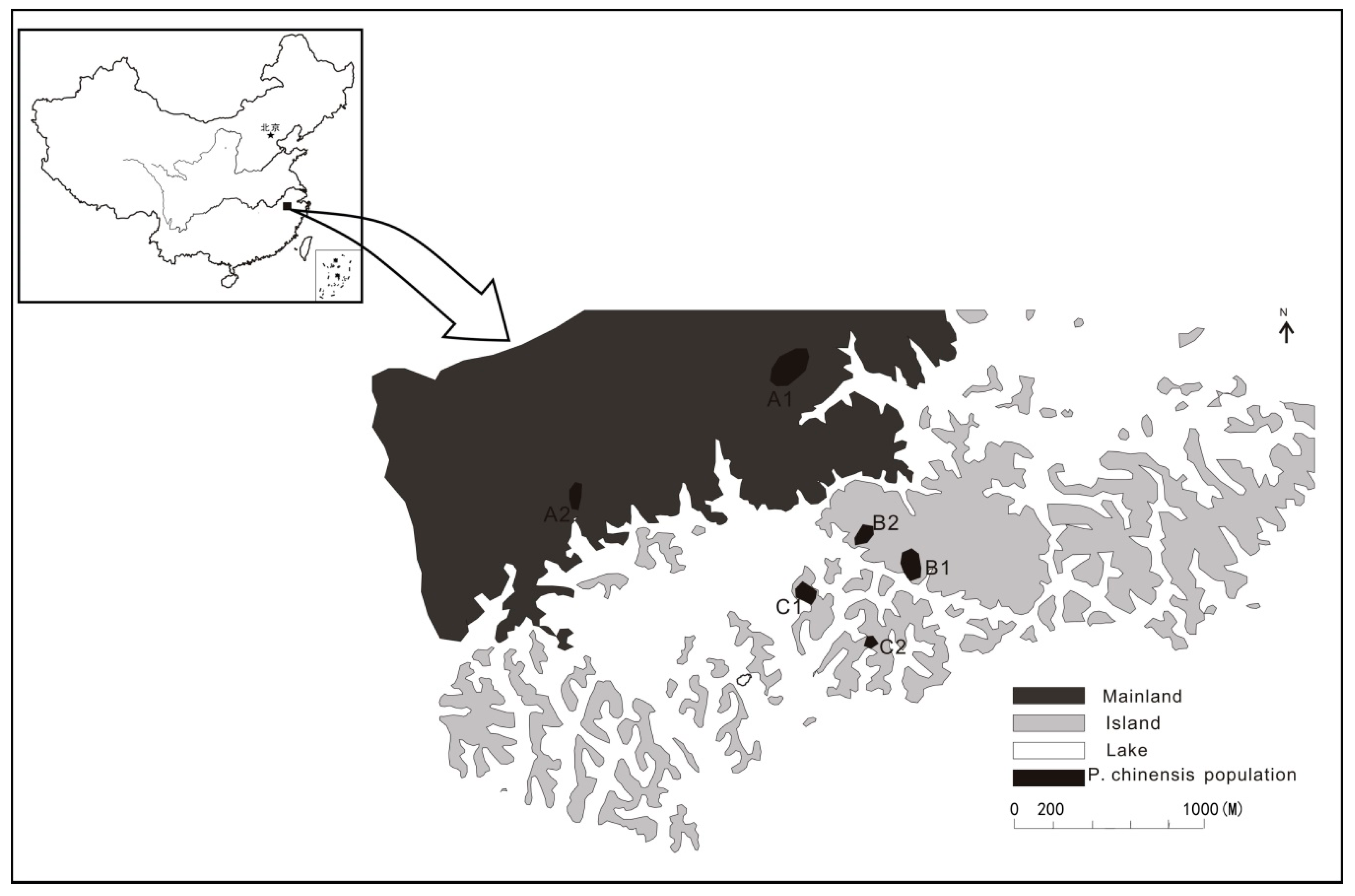

| Population | GPS | Population Size | M/T | Sampling Area (ha) | Sample Size | Pre | Post | Male | Female | Seedling |
|---|---|---|---|---|---|---|---|---|---|---|
| Pop A1 | 29°38′02.022″ N 118°57′20.046″ E | 500 | 0.525 | 2.574 | 40 | 23 | 17 | 20 | 18 | 2 |
| Pop A2 | 29°35′32.850″ N 118°54′13.266″ E | 85 | 0.538 | 0.684 | 20 | |||||
| Pop B1 | 29°35′07.554″ N 118°55′30.702″ E | 162 | 0.626 | 1.207 | 40 | 21 | 19 | 23 | 14 | 3 |
| Pop B2 | 29°34′13.387″ N 118°55′19.549″ E | 61 | 0.6 | 0.685 | 20 | |||||
| Pop C1 | 29°35′01.591″ N 118°55′07.459″ E | 163 | 0.675 | 0.802 | 40 | 16 | 24 | 23 | 11 | 6 |
| Pop C2 | 29°34′50.882″ N 118°55′21.380″ E | 30 | 0.7 | 0.278 | 20 |
| Locus | Na | Ho | He | HWE |
|---|---|---|---|---|
| Ptms3 | 10 | 0.533 | 0.720 | ns |
| Ptms7 | 21 | 0.738 | 0.736 | ns |
| Ptms9 | 16 | 0.483 | 0.490 | ns |
| Ptms10 | 18 | 0.583 | 0.628 | ns |
| Ptms33 | 12 | 0.883 | 0.747 | ** |
| Ptms40 | 12 | 0.521 | 0.438 | ns |
| Ptms41 | 25 | 0.713 | 0.780 | ** |
| Ptms42 | 13 | 0.721 | 0.661 | ns |
| Ptms45 | 13 | 0.846 | 0.702 | * |
| Mean/Total | 15.6 | 0.669 | 0.656 | ns |
| Population | Na | Ne | AR | PAR | Ho | He | FIS |
|---|---|---|---|---|---|---|---|
| Pop A1 | 7.889 | 3.879 | 5.57 | 0.77 | 0.769 | 0.725 | −0.048 |
| Pop A2 | 6.333 | 3.670 | 5.15 | 0.88 | 0.600 | 0.604 | 0.033 |
| Pop B1 | 7.556 | 3.937 | 5.56 | 1.12 | 0.725 | 0.707 | −0.013 |
| Pop B2 | 7.556 | 4.752 | 6.23 | 3.15 | 0.889 | 0.746 | −0.166 |
| Pop C1 | 4.889 | 2.849 | 3.74 | 0.16 | 0.519 | 0.581 | 0.118 * |
| Pop C2 | 4.111 | 2.872 | 3.78 | 0.25 | 0.511 | 0.570 | 0.129 * |
| Population | Ne | AR | PAR | Ho | He | FIS |
|---|---|---|---|---|---|---|
| PreA1 | 4.091 | 5.75 | 0.44 | 0.783 | 0.740 | −0.033 |
| Post A1 | 3.452 | 5.31 | 0.35 | 0.756 | 0.689 | −0.071 |
| Pre B1 | 3.947 | 5.67 | 0.40 | 0.744 | 0.716 | −0.014 |
| Post B1 | 3.854 | 5.46 | 0.35 | 0.717 | 0.679 | −0.029 |
| Pre C1 | 2.874 | 3.80 | 0.27 | 0.567 | 0.591 | 0.066 |
| Post C1 | 2.673 | 3.69 | 0.19 | 0.472 | 0.549 | 0.166 * |
| Pre A1 | Post A1 | Pre B1 | PostB1 | Pre C1 | Post C1 | |
|---|---|---|---|---|---|---|
| PreA1 | 0.000 | 45.385 | 9.563 | 7.668 | 1.932 | 1.786 |
| Post A1 | 0.005 | 0.000 | 6.102 | 8.785 | 2.099 | 1.920 |
| Pre B1 | 0.025 | 0.039 | 0.000 | 24.509 | 1.176 | 1.132 |
| Post B1 | 0.032 | 0.028 | 0.010 | 0.000 | 1.505 | 1.483 |
| Pre C1 | 0.115 | 0.106 | 0.175 | 0.142 | 0.000 | 29.822 |
| Post C1 | 0.123 | 0.115 | 0.181 | 0.144 | 0.008 | 0.000 |
| Population | AR | PAR | Ho | He | FIS |
|---|---|---|---|---|---|
| AM | 5.50 | 0.49 | 0.746 | 0.719 | −0.034 |
| AF | 5.67 | 0.55 | 0.795 | 0.721 | −0.105 |
| BM | 5.59 | 0.43 | 0.716 | 0.716 | −0.005 |
| BF | 5.43 | 0.53 | 0.741 | 0.766 | −0.114 |
| CM | 3.72 | 0.22 | 0.502 | 0.571 | 0.127 |
| CF | 3.79 | 0.29 | 0.556 | 0.575 | 0.011 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.-T.; Qiu, Y.-H.; Lu, J.-B. Effects of Landscape Fragmentation on Genetic Diversity of Male-Biased Dioecious Plant Pistacia chinensis Bunge Populations. Forests 2019, 10, 792. https://doi.org/10.3390/f10090792
Lu J-T, Qiu Y-H, Lu J-B. Effects of Landscape Fragmentation on Genetic Diversity of Male-Biased Dioecious Plant Pistacia chinensis Bunge Populations. Forests. 2019; 10(9):792. https://doi.org/10.3390/f10090792
Chicago/Turabian StyleLu, Jun-Ting, Ya-Hui Qiu, and Jian-Bo Lu. 2019. "Effects of Landscape Fragmentation on Genetic Diversity of Male-Biased Dioecious Plant Pistacia chinensis Bunge Populations" Forests 10, no. 9: 792. https://doi.org/10.3390/f10090792
APA StyleLu, J.-T., Qiu, Y.-H., & Lu, J.-B. (2019). Effects of Landscape Fragmentation on Genetic Diversity of Male-Biased Dioecious Plant Pistacia chinensis Bunge Populations. Forests, 10(9), 792. https://doi.org/10.3390/f10090792




