Ophelimus migdanorum Molina-Mercader sp. nov. (Hymenoptera: Eulophidae): Application of Integrative Taxonomy for Disentangling a Polyphenism Case in Eucalyptus globulus Labill Forest in Chile
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Sampling
2.2. Laboratory Emergence Stimulation
2.3. Morphological Description
2.4. Molecular Protocols and Sequence Editing
which recover a 658 bp region near the 5´ ends of COI, including the 648 bp barcode region for the animal kingdom [38]. Sequence editing and alignment were automatically done by mapping Sanger sequencing reads to a reference with Unipro UGENE [39] and manual corrections. DNA sequences have been submitted to GenBank (see Table S5 for accession numbers) and BoldSystem (BIN: ADP0823). DNA voucher specimens were deposited at the Museo de Zoología, Universidad de Concepción, Chile.LepF1 (ATTCAACCAATCATAAAGATATTGG) andLepR1 (TAAACTTCTGGATGTCCAAAA AATCA) [29],
2.5. Identity of Obtained COI Sequences
2.6. Behavioral Data
3. Results
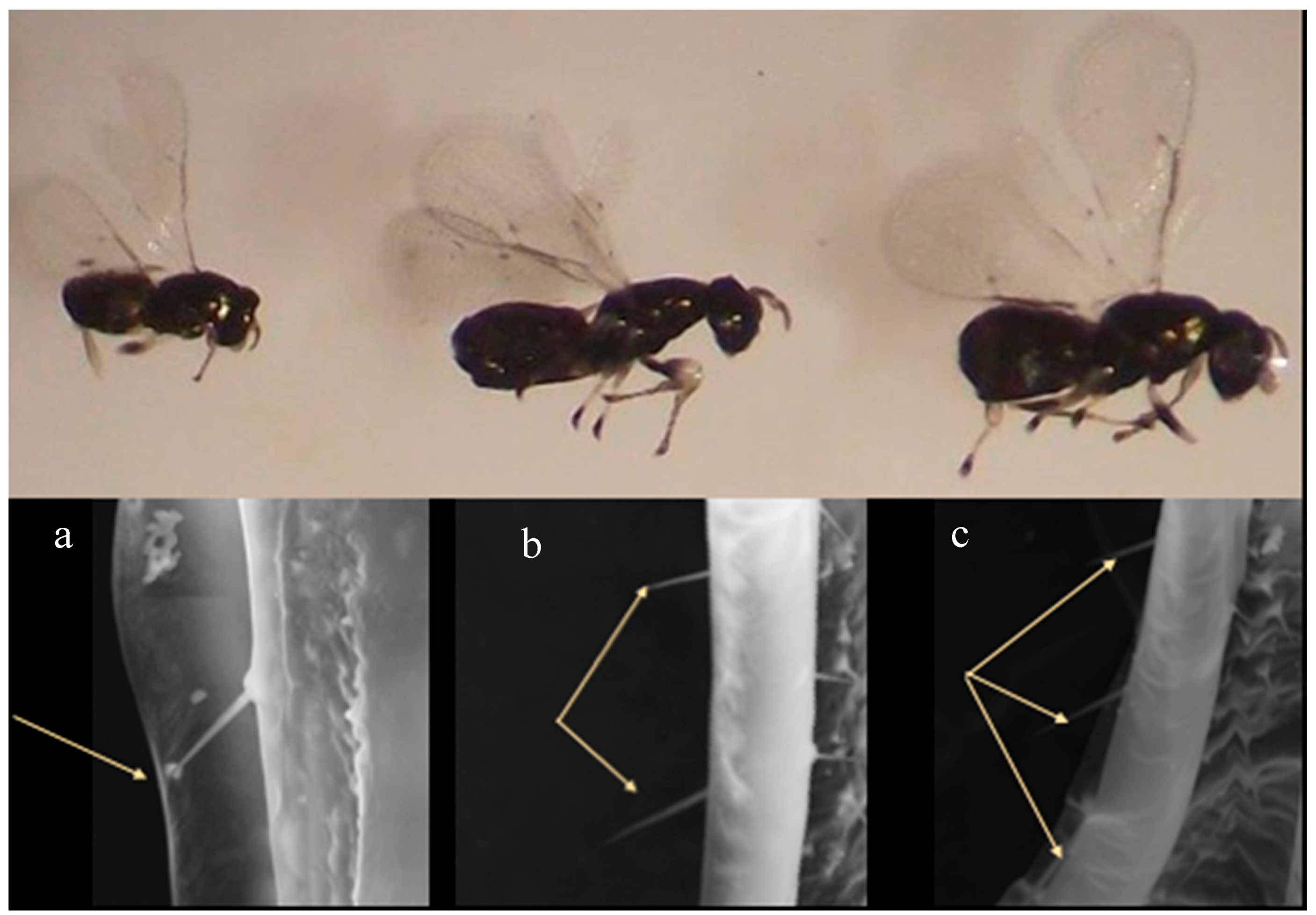
3.1. Morphology
New Ophelimus Species Diagnosis
3.2. Galls
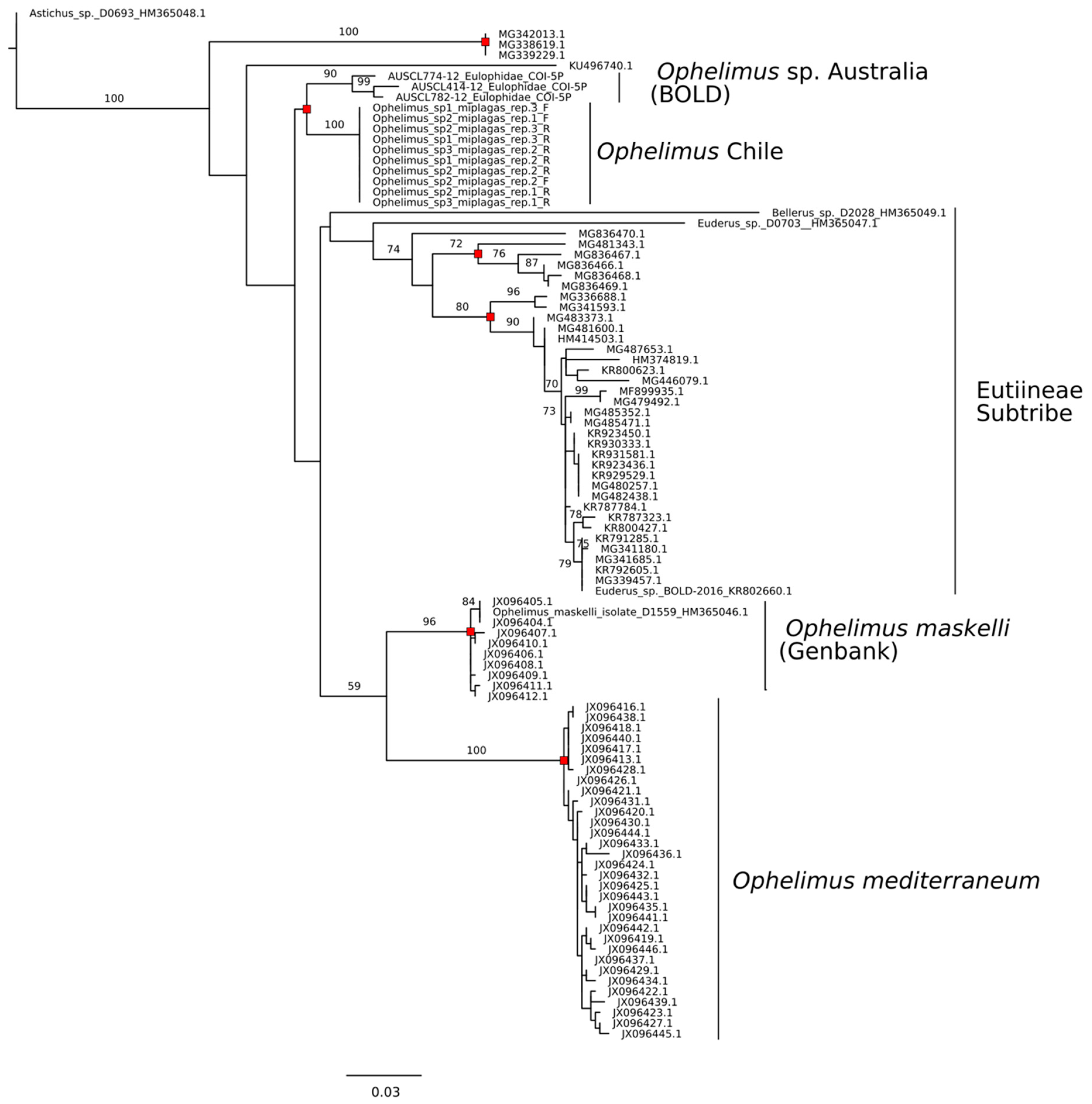
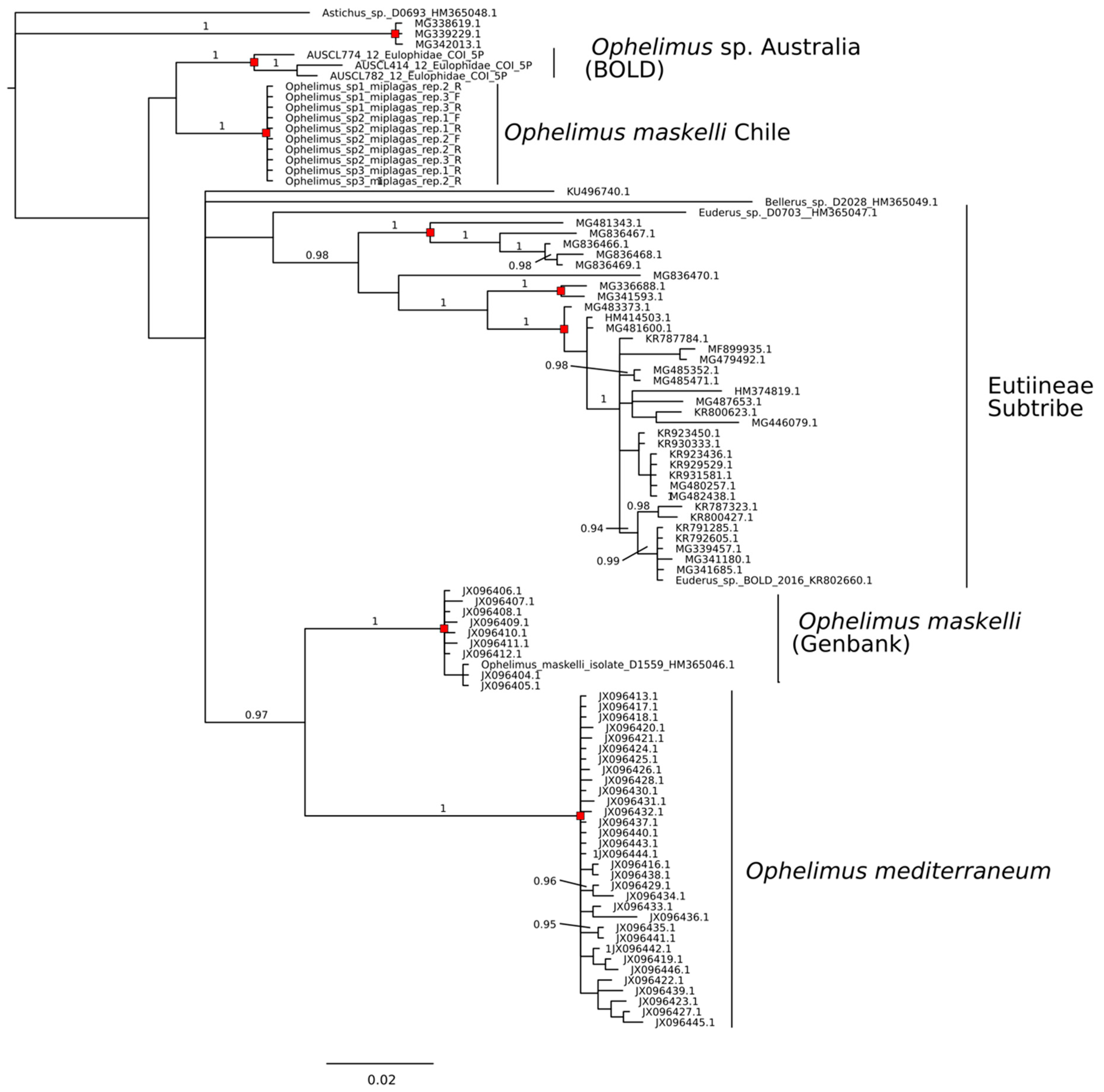
3.3. Sequence Identity COI
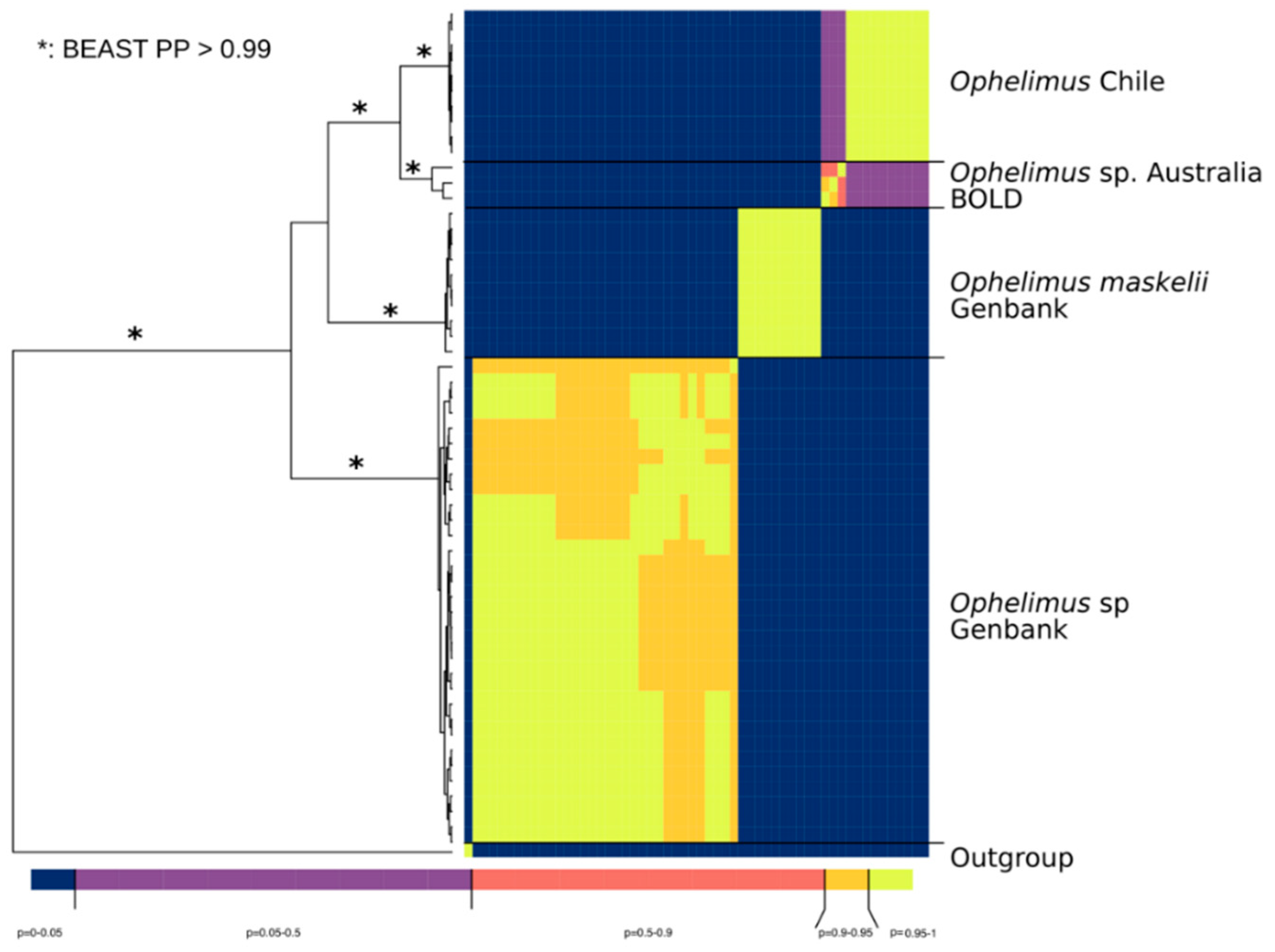
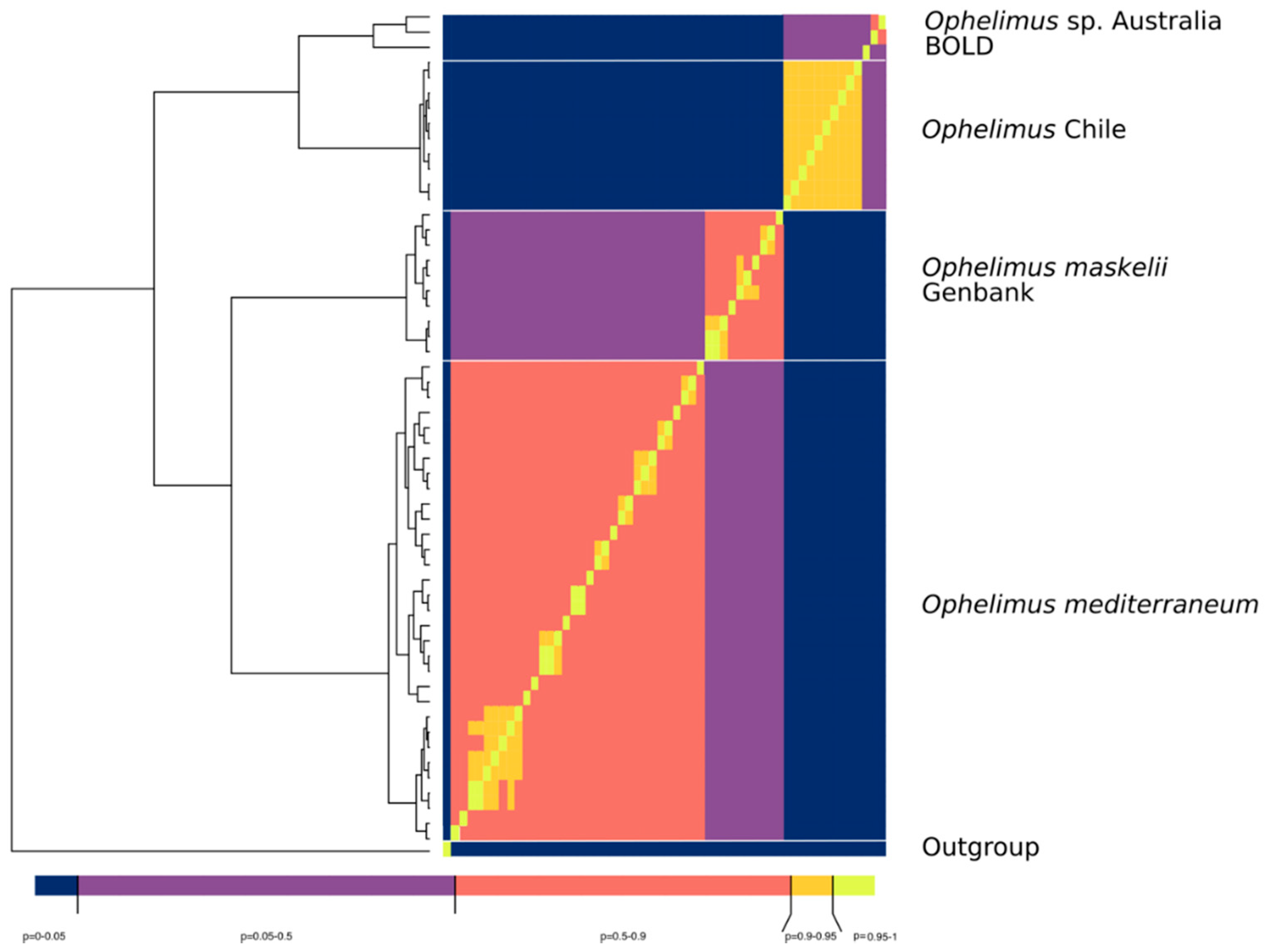
3.4. Sequence Delimitation COI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Withers, T.M. Colonization of eucalyptus in New Zealand by Australian insects. Austral Ecol. 2001, 26, 467–476. [Google Scholar] [CrossRef]
- Hodkinson, I.D. Biocontrol of eucalyptus psyllid Ctenarytaina eucalypti by the Australian parasitoid Psyllaephagus pilosus: A review of current programmes and their success. Biocontrol News Inf. 1999, 20, 129N–134N. [Google Scholar]
- Pujade-Villar, J.; Riba-Flinch, J.M. Dos especies australianas de eulófidos, muy dañinas para Eucalyptus spp. introducidas en el nordeste ibérico (Hymenoptera: Eulophidae). Boletín Soc. Entomol. Aragonesa 2004, 35, 299–301. [Google Scholar]
- Wingfield, M.; Roux, J.; Slippers, B.; Hurley, B.; Garnas, J.; Myburg, A.; Wingfield, B. Established and new technologies reduce increasing pest and pathogen threats to Eucalypt plantations. For. Ecol. Manag. 2013, 301, 35–42. [Google Scholar] [CrossRef]
- Wool, D. Galling aphids: Specialization, biological complexity, and variation. Annu. Rev. Entomol. 2004, 49, 175–192. [Google Scholar] [CrossRef]
- Protasov, A.; La Salle, J.; Blumberg, D.; Brand, D.; Saphir, N.; Assael, F.; Fisher, N.; Mendel, Z. Biology, revised taxonomy and impact on host plants of Ophelimus maskelli, an invasive gall inducer on Eucalyptus sp. in the Mediterranean Area. Phytoparasitica 2007, 35, 50–76. [Google Scholar] [CrossRef]
- La Salle, J.; Arakelian, G.; Garrison, R.; Gates, M. A new species of invasive gall wasp (Hymenoptera: Eulophidae: Tetrastichinae) on blue gum (Eucalyptus globulus) in California. Zootaxa 2009, 2121, 35–43. [Google Scholar] [CrossRef]
- La Salle, J. Biology of gall inducers and evolution of gall induction in chalcidoidea (Hymenoptera: Eulophidae, Eurytomidae, Pteromalidae, Tanaostigmatidae, Torymidae). In Biology, Ecology, and Evolution of Gall-inducing Arthropods; CRC PRESS: Boca Raton, FL, USA, 2005; pp. 507–537. [Google Scholar]
- Austin, A.D.; Yeates, D.K.; Cassis, G.; Fletcher, M.J.; La Salle, J.; Lawrence, J.F.; Mc Quillan, P.B.; Mound, L.A.; Bickel, D.J.; Gullan, P.J.; et al. Insects ‘Down Under’—Diversity, endemism and evolution of the Australian insect fauna: Examples from select orders. Aust. J. Entomol. 2004, 43, 216–234. [Google Scholar] [CrossRef]
- Burks, R.; Heratya, J.; Gebiolab, M.; Hansson, C. Combined molecular and morphological phylogeny of Eulophidae (Hymenoptera: Chalcidoidea), with focus on the subfamily Entedoninae. The Willi Hennig Society. Cladistics 2011, 27, 581–605. [Google Scholar] [CrossRef]
- Gauthier, N.; LaSalle, J.; Quicke, D.L.J.; Godfray, H.C. Phylogeny of Eulophidae (Hymenoptera: Chalcidoidea), with a reclassification of Eulophidae and the recognition that Elasmidae are derived eulophids. Syst. Entomol. 2000, 25, 521–539. [Google Scholar] [CrossRef]
- Boucek, Z. Australasian Chalcidoidea (Hymenoptera). A Biosystematic Revision of Genera of Fourteen Families, with a Reclassifcation of Species; CAB International: Wallingford, UK, 1988. [Google Scholar]
- Noyes, J.S. Universal Chalcidoidea Data Base. 2003. Available online: http://www.nhm.ac.uk/our-science/data/chalcidoids/ (accessed on 14 November 2018).
- La Salle, J.; Polaszek, A.; Noyes, J.S.; Zolnerowich, G. A new whitefly parasitoid (Hymenoptera: Pteromalidae: Eunotinae) with comments on its placement, an implication for classification of Chalcidoidea with reference to the Eriaporinae (Hymenoptera: Aphelinidae). Syst. Entomol. 1997, 22, 131–150. [Google Scholar] [CrossRef]
- Gibson, G.A.P.; Heraty, J.M.; Woolley, J.B. Phylogenetics and classification of Chalcidoidea and Mymarommatoidea—A review of current concepts (Hymenoptera, Apocrita). Zool. Scripta 1999, 28, 87–124. [Google Scholar] [CrossRef]
- Eulophidae, Opheliminae, Género Ophelimus, Ophelimus maskelli. Available online: http://ponent.atspace.org/fauna/ins/fam/eulophidae/ophelimus_oph.htm (accessed on 14 November 2018).
- Arzone, A.; Alma, A. Eulofide galligeno dell’Eucalipto in Italia. Inf. Fitopatol. 2000, 12, 43–46. [Google Scholar]
- Sánchez, I. Descubiertas dos nuevas plagas del eucalipto en España. Quercus 2003, 214, 32–33. [Google Scholar]
- European and Mediterranean Plant Protection Organization. First Report of Two New Eucalyptus Pests in the South of France Ophelimus Maskelli and Leptocybe Invasa; Reporting Service 9.9; EPPO: Paris, France, 2006. [Google Scholar]
- Doganlar, M.; Mendel, Z. First record of the eucalyptus gall wasp, Ophelimus maskelli, (Hymenoptera: Chalcidoidea: Eulophidae: Eulophinae: Ophelimini) and its parasitoid, Closterocerus chamaeleon (Hymenoptera: Chalcidoidea: Eulophidae: Entedoninae), in Turkey. Phytoparasitica 2007, 35, 333–335. [Google Scholar] [CrossRef]
- Branco, M.; Boavida, C.; Durand, N.; Franco, J.; Mendel, Z. Presence of the Eucalyptus gall wasp Ophelimus maskelli and its parasitoid Closterocerus chamaeleon in Portugal: First record, geographic distribution and host preference. Phytoparasitica 2009, 37, 51–54. [Google Scholar] [CrossRef]
- Aquino, D.A.; Hernández, C.M.; Cuello, M.; Andorno, A.V.; Botto, E.N. Primera cita de la Argentina de Ophelimus maskelli (Ashmead) (Hymenoptera: Eulophidae) y su parasitoide, Closterocerus chamaeleon (Girault) (Hymenoptera: Eulophidae). Rev. Soc. Entomológica Argent. 2014, 73, 179–182. [Google Scholar]
- Burks, R.A.; Mottern, J.L.; Waterworth, R.; Paine, T. First report of the Eucalyptus gall wasp, Ophelimus maskelli (Hymenoptera: Eulophidae), an invasive pest on Eucalyptus, from the Western Hemisphere. Zootaxa 2015, 3926, 448–450. [Google Scholar] [CrossRef]
- Paine, T.D.; Lieutier, F. (Eds.) Insects and Diseases of Mediterranean Forest Systems; Springer: Cham, Switzerland, 2016; Chapter 8; pp. 222–238. [Google Scholar]
- Withers, T.M.; Raman, A.; Berry, J.A. Host range and biology of Ophelimus eucalypti (Gahan) (Hym.: Eulophidae), a pest of New Zealand Eucalypts. N. Z. Plant Prot. 2000, 53, 339–344. [Google Scholar]
- Maina, P. New Pest Poses a Threat to Eucalyptus. 2003. Available online: http://www.nationaudio.com/News/Daily(Nation/supplements/horizon/21082003/story21 (accessed on 30 June 2019).
- Kavallieratos, N.G.; Kontodimas, D.C.; Anagnou-Veroniki, M.; Emmanouel, N.G. First record of the gall inducing insect Ophelimus eucalypti (Gahan) (Hymenoptera: Chalcidoidea: Eulophidae) in Greece. Ann. Benaki Phytopathol. Inst. 2006, 20, 125–128. [Google Scholar]
- CABI/EPPO. Ophelimus eucalypti. [Distribution map]. Distribution Maps of Plant Pests; Map 756; Ophelimus maskelli. [Distribution map]. Distribution Maps of Plant Pests; Map 757; CABI: Wallingford, UK, 2011. [Google Scholar]
- Mendel, Z.; Protasov, A.; Blumberg, D.; Brand, D.; Saphir, N.; Madar, Z.; La Salle, J. Release and recovery of parasitoids of the Eucalyptus gall wasp Ophelimus maskelli in Israel. Phytoparasitica 2007, 35, 330–332. [Google Scholar] [CrossRef]
- Wilson, F. Australia as source of beneficial insects for biological control. Tech. Bull. Commonw. Inst. Biol. Control 1963, 3, 1–28. [Google Scholar]
- Lin, X.; Xua, Y.; Jianga, J.; Lavineb, M.; Corley, L. Host quality induces phenotypic plasticity in a wing polyphenic insect. Proc. Natl. Acad. Sci. USA 2018, 115, 7563–7568. [Google Scholar] [CrossRef]
- Simpson, S.; Sword, G.; Lo, N. Polyphenism in insects. Curr. Biol. 2001, 21, R738–R749. [Google Scholar] [CrossRef]
- Cridge, A.; Leask, M.; Duncan, E.; Dearden, P. What do studies of insect polyphenisms tell us about nutritionally—Triggered epigenomic changes and their consequences? Nutrients 2015, 7, 1787–1797. [Google Scholar] [CrossRef]
- Srinivasan, D.; Brisson, J. Aphids: A model for polyphenism and epigenetics. Genet. Res. Int. 2012, 2012, 431531. [Google Scholar] [CrossRef]
- Servicio Agrícola Y Ganadero. Informativo Fitosanitario Forestal AÑO 2, No3, Informativo de la Unidad de Vigilancia y Control de Plagas Forestales y Exóticas Invasoras del Servicio Agrícola y Ganadero; Comité Técnico de la Unidad, Ed.; Servicio Agrícola Y Ganadero: Santiago, Chile, 2006. [Google Scholar]
- Gibson, G.A.P.; Huber, J.T.; Woolley, J.B. Annotated Keys to the Genera of Neartic Chalcidoidea (Hymenoptera); NRC Research Press: Ottawa, ON, Canada, 1997; 794p. [Google Scholar]
- Hebert, P.; Penton, E.; Burns, J.; Janzen, D.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the Neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Hebert, P.; Cywinska, A.; Ball, S.L.; De Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The Ugene Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Benson, D.A.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2014, 42, 32–37. [Google Scholar] [CrossRef]
- Katoh, S. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. Model Finder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mr Bayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Larget, B.; Alfaro, M.E. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol. Biol. Evol. 2004, 21, 1123–1133. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Warren, D.L.; Geneva, A.J.; Lanfear, R. RWTY (R We There Yet): An R package for examining convergence of bayesian phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Kapli, P.; Lutteropp, S.; Zhang, J.; Kobert, K.; Pavlidis, P.; Stamatakis, A.; Flouri, T. Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 2017, 33, 1630–1638. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.; Barraclough, T.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.; Hazell, S.; Kamoun, S.; Sumlin, W.; Vogler, A. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- Reids, N.M.; Carstens, B.C. Phylogenetic estimation error can decrease the accuracy of species delimitation: A Bayesian implementation of the General Mixed Yule Coalescent model. BMC Evol. Biol. 2013, 12, 196. [Google Scholar] [CrossRef] [PubMed]
- Protasov, A.; Blumberg, D.; Brand, D.; La Salle, J.; Mendel, Z. The basis for biological control of the eucalyptus gall wasp Ophelimus maskelli (Ashmead): Taxonomy and biology of the parasitoid Closterocerus chamaeleon (Girault), with information on its establishment in Israel. Biol. Control 2007, 42, 196–206. [Google Scholar] [CrossRef]
- Bain, J. Rhicnopeltella eucalypti Gahan (Hymenoptera: Chalcidoidea: Eulophidae), Blue-gum Chalcid (Forest and Timber Insects in New Zealand, 15). New Zealand Forest Service. 2003. Available online: http://www.forestre search.co.nz/PDF/Ent15Ophelimussp.pdf (accessed on 3 March 2019).
- Borowiec, N.; Thaon, M.; Brancaccio, L.; Warot, S.; Ris, N.; Malausa, J.C. L’eucalyptus menacé par une nouvelle espece d’Ophelimus en France. Ophelimus maskelli, espece répertoriée et contre laquelle il existe un auxiliaire, est accompagnée d’une espece inconnue d’Ophelimus. Phytoma 2012, 656, 42–44. [Google Scholar]
- Gahan, A.B. A List of Phytophagous Chalcidoidea with descriptions of two new species. Rhicnopeltella eucalypti. Entomol. Soc. Wash. 1922, 4, 54–55. [Google Scholar]
- INFOR. Anuario Forestal. In Boletín Estadístico; Instituto Forestal: Santiago, Chile, 2018; pp. 1–19. [Google Scholar]
- Beéche, M. Programa de detección y control del gorgojo del eucalipto, Gonipterus scutellatus (Gyll.) (Coleoptera: Curculionidae). In Proceedings of the XXI Congreso Nacional de Entomología, Arica, Chile, 3–5 November 1999; pp. 33–34. [Google Scholar]
- Molina-Mercader, G.; Angulo, A.; Sanfuentes, E.; Hasbún, R.; Olivares, T.; Castillo-Salazar, M.; Goycoolea, C. Detection and distribution of Ophelimus migdanorum and its possible biocontroller Closterocerus chamaeleon in productive areas of Eucalyptus globulus in Chile. Chil. J. Agric. Res. 2019, 79, 337–346. [Google Scholar] [CrossRef]



| Location | Region | Coordinates S | Coordinates O |
|---|---|---|---|
| Casa Blanca | Valparaíso | 33°29′3.2″ | 71°31′47.8″ |
| Litueche | O′Higgins | 34°04′33.8″ | 71°48′12.2″ |
| Cauquenes | Maule | 36°04′27.9″ | 72°00′31.0″ |
| Chillán | Biobío | 36°39′21.4″ | 72°23′27.2″ |
| Nueva Imperial | La Araucanía | 38°43′18.8″ | 72°54′47.3″ |
| Fresia | Los Lagos | 41°15′36.0″ | 73°31′51.0″ |
| Ophelimus maskelli | Ophelimus migdanorum One Setae | Ophelimus migdanorum Two Setae | Ophelimus migdanorum Three Setae | |
|---|---|---|---|---|
| Central Rib | √ | √ | ||
| Petiole | √ | √ | √ | |
| Leaf Blade | √ | √ | √ | √ |
| Stems | √ | √ |
| Character | Zoom | O. maskelli (um) | O. migdanorum (um) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 Setae | 2 Setae | 3 Setae | ||||||||||||
| Female (n) | Male (n) | Female (n) | Male (n) | Female (n) | Male (n) | |||||||||
| Long of the Adult | 4X | 1.026.1 (*) | 865.5 | (± 26.0) | 923.0 | (± 27.9) | 1.0599 | (± 30.6) | 1.1309 | (±33.1) | 1.1569 | (±28.9) | 1.2217 | (±46.7) |
| Width of the Adult | 4X | 234.1 (*) | 249.8 | (± 06.2) | 250 | (± 06.2) | 276.3 | (± 06.8) | 285.9 | (±07.6) | 295.9 | (±05.2) | 319.8 | (±08.0) |
| Long of the Wing | 10X | 900.4 (*) | 768.5 | (± 19.2) | 788 | (± 10.6) | 875 | (± 17.0) | 900.5 | (±25.7) | 934.0 | (±26.6) | 1.0750 | (±49.5) |
| Width of the Wing | 10X | 306.4 (*) | 347.1 | (± 11.4) | 348 | (± 10.8) | 297.3 | (± 10.5) | 413.2 | (±12.6) | 422.1 | (±19.4) | 494.3 | (±25.2) |
| Submarginal | 10X | 304.0 (*) | 213.5 | (± 06.4) | 226 | (± 04.4) | 241.2 | (± 12.7) | 245.3 | (±09.0) | 245.6 | (±13.3) | 300.1 | (±14.7) |
| Marginal | 10X | 153.2 (*) | 157.0 | (± 10.1) | 176 | (± 07.5) | 190.6 | (± 11.5) | 207.7 | (±12.7) | 220.3 | (±21.8) | 255.7 | (±21.2) |
| Postmarginal | 10X | 210.1 (*) | 209.7 | (± 06.1) | 221 | (± 05.4) | 261 | (± 09.5) | 274.2 | (±11.4) | 283.4 | (±13.3) | 315.0 | (±16.6) |
| Stigma | 10X | 99.4 (*) | 64.5 | (± 02.2) | 64.6 | (± 01.7) | 73.8 | (± 01.9) | 74.7 | (±02.1) | 74.6 | (±01.9) | 90.9 | (±04.1) |
| Uncus | 10X | 0.0214 (*) | 18.7 | (± 00.7) | 19.6 | (± 00.7) | 32.9 | (± 11.9) | 22.4 | (±00.9) | 22.0 | (±00.8) | 24.5 | (±01.6) |
| Width/Long of the Adult | 4.4x (*) | 3.5 | x | 3,7 | x | 3.8 | x | 4.0 | x | 3.9 | x | 3.8 | x | |
| Width/Long of the Wing | 2.9x (*) | 2.2 | x | 2,3 | x | 2.9 | x | 2.2 | x | 2.2 | x | 2.2 | x | |
| Submarginal/Marginal | 2.0x (*) | 1.4 | x | 1.3 | x | 1.3 | x | 1.2 | x | 1.1 | x | 1.2 | x | |
| Marginal/Postmarginal | 0.7x (*) | 0.7 | x | 0.8 | x | 0.7 | x | 0.8 | x | 0.8 | x | 0.8 | x | |
| Abdominal Long | 10X | 512.7 (*) | 409.5 | (±14.8) | 430.1 | (±11.7) | 512.0 | (±15.2) | 547.1 | (±17.1) | 568.7 | (±21.2) | 627.1 | (±19.5) |
| Color | 4X | Clear metal green | Dark metalic green | |||||||||||
| Number of Setaes in SV | 10X | 1 | 1 a 3 | |||||||||||
| Break in Wing Venation | 4X | Yes | No | |||||||||||
| Antennae Color | 10X | Dark yellow | light brown | |||||||||||
| Number of Rings in the Funicle | 10X | 4 | 3 | |||||||||||
| Number of Artejos in Funicle | 10X | 1 | 2 | |||||||||||
| Galls | 4X | Leaf blade | Leaf blade, midrib, secondary rib, petiole, twigs, flower cones | |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ophelimus maskelli | 8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 1 | 0 | 1 | 2 | 4 | 1 |
| Ophelimus nov. sp. 1 SVS | 8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 1 | 0 | 1 | 2 | 3 | 2 |
| Ophelimus nov. sp. 2 SVS | 8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 2 | 0 | 1 | 2 | 3 | 2 |
| Ophelimus nov. sp. 3 SVS | 8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 3 | 0 | 1 | 2 | 3 | 2 |
| Entiinae | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | ||
| Astichus n. sp | 8 | 3 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 4 | 0 | 3 | 0 | 0 | 2 | ||
| A.mirissimus | 8 | 3 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 4 | 0 | 3 | 0 | 0 | 2 | ||
| Bellerus sp. | 8 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 3 | 0 | 0 | 2 | ||
| Euderus sp. | 8 | 3 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 3 | 0 | 0 | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molina-Mercader, G.; Angulo, A.O.; Olivares, T.S.; Sanfuentes, E.; Castillo-Salazar, M.; Rojas, E.; Toro-Núñez, O.; Benítez, H.A.; Hasbún, R. Ophelimus migdanorum Molina-Mercader sp. nov. (Hymenoptera: Eulophidae): Application of Integrative Taxonomy for Disentangling a Polyphenism Case in Eucalyptus globulus Labill Forest in Chile. Forests 2019, 10, 720. https://doi.org/10.3390/f10090720
Molina-Mercader G, Angulo AO, Olivares TS, Sanfuentes E, Castillo-Salazar M, Rojas E, Toro-Núñez O, Benítez HA, Hasbún R. Ophelimus migdanorum Molina-Mercader sp. nov. (Hymenoptera: Eulophidae): Application of Integrative Taxonomy for Disentangling a Polyphenism Case in Eucalyptus globulus Labill Forest in Chile. Forests. 2019; 10(9):720. https://doi.org/10.3390/f10090720
Chicago/Turabian StyleMolina-Mercader, Gloria, Andrés O. Angulo, Tania S. Olivares, Eugenio Sanfuentes, Miguel Castillo-Salazar, Eladio Rojas, Oscar Toro-Núñez, Hugo A. Benítez, and Rodrigo Hasbún. 2019. "Ophelimus migdanorum Molina-Mercader sp. nov. (Hymenoptera: Eulophidae): Application of Integrative Taxonomy for Disentangling a Polyphenism Case in Eucalyptus globulus Labill Forest in Chile" Forests 10, no. 9: 720. https://doi.org/10.3390/f10090720
APA StyleMolina-Mercader, G., Angulo, A. O., Olivares, T. S., Sanfuentes, E., Castillo-Salazar, M., Rojas, E., Toro-Núñez, O., Benítez, H. A., & Hasbún, R. (2019). Ophelimus migdanorum Molina-Mercader sp. nov. (Hymenoptera: Eulophidae): Application of Integrative Taxonomy for Disentangling a Polyphenism Case in Eucalyptus globulus Labill Forest in Chile. Forests, 10(9), 720. https://doi.org/10.3390/f10090720





