Characterization of Two AGL6–Like Genes from a Chinese Endemic Woody Tree, Manglietia patungensis (Magnoliaceae) Provides Insight into Perianth Development and Evolution in Basal Angiosperms
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
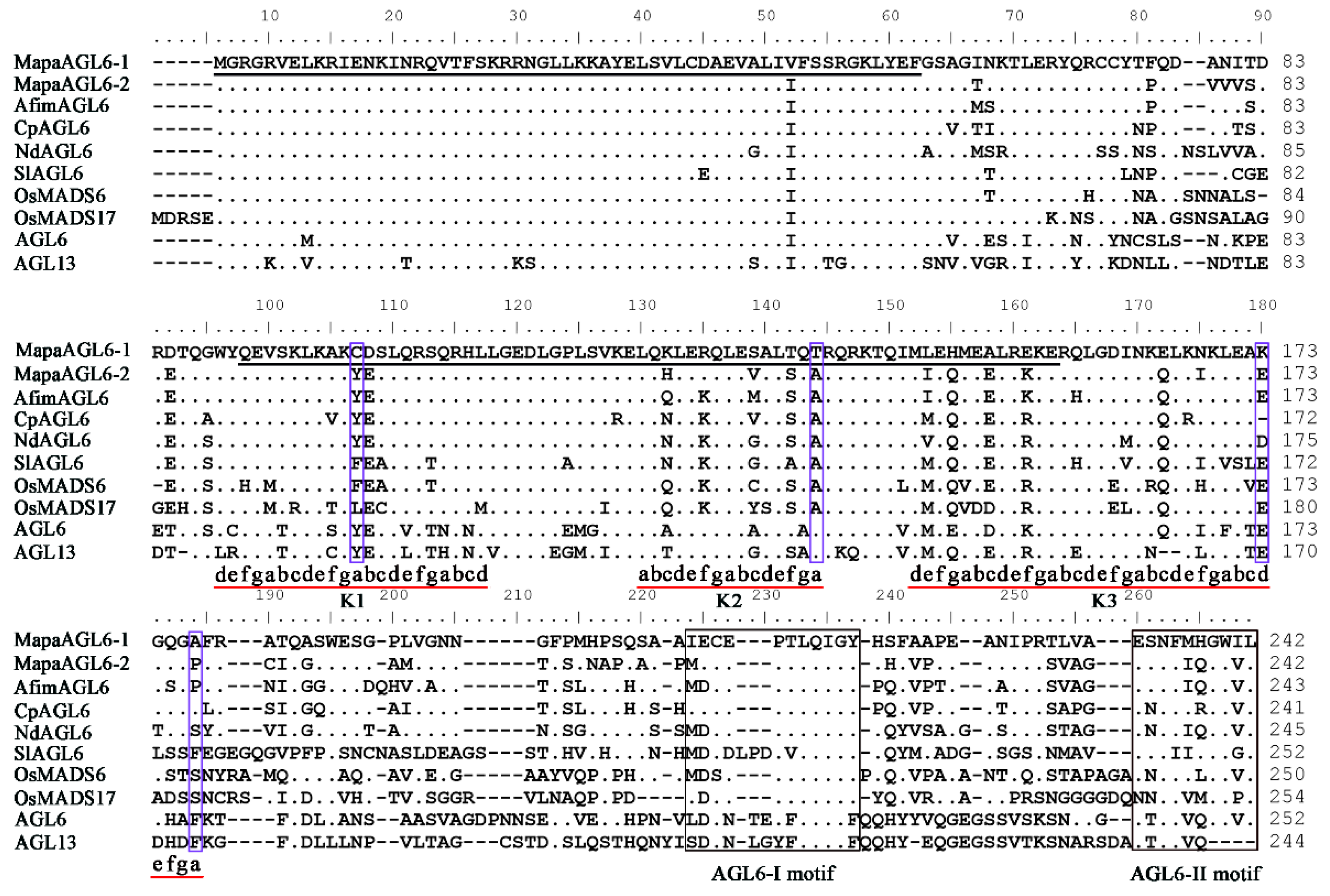
2.2. Isolation and Identification of AGL6 Homologs from M. patungensis
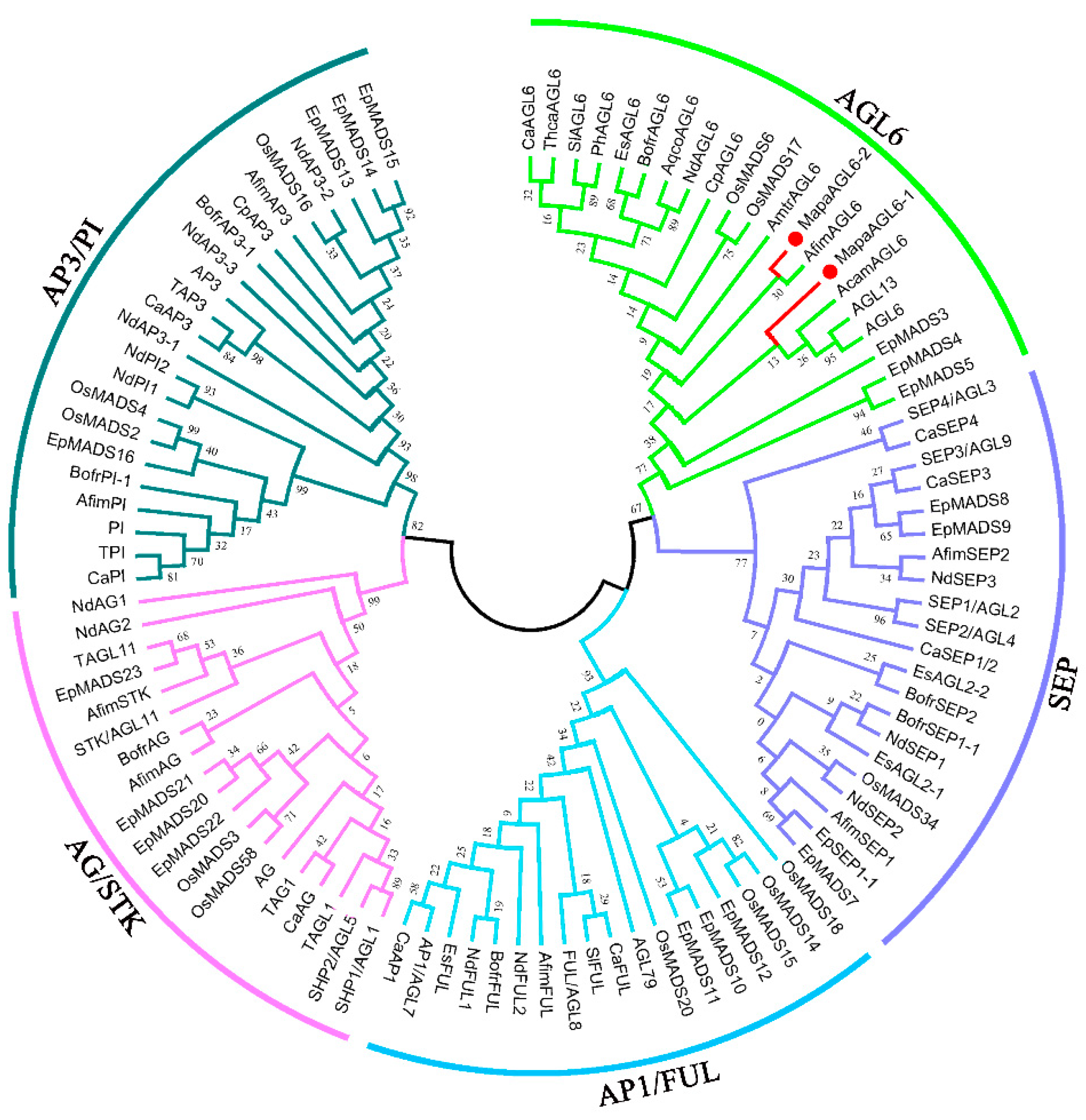
2.3. Sequence Alignments and Phylogenetic Analysis
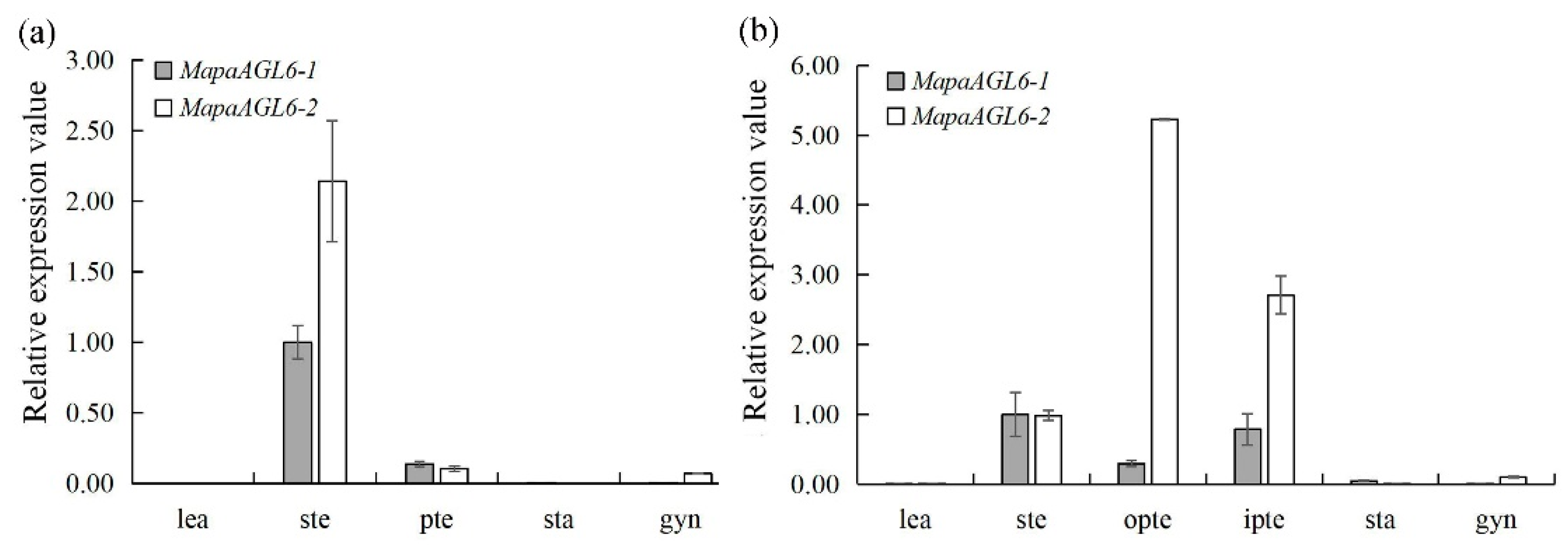
2.4. Expression Analysis of MapaAGL6-1 and MapaAGL6-2
2.5. Vector Construction and Arabidopsis Transformation
2.6. Statistical Analysis
3. Results
3.1. Cloning and Sequence Alignments of MapaAGL6-1 and MapaAGL6-2
3.2. Expression Analyses of MapaAGL6-1 and MapaAGL6-2 in M. patungensis
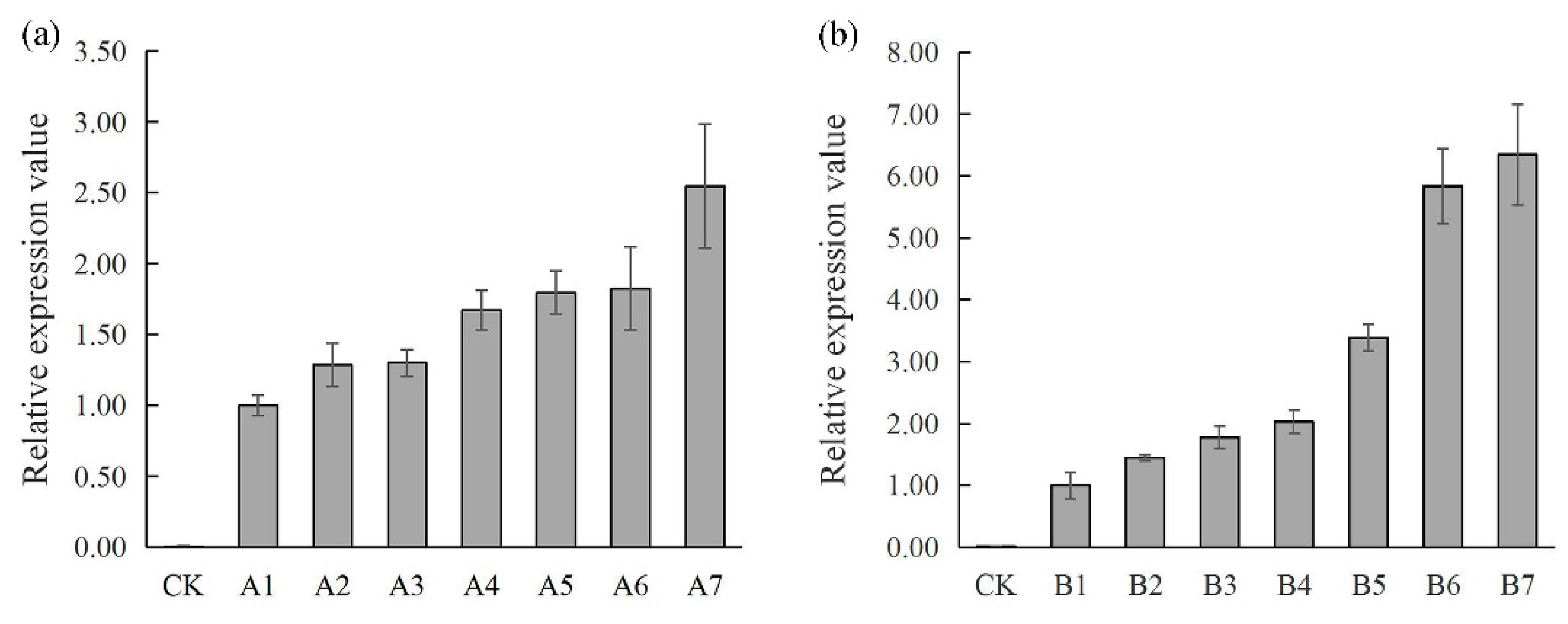
3.3. Ectopic Expression of MapaAGL6-1 and MapaAGL6-2 in ap1-10 Arabidopsis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| LSD | Least Significant Difference |
| ORF | Open Reading Frame |
| PCR | Polymerase Chain Reaction |
| qPCR | Quantitative real-time PCR |
References
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Fletcher, J.C. Molecular mechanisms of flower development: An armchair guide. Nat. Rev. Genet. 2005, 6, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Zahn, L.M.; Feng, B.M.; Ma, H. Beyond the ABC-Model: Regulation of floral homeotic genes. Adv. Bot. Res. 2006, 44, 164–196. [Google Scholar]
- Davies, B.; Cartolano, M.; Schwarz-Sommer, Z. Flower development: The Antirrhinum perspective. Adv. Bot. Res. 2006, 44, 279–320. [Google Scholar]
- Soltis, P.S.; Soltis, D.E.; Kim, S.; Chanderbali, A.; Buzgo, M. Expression of floral regulators in basal angiosperms and the origin and evolution of ABC function. Adv. Bot. Res. 2006, 44, 483–506. [Google Scholar]
- Wang, P.; Liao, H.; Zhang, W.; Yu, X.; Zhang, R.; Shan, H.; Duan, X.; Yao, X.; Kong, H. Flexibility in the structure of spiral flowers and its underlying mechanisms. Nat. Plants 2015, 2, 15188. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Koh, J.; Yoo, M.J.; Kong, H.; Hu, Y.; Ma, H.; Soltis, P.S.; Soltis, D.E. Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. 2005, 43, 724–744. [Google Scholar] [CrossRef] [PubMed]
- Pabón-Mora, N.; Suárez-Baron, H.; Ambrose, B.A.; González, F. Flower Development and Perianth Identity Candidate Genes in the Basal Angiosperm Aristolochia fimbriata (Piperales: Aristolochiaceae). Front. Plant Sci. 2015, 6, 1095. [Google Scholar] [CrossRef]
- Peréz-Mesa, P.; Suárez-Baron, H.; Ambrose, B.A.; González, F.; Pabón-Mora, N. Floral MADS-box protein interactions in the early diverging angiosperm Aristolochia fimbriata Cham. (Aristolochiaceae: Piperales). Evol. Dev. 2019, 21, 96–110. [Google Scholar] [CrossRef]
- Li, H.; Liang, W.; Jia, R.; Yin, C.; Zong, J.; Kong, H.; Zhang, D. The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 2010, 20, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Liang, W.; An, G.; Zhang, D. OsMADS6 Controls Flower Development by Activating Rice FACTOR OF DNA METHYLATION LIKE1. Plant Physiol. 2018, 177, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Dirks-Mulder, A.; Butôt, R.; van Schaik, P.; Wijnands, J.W.; van den Berg, R.; Krol, L.; Doebar, S.; van Kooperen, K.; de Boer, H.; Kramer, E.M.; et al. Exploring the evolutionary origin of floral organs of Erycina pusilla, an emerging orchid model system. BMC Evol. Biol. 2017, 17, 89. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Chen, Y.; Chen, L.; Fu, X.; Zhao, K.; Zhang, J.; Sun, C. B and E MADS-box genes determine the perianth formation in Cymbidium goeringii Rchb.f. Physiol. Plant. 2018, 162, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, G.; Guo, X.; Lu, Y.; Zhang, J.; Hu, J.; Tian, S.; Hu, Z. Silencing SlAGL6, a tomato AGAMOUS-LIKE6 lineage gene, generates fused sepal and green petal. Plant Cell Rep. 2017, 36, 959–969. [Google Scholar] [CrossRef] [PubMed]
- Rijpkema, A.S.; Zethof, J.; Gerats, T.; Vandenbussche, M. The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J. 2009, 60, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sauquet, H.; von Balthazar, M.; Magallón, S.; Doyle, J.A.; Endress, P.K.; Bailes, E.J.; Barroso de Morais, E.; Bull-Hereñu, K.; Carrive, L.; Chartier, M.; et al. The ancestral flower of angiosperms and its early diversification. Nat. Commun. 2017, 8, 16047. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Soltis, P.S.; Soltis, D.E. AGL6-like MADS-box genes are sister to AGL2-like MADS-box genes. J. Plant Biol. 2013, 56, 315–325. [Google Scholar] [CrossRef]
- Wang, B.G.; Zhang, Q.; Wang, L.G.; Duan, K.; Pan, A.H.; Tang, X.M.; Sui, S.Z.; Li, M.Y. The AGL6-like Gene CpAGL6, a Potential Regulator of Floral Time and Organ Identity in Wintersweet (Chimonanthus praecox). J. Plant Growth Regul. 2011, 30, 343–352. [Google Scholar] [CrossRef]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.; Salts, Y.; Barg, R. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 2017, 15, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, S.; Kimizu, M.; Sugita, M.; Miyao, A.; Hirochika, H.; Uchida, E.; Nagato, Y.; Yoshida, H. MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 2009, 21, 3008–3025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, Z.X.; Ma, J.; Song, Y.; Chen, F.J. Alternative splicing of the AGAMOUS orthologous gene in double flower of Magnolia stellata (Magnoliaceae). Plant Sci. 2015, 241, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Y.; Fang, Z.W.; Li, X.F.; Liu, Z.X. Isolation and Characterization of the C-class MADS-box Gene from the Distylous Pseudo-cereal Fagopyrum esculentum. J. Plant Biol. 2017, 60, 189–198. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Krizek, B.A.; Meyerowitz, E.M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 1996, 93, 4793–4798. [Google Scholar] [CrossRef]
- Yang, Y.; Jack, T. Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol. Biol. 2004, 55, 45–59. [Google Scholar] [CrossRef]
- Sun, W.; Huang, W.; Li, Z.; Song, C.; Liu, D.; Liu, Y.; Hayward, A.; Liu, Y.; Huang, H.; Wang, Y. Functional and evolutionary analysis of the AP1/SEP/AGL6 superclade of MADS-box genes in the basal eudicot Epimedium sagittatum. Ann. Bot. 2014, 113, 653–668. [Google Scholar] [CrossRef]
- Endress, P.K. Angiosperm Floral Evolution: Morphological Developmental Framework. Adv. Bot. Res. 2006, 44, 1–61. [Google Scholar]
- Monniaux, M.; Vandenbussche, M. How to Evolve a Perianth: A Review of Cadastral Mechanisms for Perianth Identity. Front. Plant. Sci. 2018, 9, 1573. [Google Scholar] [CrossRef]
- Endress, P.K. Evolutionary diversification of the flowers in angiosperms. Am. J. Bot. 2011, 98, 370–396. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Zhang, K.; Li, L.; Fei, Y.; Chen, F. Characterization of Two AGL6–Like Genes from a Chinese Endemic Woody Tree, Manglietia patungensis (Magnoliaceae) Provides Insight into Perianth Development and Evolution in Basal Angiosperms. Forests 2019, 10, 669. https://doi.org/10.3390/f10080669
Liu Z, Zhang K, Li L, Fei Y, Chen F. Characterization of Two AGL6–Like Genes from a Chinese Endemic Woody Tree, Manglietia patungensis (Magnoliaceae) Provides Insight into Perianth Development and Evolution in Basal Angiosperms. Forests. 2019; 10(8):669. https://doi.org/10.3390/f10080669
Chicago/Turabian StyleLiu, Zhixiong, Kebin Zhang, Laiyun Li, Yue Fei, and Faju Chen. 2019. "Characterization of Two AGL6–Like Genes from a Chinese Endemic Woody Tree, Manglietia patungensis (Magnoliaceae) Provides Insight into Perianth Development and Evolution in Basal Angiosperms" Forests 10, no. 8: 669. https://doi.org/10.3390/f10080669
APA StyleLiu, Z., Zhang, K., Li, L., Fei, Y., & Chen, F. (2019). Characterization of Two AGL6–Like Genes from a Chinese Endemic Woody Tree, Manglietia patungensis (Magnoliaceae) Provides Insight into Perianth Development and Evolution in Basal Angiosperms. Forests, 10(8), 669. https://doi.org/10.3390/f10080669



