Selection of Suitable Reference Genes in Pinus massoniana Lamb. Under Different Abiotic Stresses for qPCR Normalization
Abstract
1. Introduction
2. Methods
2.1. Plant Materials and Treatments
2.2. RNA Isolation and cDNA Reverse Transcription
2.3. Candidate Reference Gene Selection, Primer Design, and Gene Cloning
2.4. Quantitative Real-Time PCR Assay
2.5. Data Analysis
2.6. Validation of Reference Genes
3. Results
3.1. Selection of Reference Genes, Amplification Specificity, and Efficiency
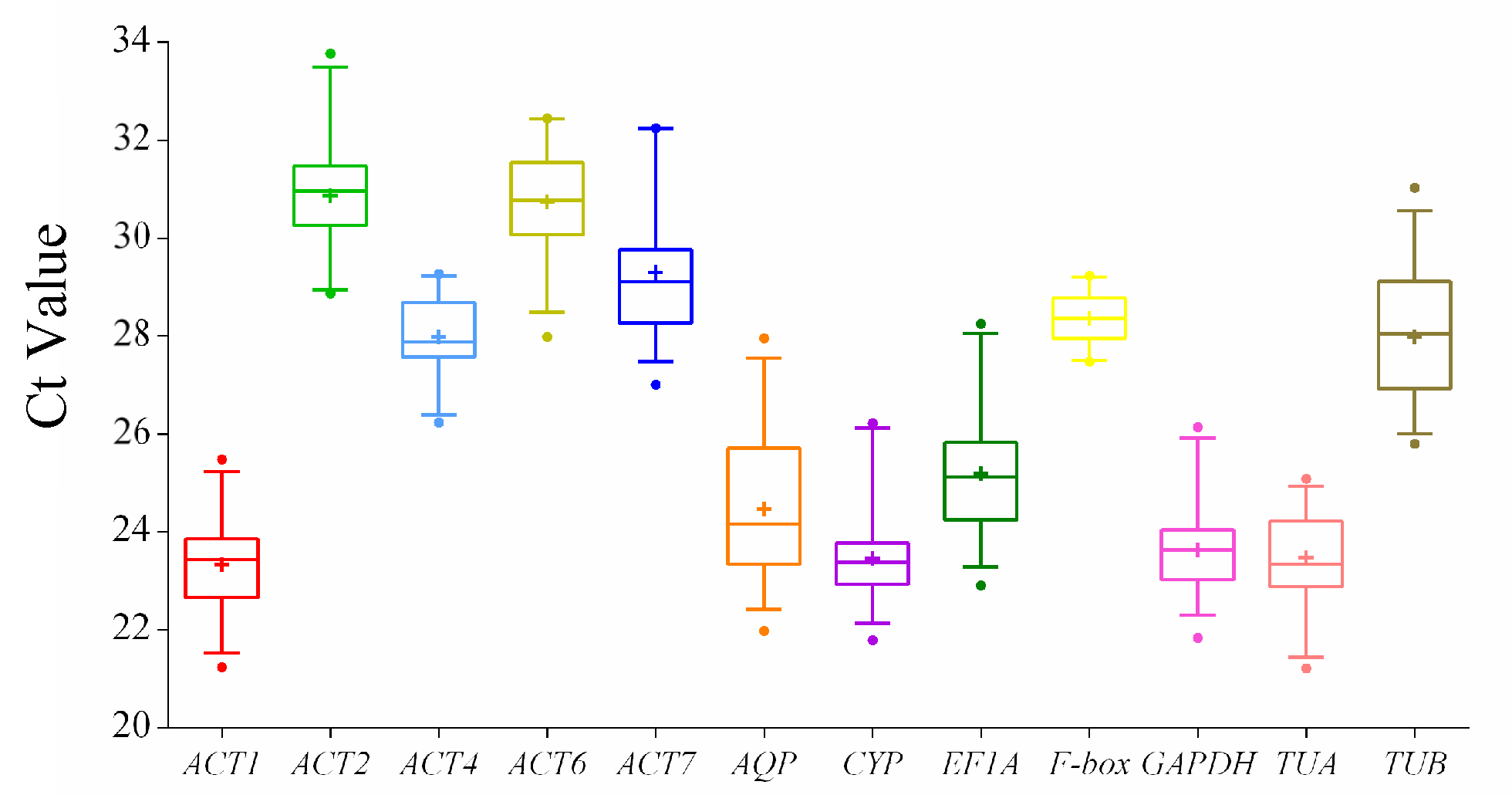
3.2. Expression Profiles of the Reference Genes
3.3. Expression Stability Analysis of the Reference Genes
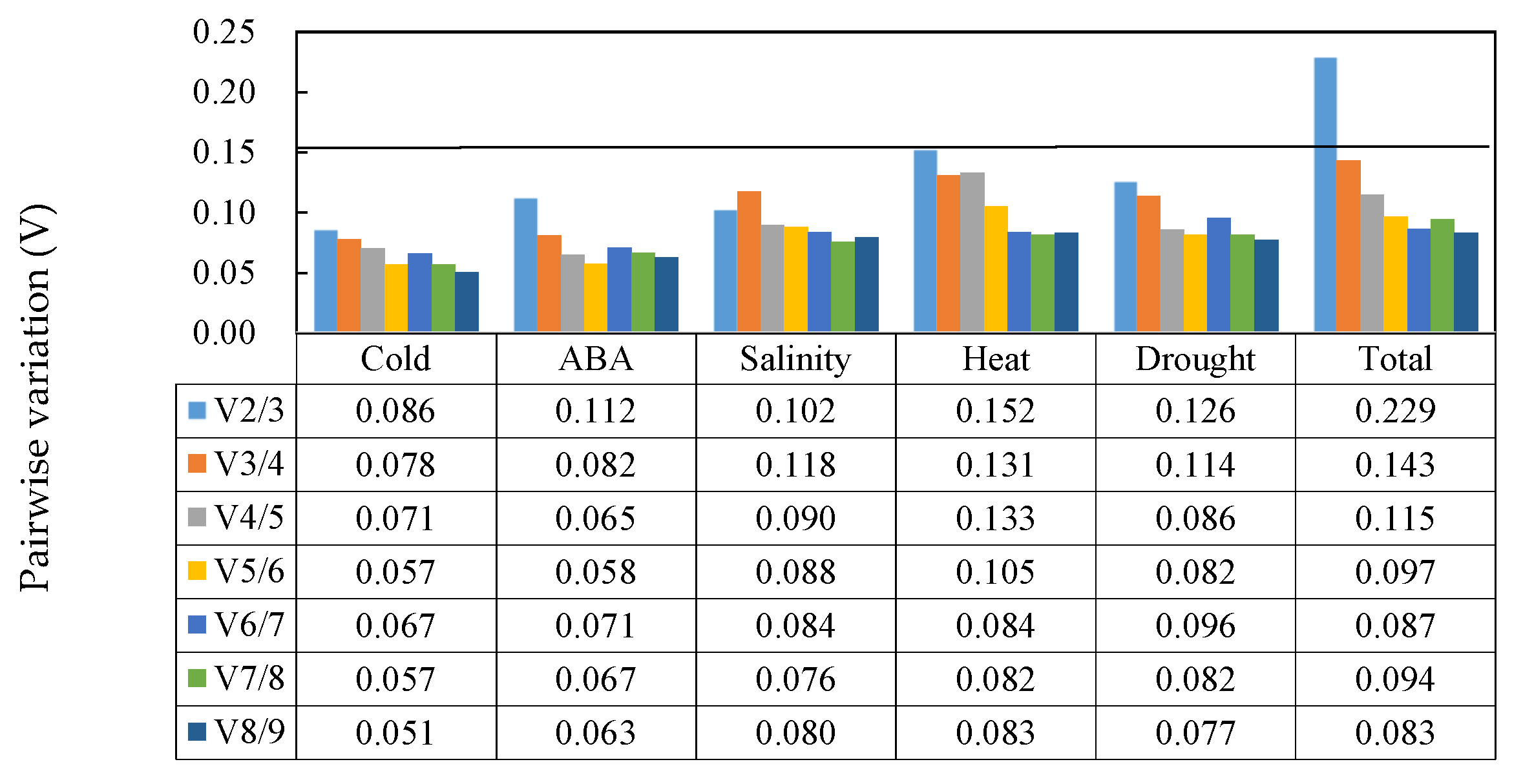
3.4. geNorm Analysis
3.5. NormFinder Analysis
3.6. BestKeeper Analysis
3.7. RefFinder Analysis
3.8. Reference Gene Validation
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
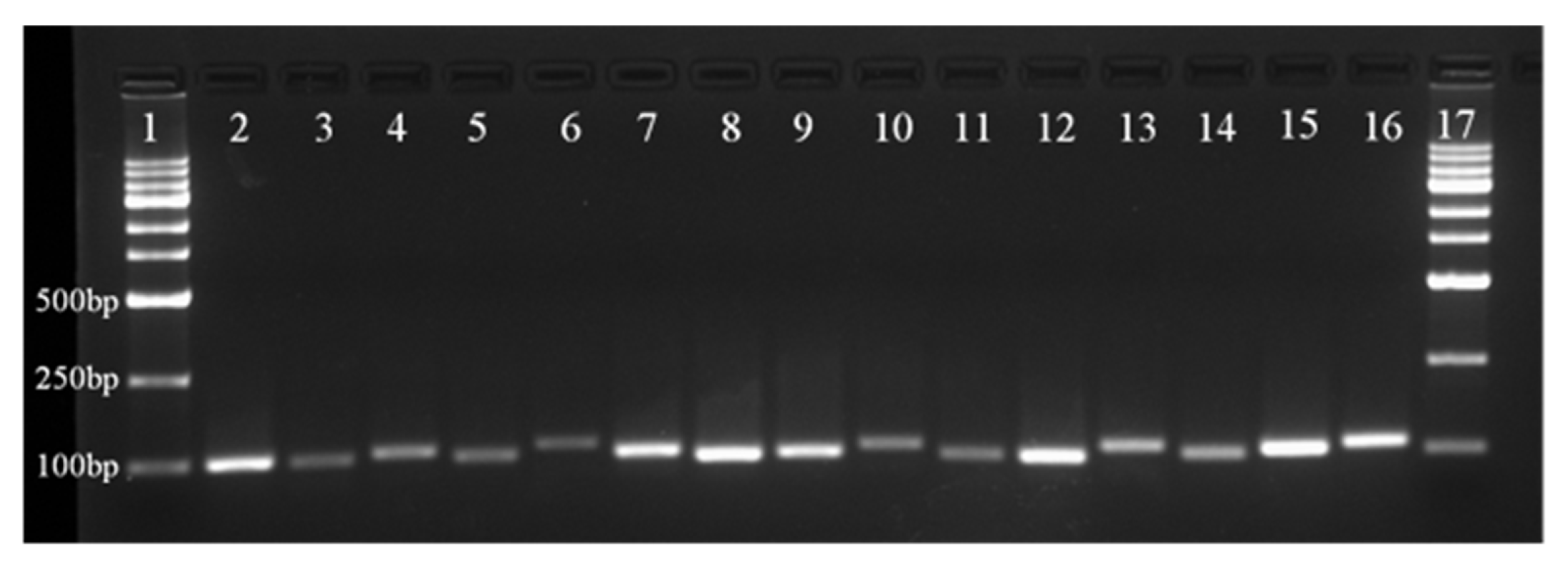
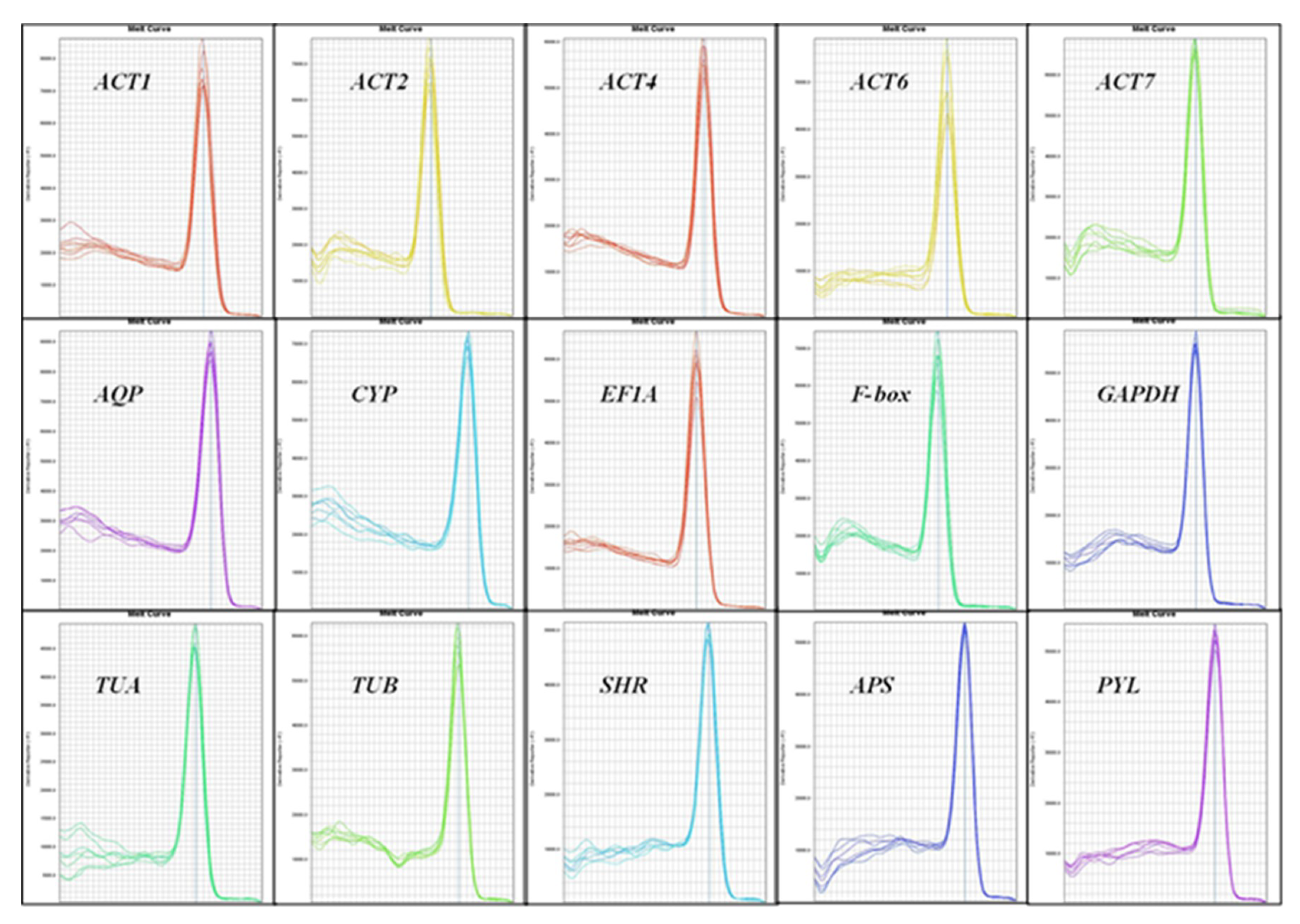
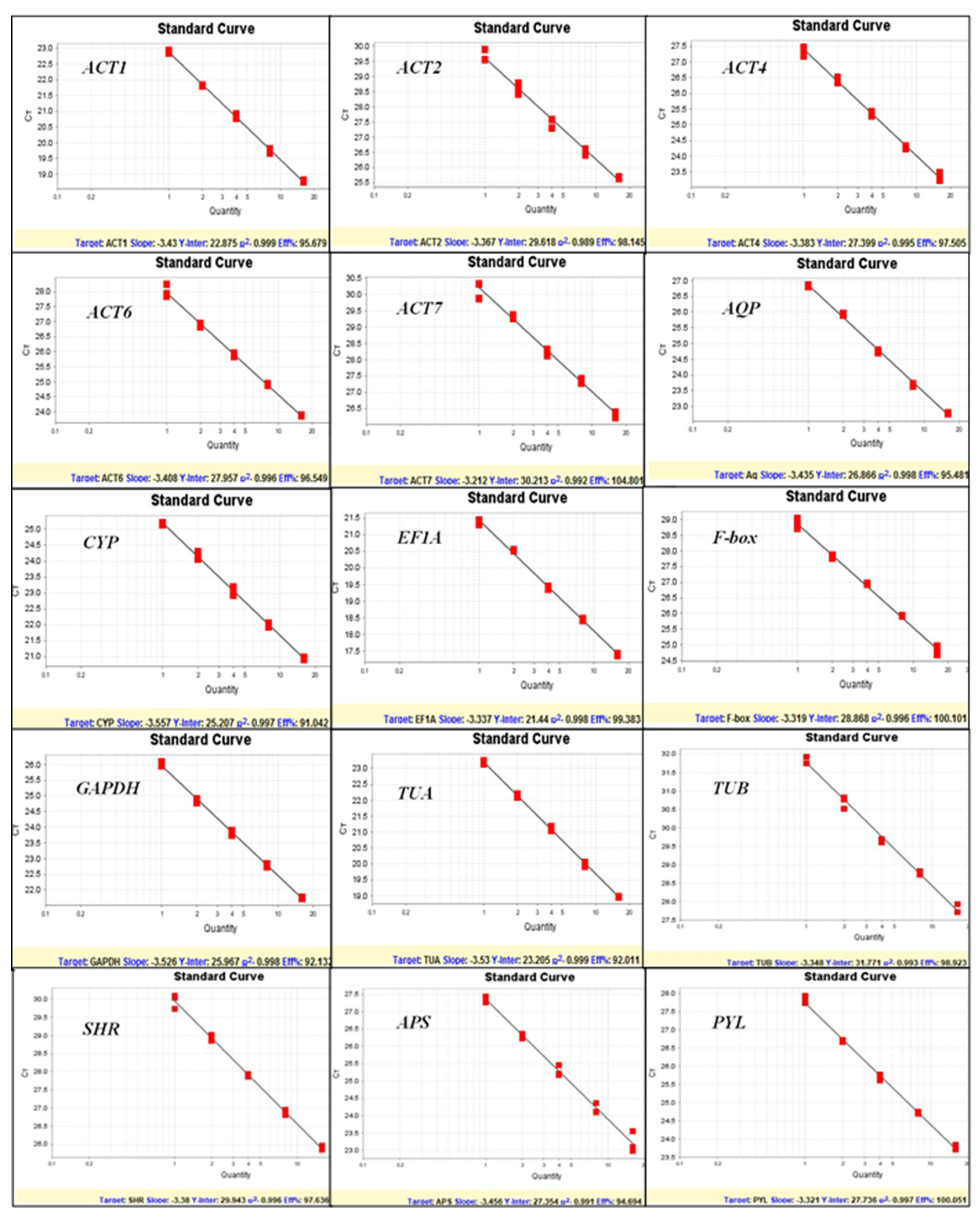

| ACT-1 | ACT-2 | ACT-4 | ACT-6 | ACT-7 | AQP | CYP | EF1A | F-box | GAPDH | TUA | TUB | SHR | APS | PYL | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 0h | 23.93 | 30.57 | 28.69 | 31.83 | 29.96 | 24.71 | 23.93 | 22.77 | 28.81 | 23.84 | 23.87 | 27.82 | 30.40 | 27.88 | 27.86 |
| 23.83 | 30.49 | 28.79 | 31.63 | 29.76 | 24.92 | 23.81 | 22.90 | 28.40 | 24.08 | 23.99 | 27.93 | 30.27 | 27.87 | 27.62 | ||
| 23.78 | 30.86 | 28.54 | 31.72 | 29.80 | 22.84 | 23.79 | 24.90 | 28.38 | 23.87 | 23.80 | 28.52 | 30.17 | 27.72 | 27.54 | ||
| ABA | 0.5h | 22.55 | 29.77 | 27.96 | 29.97 | 27.74 | 23.50 | 22.84 | 23.86 | 28.85 | 23.93 | 23.41 | 25.76 | 29.62 | 24.55 | 27.83 |
| 22.88 | 30.61 | 28.16 | 30.29 | 28.19 | 23.80 | 23.31 | 24.40 | 28.79 | 24.34 | 23.54 | 26.84 | 28.84 | 25.86 | 27.48 | ||
| 22.94 | 30.39 | 27.49 | 30.46 | 28.38 | 23.93 | 23.44 | 24.41 | 28.31 | 23.63 | 22.96 | 26.53 | 28.79 | 24.74 | 27.02 | ||
| 3h | 23.22 | 31.19 | 26.75 | 30.49 | 28.27 | 24.27 | 23.23 | 24.57 | 28.07 | 23.29 | 22.89 | 26.56 | 28.90 | 24.36 | 27.90 | |
| 22.90 | 28.40 | 25.85 | 30.81 | 28.63 | 23.01 | 22.76 | 24.29 | 27.94 | 22.91 | 22.86 | 26.42 | 28.36 | 23.70 | 27.58 | ||
| 23.18 | 28.87 | 26.10 | 30.68 | 28.86 | 23.29 | 22.98 | 24.51 | 28.01 | 23.07 | 22.88 | 26.52 | 28.55 | 23.90 | 27.72 | ||
| 6h | 22.76 | 28.47 | 26.27 | 30.46 | 28.41 | 23.25 | 22.66 | 23.67 | 28.16 | 22.40 | 21.94 | 26.37 | 28.91 | 23.65 | 26.95 | |
| 22.84 | 28.89 | 26.83 | 30.34 | 28.83 | 23.12 | 22.58 | 24.14 | 28.66 | 23.69 | 22.64 | 26.83 | 29.30 | 24.07 | 27.61 | ||
| 21.84 | 29.26 | 26.66 | 29.39 | 27.08 | 22.30 | 21.80 | 22.94 | 28.24 | 22.92 | 22.29 | 25.78 | 29.02 | 23.88 | 27.26 | ||
| 12h | 21.54 | 29.02 | 26.57 | 28.84 | 26.93 | 21.89 | 21.64 | 22.72 | 27.86 | 22.86 | 21.91 | 25.85 | 28.68 | 23.81 | 26.92 | |
| 21.89 | 29.04 | 26.01 | 29.04 | 27.09 | 22.07 | 21.91 | 23.07 | 27.52 | 22.43 | 21.34 | 25.83 | 27.90 | 22.80 | 26.40 | ||
| 21.70 | 28.92 | 26.86 | 28.95 | 27.01 | 21.96 | 21.82 | 22.92 | 27.14 | 22.49 | 21.54 | 25.73 | 28.25 | 23.06 | 26.72 | ||
| 24h | 22.85 | 29.80 | 27.66 | 30.75 | 27.96 | 22.68 | 22.92 | 24.05 | 27.85 | 23.19 | 23.44 | 26.83 | 29.01 | 24.89 | 26.87 | |
| 22.65 | 29.62 | 27.85 | 30.61 | 27.88 | 22.36 | 22.41 | 23.97 | 27.76 | 23.52 | 23.62 | 26.57 | 28.97 | 25.42 | 27.00 | ||
| 23.60 | 30.47 | 27.18 | 31.50 | 28.91 | 23.48 | 23.55 | 25.05 | 27.22 | 22.78 | 22.96 | 27.38 | 28.33 | 24.75 | 26.39 | ||
| 48h | 23.00 | 31.88 | 27.99 | 31.76 | 29.23 | 23.94 | 23.76 | 24.95 | 27.85 | 23.66 | 24.51 | 29.42 | 29.12 | 28.27 | 26.80 | |
| 23.50 | 31.65 | 27.44 | 31.98 | 29.79 | 24.19 | 23.90 | 25.65 | 27.04 | 23.41 | 23.84 | 29.82 | 28.83 | 26.09 | 26.20 | ||
| 23.05 | 31.40 | 27.78 | 31.53 | 29.18 | 23.70 | 23.54 | 25.10 | 27.53 | 23.55 | 24.21 | 29.39 | 29.04 | 26.90 | 26.56 | ||
| Cold | 0.5h | 22.96 | 29.99 | 28.37 | 31.67 | 30.30 | 22.90 | 23.45 | 24.76 | 27.88 | 22.89 | 24.56 | 27.95 | 29.73 | 28.80 | 28.15 |
| 23.59 | 30.80 | 28.68 | 30.38 | 29.01 | 23.55 | 22.34 | 25.20 | 28.52 | 23.80 | 23.17 | 27.93 | 28.55 | 26.77 | 26.79 | ||
| 23.74 | 30.93 | 28.95 | 30.73 | 29.49 | 23.65 | 22.64 | 25.41 | 28.61 | 23.89 | 23.66 | 28.23 | 28.96 | 27.42 | 27.26 | ||
| 3h | 24.89 | 32.25 | 29.87 | 31.72 | 30.25 | 23.70 | 22.86 | 25.71 | 28.71 | 24.88 | 24.95 | 29.87 | 29.01 | 29.99 | 27.58 | |
| 23.86 | 30.77 | 28.68 | 31.85 | 30.40 | 22.86 | 23.02 | 24.92 | 27.62 | 23.65 | 24.82 | 28.89 | 28.89 | 29.35 | 27.86 | ||
| 24.46 | 31.40 | 29.25 | 31.68 | 30.34 | 23.45 | 22.88 | 25.44 | 28.16 | 24.33 | 24.73 | 29.48 | 28.94 | 29.40 | 27.68 | ||
| 6h | 23.89 | 30.88 | 28.30 | 31.79 | 29.53 | 23.30 | 23.82 | 25.83 | 28.91 | 23.88 | 23.92 | 26.63 | 29.86 | 25.91 | 28.37 | |
| 23.36 | 30.64 | 27.69 | 31.75 | 29.94 | 22.77 | 23.46 | 25.40 | 28.39 | 23.55 | 23.81 | 27.83 | 29.47 | 27.88 | 27.85 | ||
| 23.55 | 30.48 | 27.87 | 31.70 | 29.45 | 22.92 | 23.34 | 25.49 | 28.48 | 23.62 | 23.71 | 27.02 | 29.36 | 26.21 | 27.66 | ||
| 12h | 23.90 | 31.01 | 28.85 | 31.91 | 30.57 | 23.88 | 23.91 | 26.30 | 28.08 | 24.38 | 24.44 | 27.93 | 29.91 | 27.87 | 27.77 | |
| 23.69 | 30.51 | 28.40 | 31.32 | 30.00 | 23.82 | 23.25 | 25.90 | 27.61 | 23.76 | 23.84 | 27.40 | 28.88 | 26.40 | 27.51 | ||
| 23.68 | 31.63 | 28.65 | 31.40 | 29.84 | 23.66 | 23.15 | 26.01 | 27.74 | 23.90 | 23.77 | 27.34 | 28.97 | 26.67 | 27.34 | ||
| 24h | 24.85 | 32.94 | 29.88 | 32.85 | 30.79 | 25.30 | 24.13 | 27.89 | 29.89 | 25.47 | 24.94 | 29.83 | 29.79 | 28.87 | 28.94 | |
| 23.65 | 31.64 | 28.55 | 31.67 | 30.47 | 23.88 | 23.17 | 27.07 | 28.75 | 24.87 | 23.76 | 28.86 | 28.84 | 26.22 | 27.65 | ||
| 23.75 | 31.36 | 28.66 | 32.10 | 30.51 | 23.90 | 23.53 | 26.99 | 28.87 | 24.87 | 24.13 | 28.85 | 29.31 | 27.06 | 28.04 | ||
| 48h | 23.94 | 31.64 | 29.35 | 30.52 | 29.73 | 23.89 | 22.75 | 26.88 | 28.82 | 23.92 | 23.89 | 28.99 | 28.86 | 29.82 | 27.80 | |
| 23.59 | 31.08 | 28.66 | 30.64 | 29.17 | 23.82 | 22.63 | 26.39 | 28.42 | 23.45 | 23.67 | 28.72 | 28.39 | 29.77 | 27.72 | ||
| 23.65 | 31.71 | 28.93 | 30.62 | 29.44 | 23.76 | 22.54 | 26.65 | 28.53 | 23.65 | 23.61 | 28.77 | 28.61 | 29.62 | 27.69 | ||
| Salinity | 0.5h | 22.62 | 30.31 | 26.61 | 30.82 | 27.58 | 22.89 | 22.66 | 23.73 | 27.86 | 23.66 | 22.84 | 25.37 | 28.65 | 24.67 | 26.60 |
| 22.82 | 30.54 | 27.84 | 30.90 | 28.61 | 23.23 | 23.01 | 24.01 | 28.21 | 24.53 | 23.15 | 27.10 | 28.82 | 25.39 | 26.47 | ||
| 22.60 | 30.08 | 26.92 | 30.58 | 27.94 | 22.70 | 22.54 | 23.50 | 27.75 | 23.92 | 22.88 | 25.94 | 28.65 | 24.90 | 26.33 | ||
| 3h | 21.73 | 30.04 | 26.68 | 29.81 | 28.40 | 22.89 | 22.89 | 24.36 | 27.81 | 22.02 | 21.62 | 26.54 | 27.76 | 23.76 | 26.35 | |
| 20.91 | 29.22 | 26.33 | 29.09 | 27.87 | 22.32 | 22.46 | 23.88 | 27.28 | 21.76 | 20.89 | 25.91 | 26.93 | 22.40 | 25.78 | ||
| 21.08 | 29.93 | 26.81 | 29.39 | 28.52 | 22.87 | 22.94 | 24.46 | 27.67 | 21.71 | 21.12 | 26.36 | 27.14 | 22.82 | 25.87 | ||
| 6h | 22.51 | 29.79 | 27.02 | 30.63 | 27.85 | 22.77 | 22.92 | 24.01 | 27.93 | 23.51 | 22.59 | 26.81 | 28.73 | 23.94 | 27.10 | |
| 22.89 | 29.78 | 26.90 | 30.75 | 27.53 | 22.85 | 22.73 | 23.94 | 27.71 | 23.88 | 22.90 | 26.90 | 29.81 | 24.54 | 27.55 | ||
| 22.57 | 30.50 | 27.49 | 30.95 | 27.99 | 23.26 | 23.43 | 24.56 | 28.29 | 23.50 | 22.62 | 27.15 | 28.97 | 24.08 | 27.21 | ||
| 12h | 22.85 | 30.57 | 27.90 | 29.87 | 28.45 | 23.92 | 23.39 | 24.80 | 27.89 | 23.28 | 22.91 | 27.38 | 28.98 | 23.95 | 26.91 | |
| 22.57 | 30.04 | 27.29 | 29.78 | 27.97 | 23.44 | 22.90 | 23.95 | 27.28 | 22.36 | 22.75 | 26.99 | 28.31 | 23.76 | 26.96 | ||
| 22.77 | 30.24 | 27.57 | 29.69 | 28.30 | 23.56 | 23.09 | 24.30 | 27.48 | 22.47 | 22.75 | 27.17 | 28.55 | 23.85 | 27.00 | ||
| 24h | 23.42 | 31.12 | 28.91 | 30.89 | 28.92 | 25.98 | 23.85 | 24.88 | 29.17 | 23.75 | 23.80 | 29.42 | 29.67 | 25.79 | 26.80 | |
| 22.55 | 30.80 | 28.49 | 29.88 | 28.70 | 26.04 | 24.19 | 24.46 | 28.83 | 22.47 | 22.92 | 28.93 | 28.59 | 23.85 | 26.11 | ||
| 22.92 | 30.95 | 28.81 | 30.26 | 28.90 | 25.88 | 24.05 | 24.63 | 28.96 | 22.84 | 23.22 | 29.00 | 28.98 | 24.45 | 26.30 | ||
| 48h | 23.45 | 30.70 | 28.69 | 31.45 | 28.77 | 25.87 | 22.95 | 23.95 | 28.79 | 22.84 | 24.55 | 28.85 | 29.81 | 26.96 | 27.71 | |
| 23.83 | 30.67 | 28.57 | 31.69 | 28.87 | 25.80 | 23.19 | 23.83 | 28.69 | 23.20 | 24.72 | 29.07 | 29.46 | 27.89 | 27.89 | ||
| 23.85 | 30.90 | 28.87 | 31.35 | 29.06 | 26.16 | 23.50 | 24.15 | 28.84 | 23.06 | 24.80 | 29.12 | 29.63 | 27.62 | 27.80 | ||
| Drought | 0.5h | 22.94 | 31.58 | 27.86 | 30.91 | 29.28 | 25.66 | 23.79 | 25.46 | 29.42 | 23.91 | 22.82 | 27.55 | 29.91 | 24.95 | 27.30 |
| 22.19 | 30.87 | 27.34 | 27.79 | 28.53 | 24.91 | 23.59 | 24.95 | 28.90 | 23.59 | 22.63 | 26.82 | 28.84 | 23.98 | 27.70 | ||
| 22.18 | 31.66 | 27.92 | 27.78 | 28.94 | 25.19 | 23.92 | 25.40 | 29.39 | 23.47 | 22.59 | 27.28 | 28.95 | 24.14 | 27.37 | ||
| 3h | 23.03 | 30.84 | 27.78 | 31.05 | 28.90 | 25.21 | 22.93 | 24.86 | 28.88 | 23.85 | 23.14 | 26.87 | 28.93 | 24.88 | 27.84 | |
| 21.85 | 28.97 | 26.08 | 30.48 | 28.02 | 24.40 | 22.40 | 23.89 | 28.17 | 22.51 | 21.87 | 26.00 | 28.83 | 23.38 | 26.88 | ||
| 22.23 | 29.87 | 26.81 | 30.71 | 28.71 | 24.89 | 22.92 | 24.63 | 28.79 | 22.90 | 22.29 | 26.59 | 28.87 | 23.83 | 27.19 | ||
| 6h | 23.37 | 30.69 | 27.82 | 30.91 | 29.12 | 25.54 | 23.90 | 25.47 | 29.04 | 24.09 | 22.96 | 26.70 | 29.80 | 24.17 | 28.25 | |
| 23.75 | 30.58 | 28.00 | 31.54 | 29.22 | 25.38 | 23.86 | 24.88 | 28.95 | 24.27 | 23.77 | 27.90 | 30.19 | 24.84 | 27.97 | ||
| 23.33 | 30.36 | 27.46 | 30.92 | 28.92 | 24.93 | 23.46 | 24.68 | 28.54 | 23.99 | 23.19 | 26.81 | 29.86 | 24.37 | 27.97 | ||
| 12h | 22.49 | 30.46 | 27.89 | 29.82 | 28.73 | 24.78 | 22.93 | 24.74 | 28.65 | 22.88 | 22.79 | 26.88 | 28.87 | 22.56 | 26.65 | |
| 22.88 | 31.28 | 28.47 | 30.16 | 29.35 | 25.28 | 23.56 | 24.91 | 28.76 | 23.10 | 23.46 | 28.32 | 28.68 | 24.94 | 26.82 | ||
| 22.46 | 31.22 | 28.50 | 29.70 | 29.25 | 25.41 | 23.61 | 24.99 | 28.88 | 22.64 | 22.88 | 27.91 | 28.59 | 23.29 | 26.45 | ||
| 24h | 22.30 | 31.22 | 28.81 | 30.24 | 29.07 | 25.66 | 23.92 | 25.56 | 28.66 | 23.05 | 22.91 | 27.89 | 28.85 | 23.64 | 26.58 | |
| 21.89 | 31.81 | 28.46 | 30.31 | 29.31 | 25.56 | 23.97 | 25.80 | 28.85 | 22.70 | 23.33 | 28.49 | 28.57 | 23.70 | 25.79 | ||
| 21.34 | 31.52 | 28.78 | 29.69 | 29.36 | 25.90 | 24.20 | 25.95 | 29.00 | 22.09 | 22.86 | 28.59 | 28.17 | 22.96 | 25.38 | ||
| 48h | 24.54 | 31.72 | 28.89 | 31.93 | 29.71 | 24.96 | 23.67 | 25.49 | 28.34 | 24.75 | 25.10 | 29.41 | 29.92 | 27.72 | 27.88 | |
| 23.86 | 30.47 | 27.88 | 31.21 | 29.24 | 24.31 | 22.96 | 24.86 | 27.93 | 24.03 | 23.76 | 27.88 | 29.61 | 25.58 | 27.75 | ||
| 23.92 | 31.43 | 28.34 | 31.39 | 29.68 | 24.81 | 23.46 | 25.22 | 28.15 | 23.97 | 23.97 | 28.38 | 29.41 | 25.94 | 27.38 | ||
| Heat | 0.5h | 23.95 | 31.52 | 27.82 | 29.53 | 29.46 | 25.92 | 23.92 | 25.90 | 29.24 | 22.94 | 22.74 | 28.40 | 28.47 | 25.47 | 26.67 |
| 23.43 | 30.66 | 27.67 | 29.49 | 28.96 | 24.74 | 23.23 | 25.01 | 28.52 | 22.92 | 22.87 | 27.84 | 29.00 | 25.36 | 26.52 | ||
| 23.58 | 30.75 | 27.98 | 29.94 | 28.84 | 24.95 | 23.39 | 25.25 | 28.72 | 23.08 | 23.04 | 27.93 | 28.93 | 25.53 | 26.86 | ||
| 3h | 24.55 | 30.89 | 28.19 | 31.92 | 29.94 | 25.51 | 23.47 | 25.87 | 28.72 | 23.87 | 24.69 | 29.72 | 29.86 | 29.25 | 26.53 | |
| 24.61 | 31.15 | 27.52 | 31.15 | 29.40 | 26.20 | 23.54 | 25.63 | 28.64 | 22.71 | 23.90 | 29.76 | 28.78 | 26.65 | 25.98 | ||
| 24.90 | 31.48 | 27.63 | 31.11 | 29.57 | 26.34 | 23.88 | 25.96 | 28.95 | 23.03 | 24.05 | 30.39 | 29.08 | 27.35 | 26.15 | ||
| 6h | 23.78 | 30.59 | 28.78 | 32.62 | 29.62 | 25.70 | 23.94 | 25.78 | 27.66 | 24.38 | 24.84 | 28.63 | 29.88 | 28.78 | 27.60 | |
| 23.88 | 31.50 | 28.43 | 32.14 | 29.79 | 25.87 | 23.78 | 25.89 | 28.13 | 23.88 | 23.85 | 29.08 | 29.40 | 29.26 | 27.00 | ||
| 23.90 | 31.06 | 28.65 | 32.58 | 29.87 | 25.67 | 23.83 | 25.90 | 27.96 | 23.98 | 24.20 | 29.08 | 29.57 | 28.95 | 27.12 | ||
| 12h | 24.32 | 31.95 | 27.65 | 28.33 | 31.81 | 26.56 | 24.02 | 27.92 | 28.38 | 23.87 | 23.88 | 29.76 | 28.59 | 26.92 | 26.72 | |
| 23.90 | 31.63 | 27.87 | 27.74 | 30.89 | 25.95 | 23.64 | 27.36 | 27.86 | 23.80 | 23.82 | 29.08 | 28.95 | 26.63 | 27.17 | ||
| 24.30 | 31.96 | 27.61 | 27.86 | 31.48 | 26.52 | 24.02 | 27.89 | 28.08 | 23.74 | 23.74 | 29.60 | 28.73 | 26.85 | 26.92 | ||
| 24h | 25.91 | 33.44 | 29.89 | 32.81 | 32.93 | 28.37 | 26.43 | 28.82 | 29.31 | 26.75 | 25.61 | 31.56 | 30.22 | 28.16 | 28.44 | |
| 25.34 | 33.59 | 28.73 | 31.96 | 31.83 | 27.90 | 25.84 | 27.99 | 28.81 | 25.79 | 24.78 | 30.61 | 29.98 | 30.09 | 27.89 | ||
| 25.20 | 32.89 | 28.96 | 32.50 | 31.97 | 27.58 | 25.85 | 27.95 | 28.67 | 25.86 | 24.87 | 30.90 | 30.00 | 28.86 | 27.94 | ||
| 48h | 24.67 | 33.64 | 28.79 | 31.05 | 32.03 | 26.87 | 25.94 | 27.86 | 28.18 | 25.86 | 24.31 | 30.01 | 29.93 | 29.32 | 27.83 | |
| 25.60 | 33.96 | 29.40 | 31.20 | 32.62 | 27.86 | 26.66 | 28.23 | 28.49 | 25.73 | 24.88 | 30.61 | 29.52 | 30.24 | 28.46 | ||
| 24.90 | 33.72 | 29.01 | 31.12 | 32.01 | 27.07 | 26.04 | 27.63 | 28.01 | 25.72 | 24.57 | 30.11 | 29.80 | 29.84 | 28.05 |
References
- Zhou, X.; Liu, J.; Zhuang, Y. Selection of appropriate reference genes in eggplant for quantitative gene expression studies under different experimental conditions. Sci. Hortic. 2014, 176, 200–207. [Google Scholar] [CrossRef]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Ginzinger, D.G. Gene quantification using real-time quantitative PCR. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef]
- Ye, J.; Jin, C.F.; Li, N.; Liu, M.H.; Fei, Z.X.; Dong, L.Z.; Li, L.; Li, Z.Q. Selection of suitable reference genes for qRT-PCR normalisation under different experimental conditions in Eucommia ulmoides Oliv. Sci. Rep. 2018, 8, 15043. [Google Scholar] [CrossRef] [PubMed]
- Leong, D.T.; Gupta, A.; Bai, H.F.; Wan, G.; Yoong, L.F.; Too, H.P.; Chew, F.T.; Hutmacher, D.W. Absolute quantification of gene expression in biomaterials research using real-time PCR. Biomaterials 2007, 28, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, Z.; Huang, H.; Wei, S.; Liu, Y.; Jiang, C.; Zhang, J.; Zhang, C. Selection of relatively exact reference genes for gene expression studies in goosegrass (Eleusine indica) under herbicide stress. Sci. Rep. 2017, 7, 46494. [Google Scholar] [CrossRef]
- Yang, Q.; Yin, J.; Qi, L.; Yang, F.; Wang, R.; Li, G. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol. Biol. Rep. 2014, 41, 2325–2334. [Google Scholar] [CrossRef] [PubMed]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Jiang, Q.; Wang, F.; Wang, G.L.; Xu, Z.S.; Xiong, A.S. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE 2015, 10, e0117569. [Google Scholar] [CrossRef] [PubMed]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Thibivilliers, S.; Bilgin, D.D.; Radwan, O.; Benitez, M.; Clough, S.J.; Stacey, G. Identification of Four Soybean Reference Genes for Gene Expression Normalization. Plant Genome J. 2008, 1, 44–54. [Google Scholar] [CrossRef]
- Fei, X.; Shi, Q.; Yang, T.; Fei, Z.; Wei, A. Expression stabilities of ten candidate reference genes for RT-qPCR in zanthoxylum bungeanum Maxim. Molecules 2018, 23, 802. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.L.; Johnson, J.C. Validation of reference genes for gene expression analysis in olive (Olea europaea) mesocarp tissue by quantitative real-time RT-PCR. BMC Res. Notes 2014, 7, 802. [Google Scholar] [CrossRef]
- Lucho, S.R.; do Amaral, M.N.; Benitez, L.C.; Milech, C.; Kleinowski, A.M.; Bianchi, V.J.; Braga, E.J.B. Validation of reference genes for RT-qPCR studies in Stevia rebaudiana in response to elicitor agents. Physiol. Mol. Biol. Plants 2018, 24, 767–779. [Google Scholar] [CrossRef]
- Li, M.-Y.; Wang, F.; Jiang, Q.; Wang, G.-L.; Tian, C.; Xiong, A.-S. Validation and Comparison of Reference Genes for qPCR Normalization of Celery (Apium graveolens) at Different Development Stages. Front. Plant Sci. 2016, 7, 313. [Google Scholar] [CrossRef]
- Wu, Z.J.; Tian, C.; Jiang, Q.; Li, X.H.; Zhuang, J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci. Rep. 2016, 6, 19748. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Zhou, P.; Xu, M.; Xu, L.A. Development and characterization of chloroplast microsatellite markers for Pinus massoniana and their application in Pinus (Pinaceae) species. J. Genet. 2018, 97, 53–59. [Google Scholar] [CrossRef]
- Maleki, S.S.; Mohammadi, K.; Ji, K.S. Study on factors influencing transformation efficiency in Pinus massoniana using Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult. 2018, 133, 437–445. [Google Scholar] [CrossRef]
- Ni, Z.X.; Ye, Y.J.; Bai, T.; Xu, M.; Xu, L.A. Complete chloroplast genome of pinus massoniana (pinaceae): Gene rearrangements, loss of ndh genes, and short inverted repeats contraction, expansion. Molecules 2017, 22, 1528. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wu, X. Physiological and Proteomic Analysis of Mycorrhizal Pinus massoniana Inoculated with Lactarius insulsus Under Drought Stress. Физиология Растений 2016, 63, 754–762. [Google Scholar] [CrossRef]
- Wei, Y.; Liu, Q.; Dong, H.; Zhou, Z.; Hao, Y.; Chen, X.; Xu, L. Selection of Reference Genes for Real-Time Quantitative PCR in Pinus massoniana Post Nematode Inoculation. PLoS ONE 2016, 11, e0147224. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, Z.; Hu, Y.; Tan, J.; Jia, J.; Xu, H.; Chen, X. Reference genes selection for quantitative gene expression studies in Pinus massoniana L. Trees-Struct. Funct. 2016, 30, 685–696. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F.; Åman, P.; Semb, H.; Powers, D.; et al. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. Genome Biol. 2002, 3, 37. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Petriccione, M.; Mastrobuoni, F.; Zampella, L.; Scortichini, M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. Actinidiae. Sci. Rep. 2015, 5, 16961. [Google Scholar] [CrossRef] [PubMed]
- Klie, M.; Debener, T. Identification of superior reference genes for data normalisation of expression studies via quantitative PCR in hybrid roses (Rosa hybrida). BMC Res. Notes 2011, 4, 518. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.-R. Eleven Golden Rules of Quantitative RT-PCR. Plant Cell Online 2008, 20, 1736–1737. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Scholtz, J.J.; Visser, B. Reference gene selection for qPCR gene expression analysis of rust-infected wheat. Physiol. Mol. Plant Pathol. 2013, 81, 22–25. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Lu, J.; Xu, Z.; Yang, W.; Wang, J.; Cheng, T.; Zhang, Q. Selection of suitable reference genes for miRNA expression normalization by qRT-PCR during flower development and different genotypes of Prunus mume. Sci. Hortic. 2014, 169, 130–137. [Google Scholar] [CrossRef]
- Barsalobres-Cavallari, C.F.; Severino, F.E.; Maluf, M.P.; Maia, I.G. Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Mol. Biol. 2008, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Han, S.; Yin, W.; Xia, X.; Liu, C. Comparison of reliable reference genes following different hormone treatments by various algorithms for qRT-PCR analysis of Metasequoia. Int. J. Mol. Sci. 2019, 20, 34. [Google Scholar] [CrossRef] [PubMed]
- De Vega-Bartol, J.J.; Santos, R.R.; Simões, M.; Miguel, C.M. Normalizing gene expression by quantitative PCR during somatic embryogenesis in two representative conifer species: Pinus pinaster and Picea abies. Plant Cell Rep. 2013, 32, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.; Sebastiana, M.; Pais, M.S.; Figueiredo, A. Reference Gene Selection and Validation for the Early Responses to Downy Mildew Infection in Susceptible and Resistant Vitis vinifera Cultivars. PLoS ONE 2013, 8, e72998. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, J.; Huang, S.; Guo, T.; Deng, L.; Hua, W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 2014, 538, 113–122. [Google Scholar] [CrossRef]
- Zhu, J.; He, F.; Song, S.; Wang, J.; Yu, J. How many human genes can be defined as housekeeping with current expression data? BMC Genomics 2008, 9, 172. [Google Scholar] [CrossRef]
- Schmid, H.; Cohen, C.D.; Henger, A.; Irrgang, S.; Schlöndorff, D.; Kretzler, M. Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies. Kidney Int. 2003, 64, 356–360. [Google Scholar] [CrossRef]
- Bao, W.; Qu, Y.; Shan, X.; Wan, Y. Screening and validation of housekeeping genes of the root and cotyledon of cunninghamia lanceolata under abiotic stresses by using quantitative real-time PCR. Int. J. Mol. Sci. 2016, 17, 1198. [Google Scholar] [CrossRef]
- Tu, Z.; Hao, Z.; Zhong, W.; Li, H. Identification of suitable reference genes for RT-qPCR assays in Liriodendron chinense (Hemsl.) Sarg. Forests 2019, 10, 441. [Google Scholar] [CrossRef]
- Mallona, I.; Lischewski, S.; Weiss, J.; Hause, B.; Egea-Cortines, M. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 2010, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Die, J.V.; Román, B.; Nadal, S.; González-Verdejo, C.I. Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 2010, 232, 145–153. [Google Scholar] [CrossRef] [PubMed]
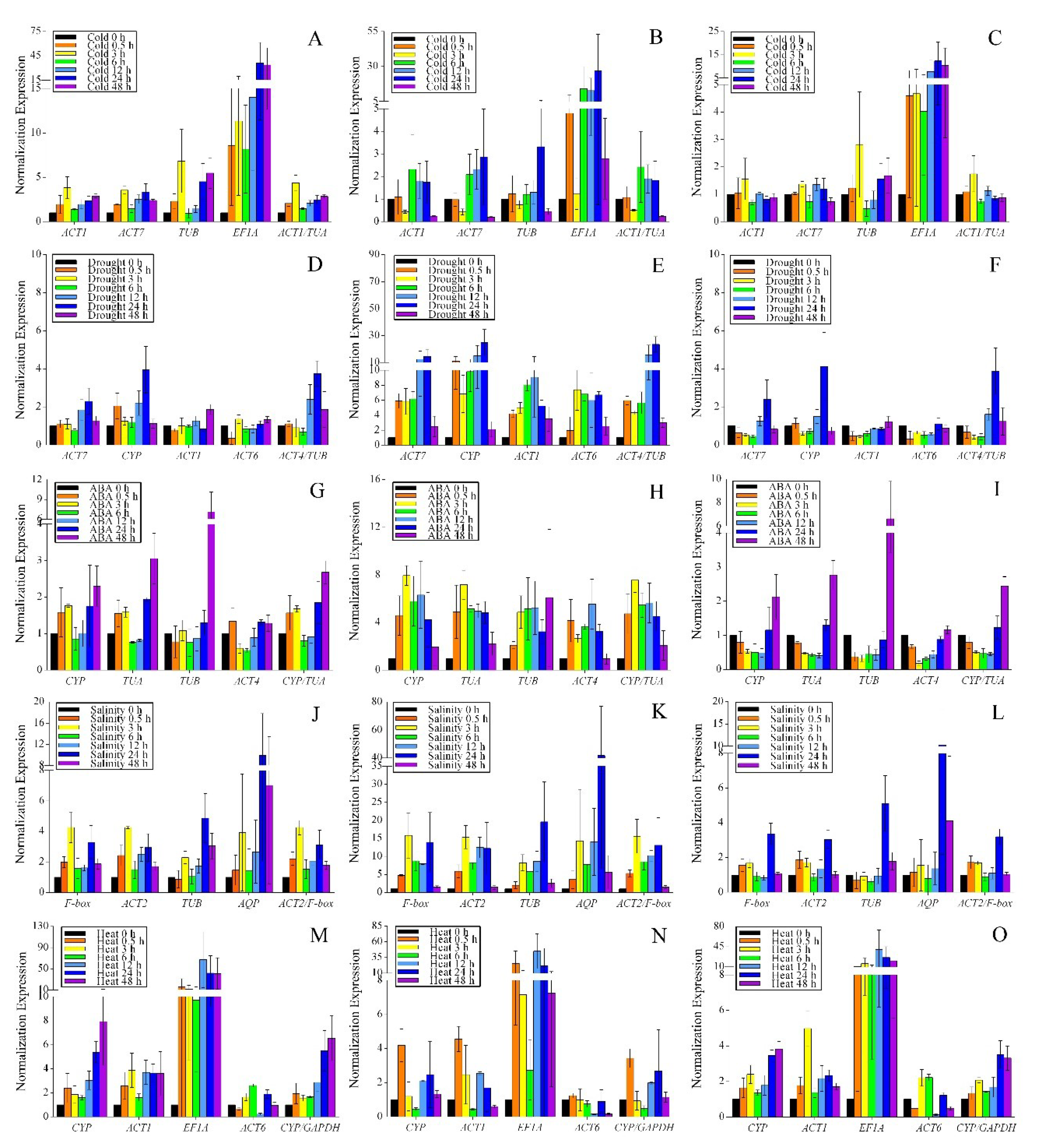
| Gene Symbol | Gene Description | Accession Number | Primer Sequence (5′-3′) | Amplicon Size (bp) | PCR Efficiency (%) | Regression Coefficient (R2) | Tm (°C) |
|---|---|---|---|---|---|---|---|
| Reference genes | |||||||
| ACT1 | Actin 1 gene | KM496527.1 | CCGTATGAGCAAGGAAATCAC | 100 | 95.679 | 0.999 | 85.0 |
| AGAACCTCCAATCCAGACACT | |||||||
| ACT2 | Actin 2 gene | KM496525.1 | CACGGAATAGGCAGAAGTTGG | 97 | 98.145 | 0.989 | 80.8 |
| TGGGCATAAAGTGTTAGAATAGC | |||||||
| ACT4 | Actin 4 gene | KM496528.1 | ATTTATGAGGGATACGCTTTG | 106 | 97.505 | 0.995 | 84.6 |
| AGGTGTACCCACGTTCTGTAA | |||||||
| ACT6 | Actin 6 gene | KM496530.1 | AACTCCTGCCATCCTCATCTT | 99 | 96.549 | 0.996 | 83.1 |
| CTGTTCCAGCCTTGCTTTCA | |||||||
| ACT7 | Actin 7 gene | KM496529.1 | TGGGATGCTATGGAAGATTTG | 114 | 104.801 | 0.992 | 82.9 |
| TACGCCCTTTGGAGTAAGAAG | |||||||
| AQP | Aquaporin protein gene | KF582038.1 | CACCTTGCCACAATTCCTATCA | 103 | 95.481 | 0.998 | 86.3 |
| TCCAATGGTCATCCCAAACAC | |||||||
| CYP | Cyclophilin gene | KM496534.1 | CGAGAAGTTTGCCGATGAGAA | 97 | 91.042 | 0.997 | 87.5 |
| GAATTGCGAGCCGTTAGTGTT | |||||||
| EF1A | Elongation factor 1-alpha gene | KM496532.1 | GGATTTGAAACGTGGGTATGT | 97 | 99.383 | 0.998 | 83.5 |
| CAGGGTGGTTCATTATGATTACT | |||||||
| F-box | F-box family protein gene | KM496542.1 | TATTATTGTTGCAGGTGGGTT | 109 | 100.101 | 0.996 | 81.6 |
| AGAATGTTGAAGTTCGGCTAT | |||||||
| GAPDH | Glyceraldehyde 3-Phosphatase gene | KM496531.1 | GGATTTGGTCGTATTGGGAGG | 96 | 92.132 | 0.998 | 82.9 |
| TTTGGCATCAATGAAAGGGTC | |||||||
| TUA | Tubulin alpha gene | KM496535.1 | CAAACTTGGTCCCGTATCCTC | 95 | 92.011 | 0.999 | 83.7 |
| CACAGAAAGCTGCTCATGGTAA | |||||||
| TUB | Tubulin beta gene | KM496536.1 | CTGCGACTATGAGTGGAGTGA | 108 | 98.923 | 0.993 | 85.6 |
| AGAAATGAAGACGAGGGAATG | |||||||
| Target genes | |||||||
| SHR | Short-Root gene | MK153765 | GCCTGTGAGGATTCTGAAGTT | 97 | 97.636 | 0.996 | 85.3 |
| CACTGAAAGCAGCATGTATGA | |||||||
| APS | Alpha-pinene synthase gene | KF547035 | TGGATCGCCAGTGGTGAGGTG | 105 | 94.694 | 0.991 | 86.2 |
| GTCGGTCGTCAGAATGGGTTG | |||||||
| PYL | Pyrabactin resistance-like gene | MK953936 | GAGTCCGAGTATGTGTGGAGGC | 110 | 100.051 | 0.997 | 86.2 |
| ACTAATGACCAAACCAGATGAA | |||||||
| Treatment | Rank | geNorm | NormFinder | BestKeeper | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | SD(±Ct) | CV(%Ct) | Gene | Stability | ||
| Salinity | 1 | ACT2 | 0.275 | F-box | 0.145 | EF1A | 0.30 | 1.24 | ACT2 | 1.41 |
| 2 | F-box | 0.275 | ACT2 | 0.164 | ACT2 | 0.34 | 1.13 | F-box | 1.68 | |
| 3 | CYP | 0.315 | ACT4 | 0.177 | CYP | 0.39 | 1.69 | CYP | 3.46 | |
| 4 | ACT4 | 0.410 | CYP | 0.264 | F-box | 0.49 | 1.74 | ACT4 | 4.36 | |
| 5 | ACT7 | 0.453 | ACT1 | 0.266 | ACT7 | 0.54 | 1.91 | ACT1 | 5.48 | |
| 6 | ACT1 | 0.502 | ACT7 | 0.347 | ACT1 | 0.58 | 2.53 | ACT7 | 5.48 | |
| 7 | ACT6 | 0.549 | TUA | 0.398 | GAPDH | 0.60 | 2.58 | EF1A | 6.04 | |
| 8 | TUA | 0.583 | ACT6 | 0.405 | ACT6 | 0.66 | 2.15 | ACT6 | 7.48 | |
| 9 | GAPDH | 0.633 | TUB | 0.501 | TUA | 0.76 | 3.28 | TUA | 7.97 | |
| 10 | TUB | 0.687 | GAPDH | 0.572 | ACT4 | 0.78 | 2.81 | GAPDH | 8.91 | |
| 11 | EF1A | 0.734 | EF1A | 0.588 | TUB | 1.03 | 3.75 | TUB | 9.72 | |
| 12 | AQP | 0.787 | AQP | 0.656 | AQP | 1.12 | 4.67 | AQP | 12.00 | |
| ABA | 1 | CYP | 0.246 | CYP | 0.085 | GAPDH | 0.42 | 1.78 | CYP | 1.41 |
| 2 | TUA | 0.246 | TUA | 0.142 | F-box | 0.42 | 1.51 | TUA | 2.38 | |
| 3 | ACT6 | 0.319 | AQP | 0.193 | ACT1 | 0.47 | 2.07 | ACT1 | 3.94 | |
| 4 | ACT7 | 0.345 | ACT1 | 0.202 | CYP | 0.53 | 2.29 | AQP | 4.24 | |
| 5 | ACT1 | 0.361 | ACT7 | 0.257 | EF1A | 0.61 | 2.52 | GAPDH | 4.30 | |
| 6 | AQP | 0.375 | ACT6 | 0.267 | AQP | 0.62 | 2.67 | ACT7 | 5.14 | |
| 7 | GAPDH | 0.423 | GAPDH | 0.282 | ACT7 | 0.69 | 2.43 | ACT6 | 5.73 | |
| 8 | ACT2 | 0.466 | ACT2 | 0.321 | TUA | 0.70 | 3.03 | F-box | 7.18 | |
| 9 | ACT4 | 0.507 | ACT4 | 0.400 | ACT2 | 0.75 | 2.49 | ACT2 | 8.24 | |
| 10 | EF1A | 0.556 | EF1A | 0.453 | ACT6 | 0.75 | 2.44 | EF1A | 8.41 | |
| 11 | F-box | 0.611 | F-box | 0.578 | ACT4 | 0.75 | 2.74 | ACT4 | 9.46 | |
| 12 | TUB | 0.663 | TUB | 0.580 | TUB | 0.99 | 3.67 | TUB | 12.00 | |
| Drought | 1 | ACT4 | 0.280 | ACT7 | 0.093 | F-box | 0.24 | 0.84 | ACT7 | 1.57 |
| 2 | TUB | 0.280 | CYP | 0.223 | ACT7 | 0.31 | 1.06 | CYP | 2.63 | |
| 3 | ACT7 | 0.360 | TUA | 0.264 | CYP | 0.34 | 1.43 | ACT4 | 3.36 | |
| 4 | CYP | 0.427 | ACT4 | 0.296 | AQP | 0.39 | 1.56 | TUA | 4.24 | |
| 5 | ACT2 | 0.458 | ACT2 | 0.328 | ACT2 | 0.45 | 1.45 | TUB | 4.36 | |
| 6 | TUA | 0.490 | TUB | 0.344 | TUA | 0.50 | 2.16 | F-box | 4.45 | |
| 7 | F-box | 0.557 | GAPDH | 0.398 | EF1A | 0.51 | 2.04 | ACT2 | 5.00 | |
| 8 | AQP | 0.600 | F-box | 0.403 | ACT4 | 0.52 | 1.84 | AQP | 7.14 | |
| 9 | EF1A | 0.640 | AQP | 0.466 | GAPDH | 0.56 | 2.36 | GAPDH | 8.43 | |
| 10 | GAPDH | 0.679 | ACT1 | 0.512 | TUB | 0.60 | 2.16 | EF1A | 9.34 | |
| 11 | ACT1 | 0.722 | EF1A | 0.555 | ACT1 | 0.74 | 3.21 | ACT1 | 10.49 | |
| 12 | ACT6 | 0.785 | ACT6 | 0.692 | ACT6 | 0.83 | 2.71 | ACT6 | 12.00 | |
| Cold | 1 | ACT1 | 0.157 | GAPDH | 0.111 | ACT1 | 0.24 | 0.99 | ACT1 | 1.57 |
| 2 | TUA | 0.157 | ACT7 | 0.133 | TUA | 0.29 | 1.20 | ACT7 | 2.34 | |
| 3 | ACT7 | 0.232 | ACT1 | 0.145 | ACT4 | 0.30 | 1.04 | TUA | 2.51 | |
| 4 | GAPDH | 0.281 | ACT2 | 0.150 | F-box | 0.30 | 1.06 | GAPDH | 2.91 | |
| 5 | ACT4 | 0.325 | TUA | 0.215 | ACT7 | 0.35 | 1.18 | ACT4 | 4.82 | |
| 6 | ACT2 | 0.350 | ACT4 | 0.224 | GAPDH | 0.37 | 1.54 | ACT2 | 5.89 | |
| 7 | ACT6 | 0.394 | AQP | 0.283 | AQP | 0.39 | 1.64 | F-box | 6.93 | |
| 8 | AQP | 0.427 | F-box | 0.284 | CYP | 0.40 | 1.72 | AQP | 7.24 | |
| 9 | F-box | 0.453 | ACT6 | 0.330 | ACT6 | 0.42 | 1.34 | ACT6 | 8.45 | |
| 10 | CYP | 0.476 | CYP | 0.395 | ACT2 | 0.45 | 1.43 | CYP | 9.46 | |
| 11 | TUB | 0.523 | TUB | 0.455 | TUB | 0.70 | 2.47 | TUB | 11.00 | |
| 12 | EF1A | 0.634 | EF1A | 0.782 | EF1A | 0.87 | 3.41 | EF1A | 12.00 | |
| Heat | 1 | CYP | 0.309 | ACT1 | 0.187 | TUA | 0.50 | 2.06 | ACT1 | 2.30 |
| 2 | GAPDH | 0.309 | TUA | 0.275 | ACT4 | 0.55 | 1.92 | CYP | 2.63 | |
| 3 | ACT2 | 0.416 | CYP | 0.284 | ACT1 | 0.58 | 2.39 | TUA | 3.13 | |
| 4 | ACT7 | 0.496 | TUB | 0.325 | TUB | 0.89 | 3.02 | GAPDH | 3.64 | |
| 5 | AQP | 0.589 | GAPDH | 0.329 | AQP | 0.94 | 3.58 | ACT2 | 5.05 | |
| 6 | TUB | 0.635 | ACT2 | 0.356 | GAPDH | 0.96 | 3.96 | TUB | 5.18 | |
| 7 | ACT1 | 0.658 | AQP | 0.405 | ACT2 | 0.99 | 3.10 | F-box | 5.62 | |
| 8 | TUA | 0.686 | ACT4 | 0.419 | F-box | 1.13 | 4.07 | AQP | 6.19 | |
| 9 | ACT4 | 0.724 | ACT7 | 0.482 | ACT6 | 1.23 | 3.96 | ACT4 | 6.45 | |
| 10 | F-box | 0.798 | F-box | 0.682 | ACT7 | 1.32 | 4.35 | ACT7 | 7.54 | |
| 11 | EF1A | 0.874 | EF1A | 0.847 | EF1A | 1.38 | 5.24 | EF1A | 11.24 | |
| 12 | ACT6 | 1.019 | ACT6 | 1.145 | CYP | 1.46 | 5.82 | ACT6 | 11.74 | |
| Total | 1 | ACT1 | 0.437 | ACT2 | 0.267 | F-box | 0.44 | 1.57 | TUA | 2.11 |
| 2 | TUA | 0.437 | TUA | 0.306 | CYP | 0.59 | 2.53 | ACT2 | 2.30 | |
| 3 | ACT7 | 0.625 | ACT1 | 0.316 | GAPDH | 0.65 | 2.76 | ACT1 | 2.71 | |
| 4 | ACT2 | 0.650 | CYP | 0.335 | ACT4 | 0.72 | 2.57 | CYP | 3.72 | |
| 5 | GAPDH | 0.671 | ACT7 | 0.358 | TUA | 0.75 | 3.19 | ACT7 | 5.10 | |
| 6 | CYP | 0.686 | ACT4 | 0.372 | ACT1 | 0.77 | 3.31 | GAPDH | 5.21 | |
| 7 | ACT4 | 0.703 | GAPDH | 0.394 | ACT2 | 0.77 | 2.50 | F-box | 5.62 | |
| 8 | TUB | 0.747 | TUB | 0.508 | ACT6 | 0.85 | 2.76 | ACT4 | 5.63 | |
| 9 | EF1A | 0.778 | EF1A | 0.541 | ACT7 | 0.89 | 3.03 | TUB | 8.66 | |
| 10 | F-box | 0.827 | F-box | 0.580 | EF1A | 1.00 | 3.96 | EF1A | 9.24 | |
| 11 | AQP | 0.882 | AQP | 0.698 | TUB | 1.16 | 4.16 | ACT6 | 10.84 | |
| 12 | ACT6 | 0.932 | ACT6 | 0.711 | AQP | 1.22 | 4.99 | AQP | 11.24 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, P.; Ma, Y.; Zhu, L.; Chen, Y.; Li, R.; Ji, K. Selection of Suitable Reference Genes in Pinus massoniana Lamb. Under Different Abiotic Stresses for qPCR Normalization. Forests 2019, 10, 632. https://doi.org/10.3390/f10080632
Zhu P, Ma Y, Zhu L, Chen Y, Li R, Ji K. Selection of Suitable Reference Genes in Pinus massoniana Lamb. Under Different Abiotic Stresses for qPCR Normalization. Forests. 2019; 10(8):632. https://doi.org/10.3390/f10080632
Chicago/Turabian StyleZhu, Peihuang, Yinyan Ma, Lingzhi Zhu, Yu Chen, Rong Li, and Kongshu Ji. 2019. "Selection of Suitable Reference Genes in Pinus massoniana Lamb. Under Different Abiotic Stresses for qPCR Normalization" Forests 10, no. 8: 632. https://doi.org/10.3390/f10080632
APA StyleZhu, P., Ma, Y., Zhu, L., Chen, Y., Li, R., & Ji, K. (2019). Selection of Suitable Reference Genes in Pinus massoniana Lamb. Under Different Abiotic Stresses for qPCR Normalization. Forests, 10(8), 632. https://doi.org/10.3390/f10080632





