Cloning, Characterization and Expression of the Phenylalanine Ammonia-Lyase Gene (PaPAL) from Spruce Picea asperata
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Isolation and cDNA Synthesis
2.3. Cloning of the Full-Length PaPAL cDNA
2.4. Bioinformatics Analysis of PaPAL
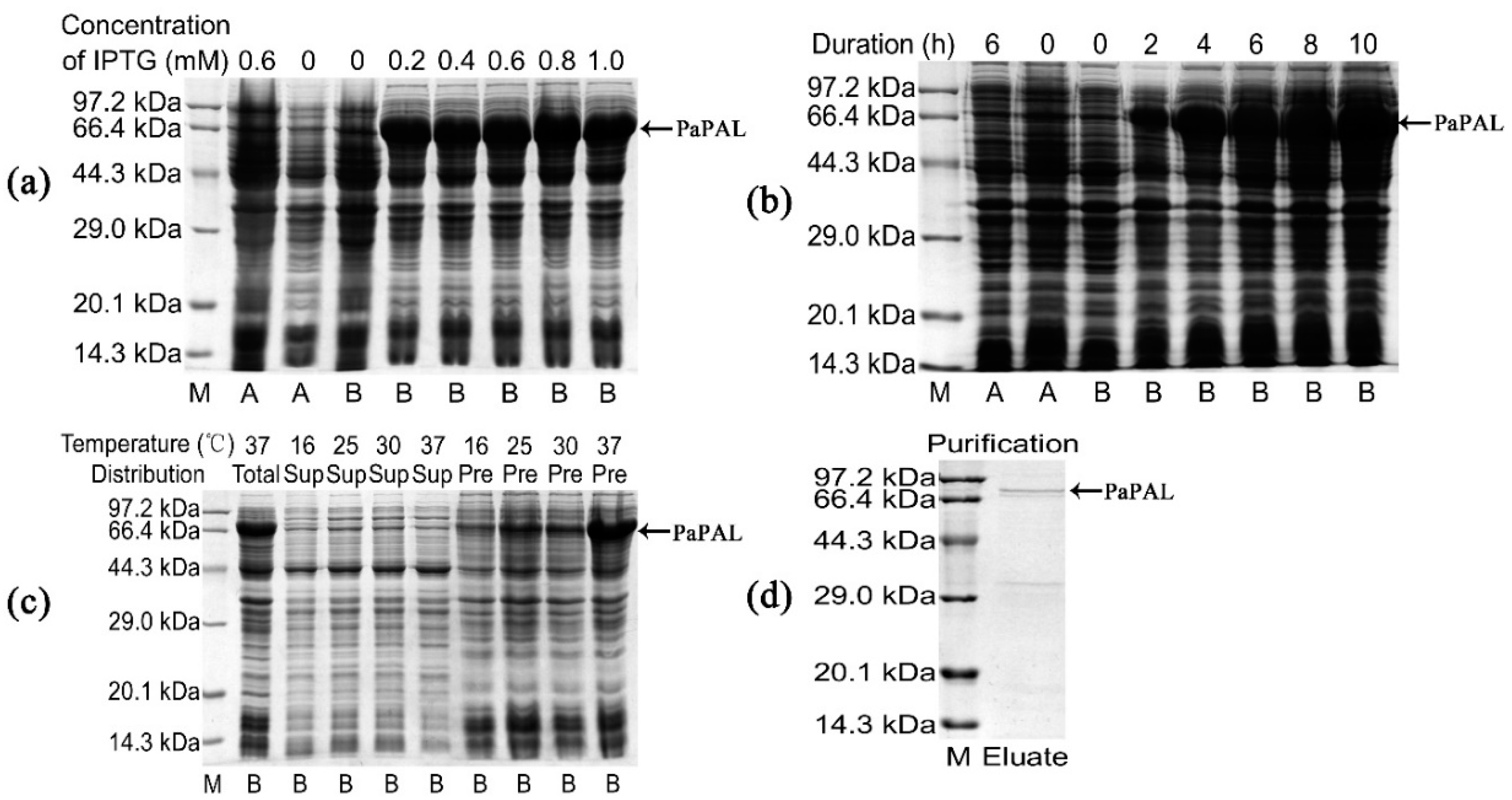
2.5. Expression and Purification of PaPAL in E. coli
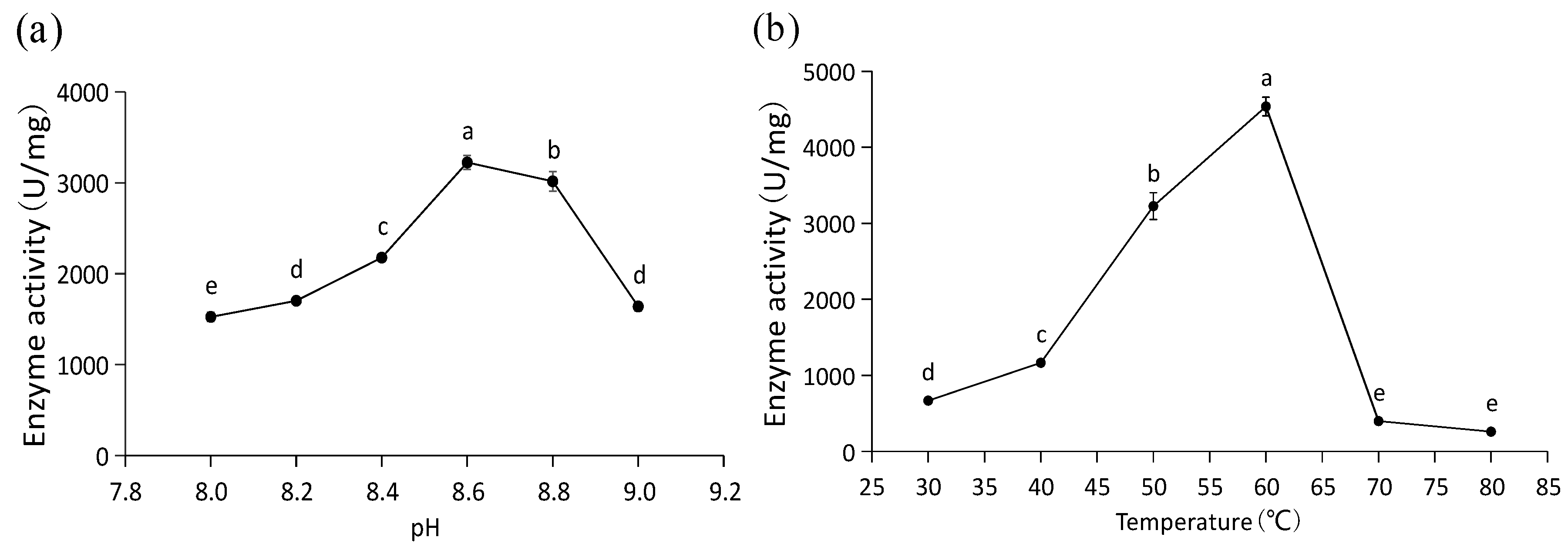
2.6. Enzyme Activity Assay for Recombinant PaPAL
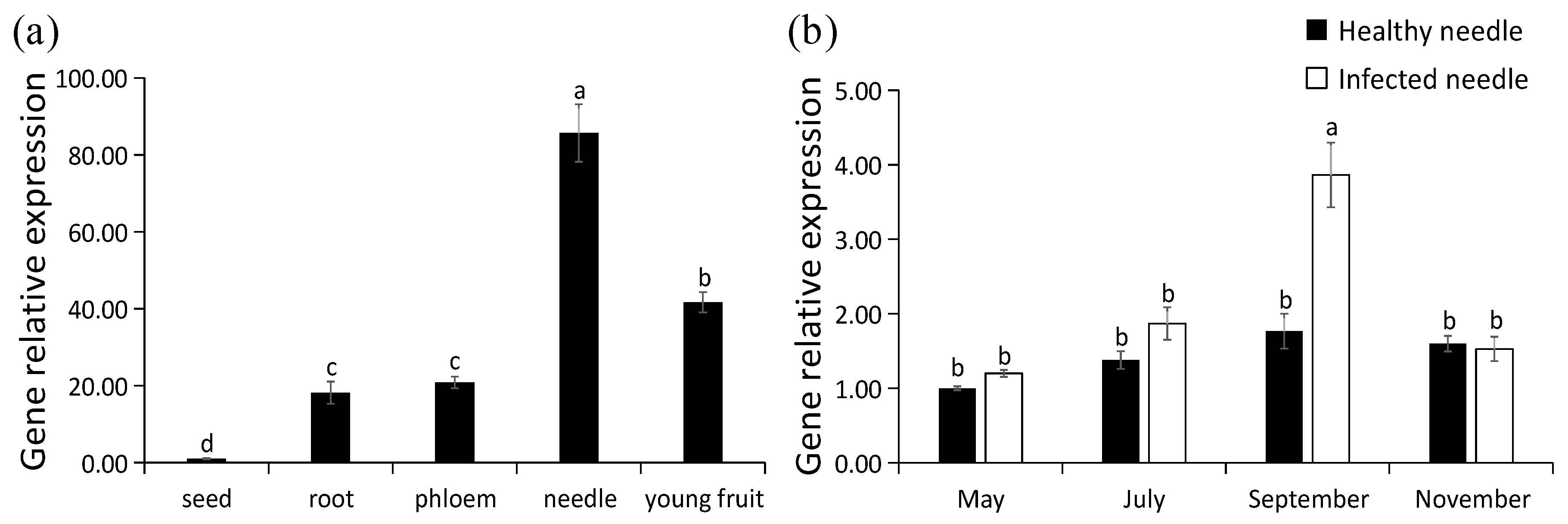
2.7. Expression Properties of PaPAL by qRT-PCR
3. Results
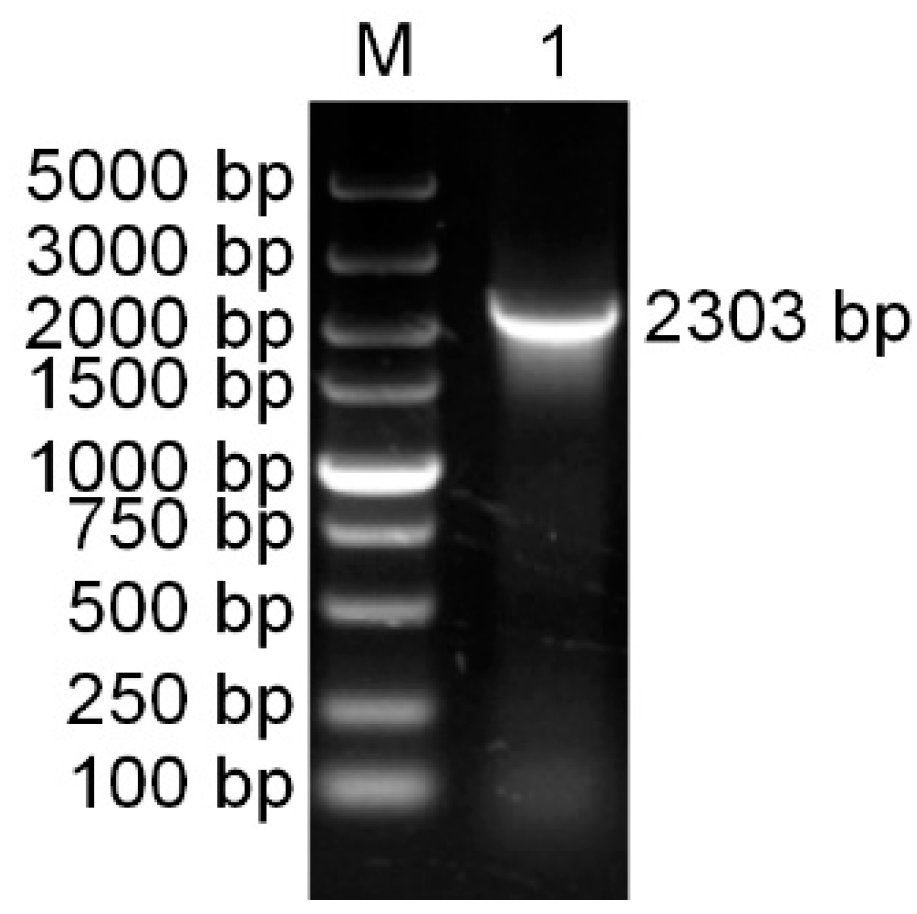
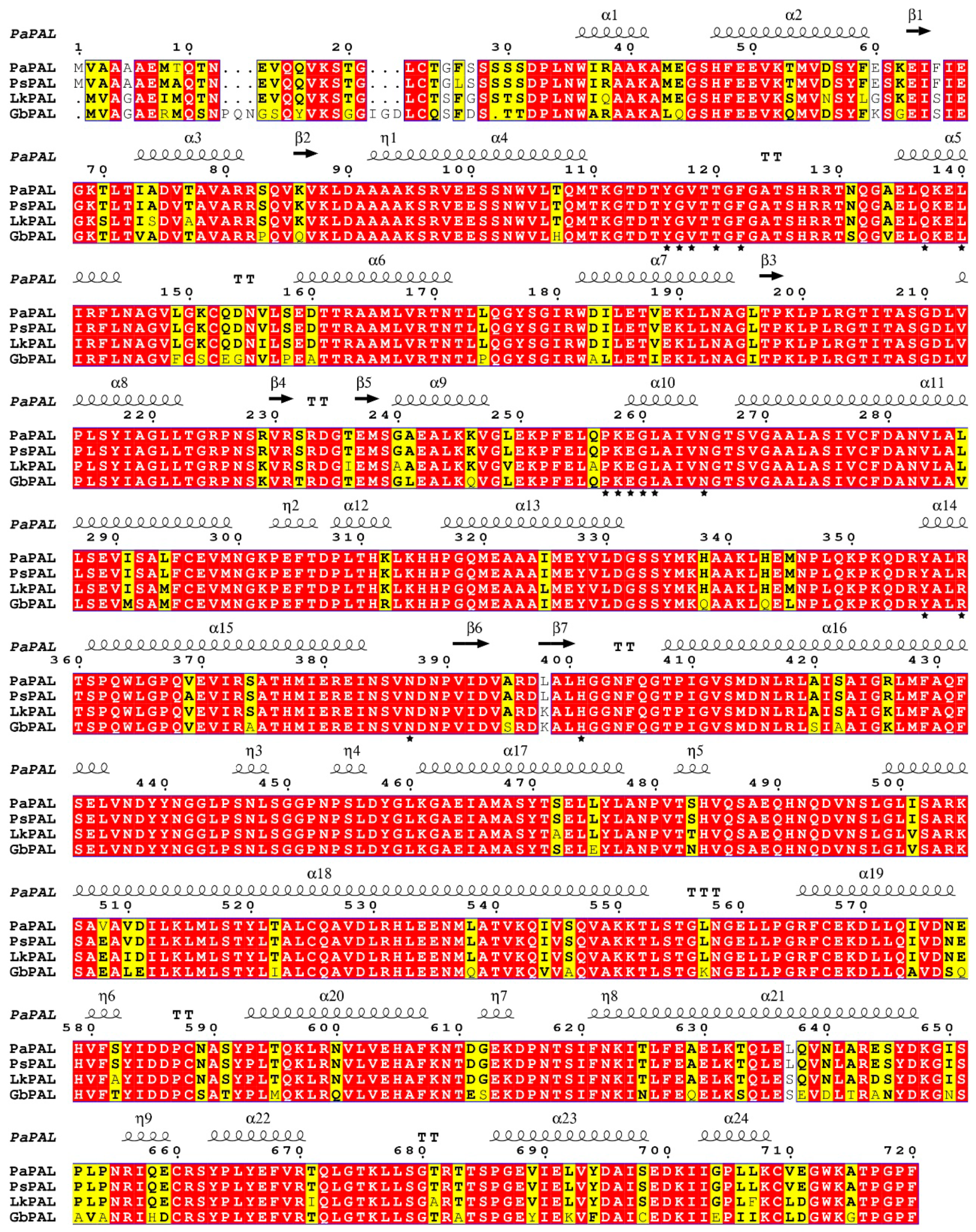
3.1. Cloning and Characterization of the PaPAL Gene
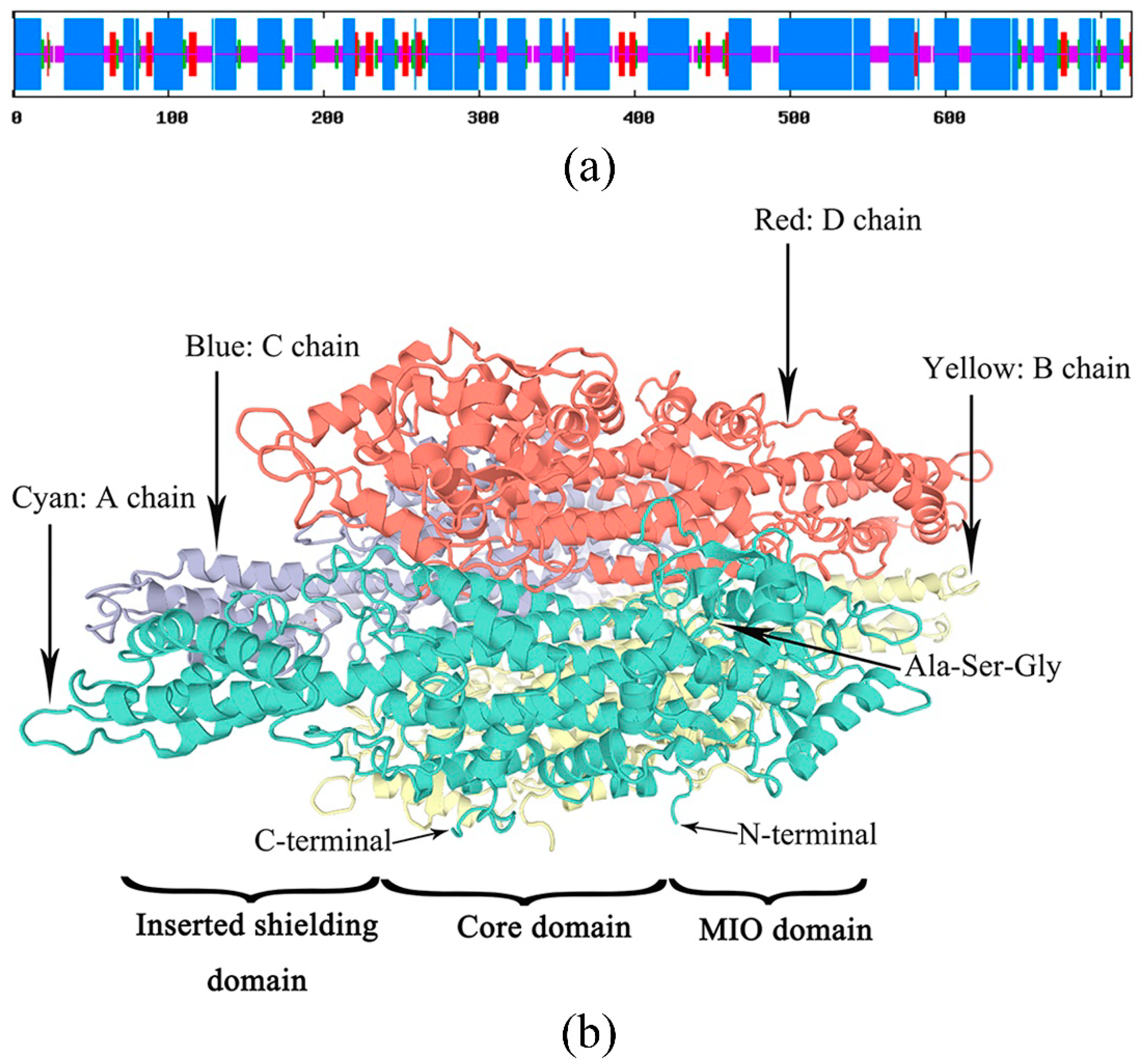
3.2. Analysis of Secondary Structure and Tertiary Structure of the PaPAL Protein
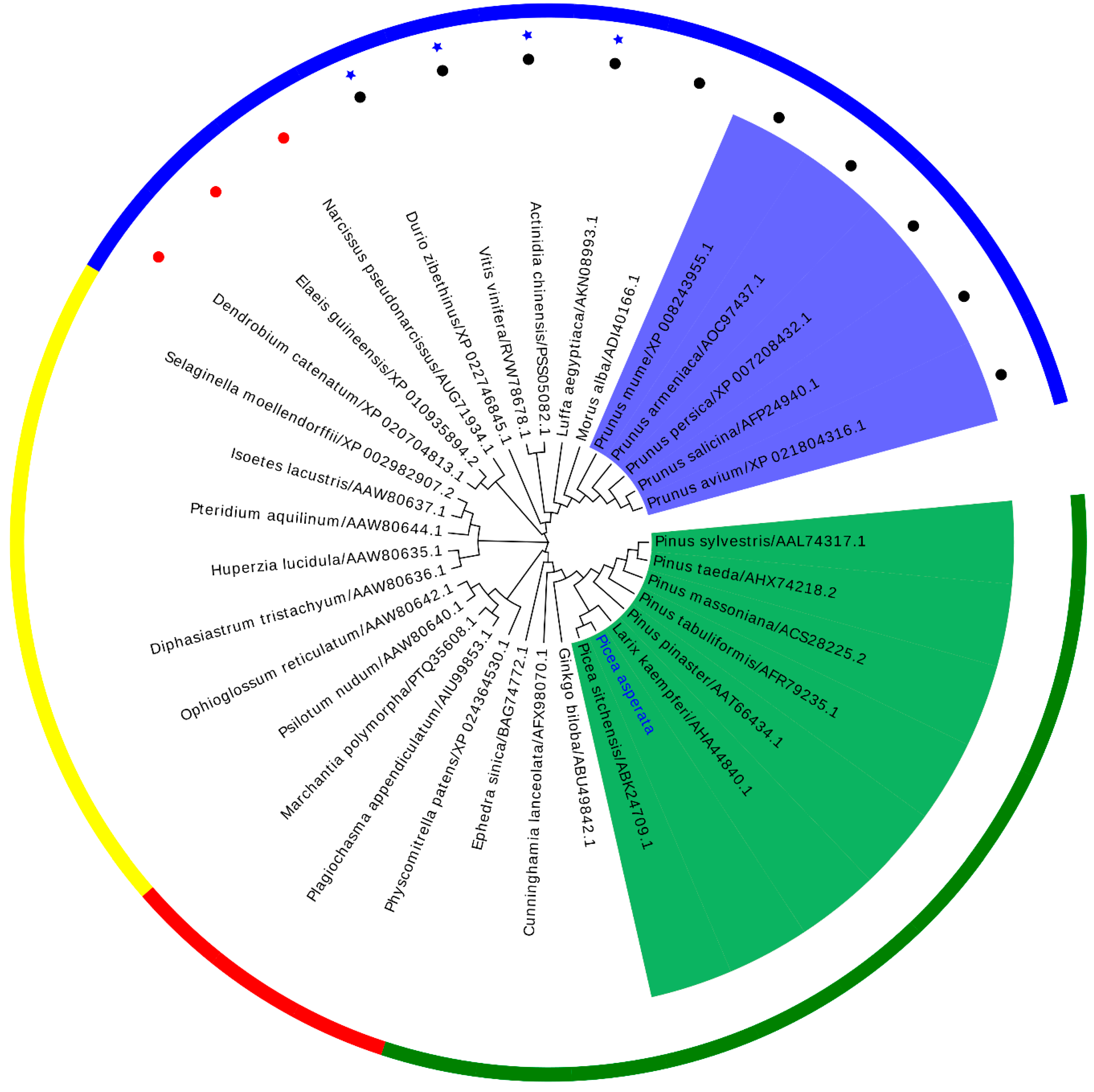
3.3. Phylogenetic Analysis of PaPAL
3.4. Expression, Purification and Functional Characterization of Recombinant PaPAL
3.5. Transcriptional Profiles of PaPAL in Different Tissues and at Different Periods during Pathogen Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bagal, U.R.; Leebens-Mack, J.H.; Lorenz, W.W.; Dean, J.F. The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage. BMC Genom. 2012, 13, S1. [Google Scholar]
- Bonello, P.; Gordon, T.R.; Herms, D.A.; Wood, D.L.; Erbilgin, N. Nature and ecological implications of pathogen-induced systemic resistance in conifers: A novel hypothesis. Physiol. Mol. Plant Pathol. 2006, 68, 95–104. [Google Scholar]
- Sangsil, P.; Nualsri, C.; Woraathasin, N.; Nakkanong, K. Characterization of the phenylalanine ammonia lyase gene from the rubber tree (Hevea brasiliensis Müll. Arg.) and differential response during Rigidoporus microporus infection. J. Plant Prot. Res. 2016, 56, 380–388. [Google Scholar]
- Wu, Z.H.; Gui, S.T.; Wang, S.Z.; Ding, Y. Molecular evolution and functional characterisation of an ancient phenylalanine ammonia-lyase gene (NnPAL1) from Nelumbo nucifera: Novel insight into the evolution of the PAL family in angiosperms. BMC Evol. Biol. 2014, 14, 100. [Google Scholar]
- Xu, F.; Deng, G.; Cheng, S.Y.; Zhang, W.Y.; Huang, X.H.; Li, L.L.; Cheng, H.; Rong, X.F.; Li, J.B. Molecular cloning, characterization and expression of the phenylalanine ammonia-lyase gene from Juglans regia. Molecules 2012, 17, 7810–7823. [Google Scholar] [PubMed]
- Bowles, D.J. Defense-related proteins in higher plants. Annu. Rev. Biochem. 1990, 59, 873–907. [Google Scholar] [PubMed]
- Cho, M.H.; Lee, S.W. Phenolic phytoalexins in rice: Biological functions and biosynthesis. Int. J. Mol. Sci. 2015, 16, 29120–29133. [Google Scholar]
- Petersen, M.; Abdullah, Y.; Benner, J.; Eberle, D.; Gehlen, K.; Hücherig, S.; Janiak, V.; Kim, K.H.; Sander, M.; Weitzel, C.; et al. Evolution of rosmarinic acid biosynthesis. Phytochemistry 2009, 70, 1663–1679. [Google Scholar]
- Clarke, J.D.; Volko, S.M.; Ledford, H.; Ausubel, F.M.; Dong, X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in arabidopsis. Plant Cell 2000, 12, 2175–2190. [Google Scholar]
- Suzuki, H.; Xia, Y.; Cameron, R.; Shadle, G.; Blount, J.; Lamb, C.; Dixon, R.A. Signals for local and systemic responses of plants to pathogen attack. J. Exp. Bot. 2004, 55, 169–179. [Google Scholar]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defense response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [PubMed]
- Iiyama, K.; Lam, T.B.; Stone, B.A. Covalent cross-links in the cell wall. Plant Physiol. 1994, 104, 315–320. [Google Scholar] [PubMed]
- Bhuiyan, N.H.; Selvaraj, G.; Wei, Y.; King, J. Role of lignification in plant defense. Plant Signal. Behav. 2009, 4, 158–159. [Google Scholar] [PubMed]
- Collinge, D.B. Cell wall appositions: The first line of defense. J. Exp. Bot. 2009, 60, 351–352. [Google Scholar] [PubMed]
- Araujo, L.; Bispo, W.M.S.; Rios, V.S.; Fernandes, S.A.; Rodrigues, F.A. Induction of the phenylpropanoid pathway by acibenzolar-s-methyl and potassium phosphite increases mango resistance to Ceratocystis fimbriata infection. Plant Dis. 2015, 99, 447–459. [Google Scholar] [PubMed]
- Kavousi, H.R.; Marashi, H.; Bagheri, A.R.; Mozafari, J. Expression of phenylpropanoid pathway genes in chickpea defense against race 3 of ascochyta rabiei. Plant Pathol. J. 2009, 8, 127–132. [Google Scholar]
- Yusuf, C.Y.L.; Abdullah, J.O.; Shaharuddin, N.A.; Abu Seman, I.; Abdullah, M.P. Characterization of promoter of EgPAL1, a novel PAL gene from the oil palm Elaeis guineensis Jacq. Plant Cell Rep. 2018, 37, 265–278. [Google Scholar]
- Koukol, J.; Conn, E.E. The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare. J. Biol. Chem. 1961, 236, 2692–2698. [Google Scholar]
- Jin, Q.; Yao, Y.; Cai, Y.P.; Lin, Y. Molecular cloning and sequence analysis of a phenylalanine ammonia-lyase gene from Dendrobium. PLoS ONE 2013, 8, e62352. [Google Scholar]
- Jiang, Y.M.; Xia, B.; Liang, L.J.; Li, X.D.; Xu, S.; Peng, F.; Wang, R. Molecular and analysis of a phenylalanine ammonia-lyase gene (LrPAL2) from Lycoris radiata. Mol. Biol. Rep. 2013, 40, 2293–2300. [Google Scholar]
- Kim, S.H.; Kronstad, J.W.; Ellis, B.E. Purification and characterization of phenylalanine ammonia-lyase from Ustilago maydis. Phytochemistry 1996, 43, 351–357. [Google Scholar]
- Hattori, T.; Nishiyama, A.; Shimada, M. Induction of l-phenylalanine ammonia-lyase and suppression of veratryl alcohol biosynthesis by exogenously added l-phenylalanine in a white-rot fungus Phanerochaete chrysosporium. FEMS Microbiol. Lett. 1999, 179, 305–309. [Google Scholar] [PubMed]
- Moffitt, M.C.; Louie, G.V.; Bowman, M.E.; Pence, J.; Noel, J.P.; Moore, B.S. Discovery of two cyanobacterial phenylalanine ammonia lyases: Kinetic and structural characterization. Biochemistry 2007, 46, 1004–1012. [Google Scholar] [PubMed]
- Xiang, L.K.; Moore, B.S. Biochemical characterization of a prokaryotic phenylalanine ammonia lyase. J. Bacteriol. 2005, 187, 4286–4289. [Google Scholar] [PubMed]
- Zhang, C.Z.; Wang, X.; Zhang, F.; Dong, L.D.; Wu, J.J.; Cheng, Q.; Qi, D.Y.; Yan, X.F.; Jiang, L.Y.; Fan, S.J.; et al. Phenylalanine ammonia-lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 2017, 7, 7142. [Google Scholar]
- Xu, F.; Cai, R.; Cheng, S.; Du, H.; Wang, Y. Molecular cloning, characterization and expression of phenylalanine ammonia-lyase gene from Ginkgo biloba. Afr. J. Biotechnol. 2008, 7, 721–729. [Google Scholar]
- Jiang, Y.M.; Xia, N.; Li, X.D.; Shen, W.B.; Liang, L.J.; Wang, C.Y.; Wang, R.; Peng, F.; Xia, B. Molecular cloning and characterization of a phenylalanine ammonia-lyase gene (LrPAL) from Lycoris radiata. Mol. Biol. Rep. 2011, 38, 1935–1940. [Google Scholar]
- Kasirajan, L.; Aruchamy, K.; Thirugnanasambandam, P.P.; Athiappan, S. Molecular cloning, characterization, and expression analysis of lignin genes from Sugarcane genotypes varying in lignin content. Appl. Biochem. Biotechnol. 2017, 181, 1270–1282. [Google Scholar] [PubMed]
- Thiyagarajan, K.; Vitali, F.; Tolaini, V.; Galeffi, P.; Cantale, C.; Vikram, P.; Singh, S.; De Rossi, P.; Nobili, C.; Procacci, S.; et al. Genomic characterization of phenylalanine ammonia lyase gene in Buckwheat. PLoS ONE 2016, 11, e151187. [Google Scholar]
- Gayoso, C.; Pomar, F.; Novo-Uzal, E.; Merino, F.; Ilárduya, O.M.D. The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 2010, 10, 232. [Google Scholar]
- Tonnessen, B.W.; Manosalva, P.; Lang, J.M.; Baraoidan, M.; Bordeos, A.; Mauleon, R.; Oard, J.; Hulbert, S.; Leung, H.; Leach, J.E. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 2015, 87, 273–286. [Google Scholar] [PubMed]
- Huang, J.L.; Gu, M.; Lai, Z.B.; Fan, B.F.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z.X. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [PubMed]
- Yaqoob, N.; Yakovlev, I.A.; Krokene, P.; Kvaalen, H.; Solheim, H.; Fossdal, C.G. Defense-related gene expression in bark and sapwood of Norway spruce in response to Heterobasidion parviporum and methyl jasmonate. Physiol. Mol. Plant Pathol. 2012, 77, 10–16. [Google Scholar]
- Koutaniemi, S.; Warinowski, T.; Kärkönen, A.; Alatalo, E.; Fossdal, C.G.; Saranpää, P.; Laakso, T.; Fagerstedt, K.V.; Simola, L.K.; Paulin, L.; et al. Expression profiling of the lignin biosynthetic pathway in Norway spruce using EST sequencing and real-time RT-PCR. Plant Mol. Biol. 2007, 65, 311–328. [Google Scholar] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [PubMed]
- Zhu, Q.L.; Xie, X.R.; Lin, H.X.; Sui, S.Z.; Shen, R.X.; Yang, Z.F.; Lu, K.; Li, M.Y.; Liu, Y.G. Isolation and functional characterization of a phenylalanine ammonia-lyase gene (SsPAL1) from Coleus (Solenostemon scutellarioides (L.) Codd). Molecules 2015, 20, 16833–16851. [Google Scholar] [PubMed]
- Zhang, T.; Zhang, D.; Liu, Y.J.; Luo, C.B.; Zhou, Y.N.; Zhang, L.Y. Overexpression of a NF-YB3 transcription factor from Picea wilsonii confers tolerance to salinity and drought stress in transformed Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 94, 153–164. [Google Scholar] [PubMed]
- Kolosova, N.; Breuil, C.; Bohlmann, J. Cloning and characterization of chitinases from interior spruce and lodgepole pine. Phytochemistry 2014, 101, 32–39. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ÄÄCT method. Methods 2001, 25, 402–408. [Google Scholar] [PubMed]
- Macdonald, M.J.; D’Cunha, G.B. A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. 2007, 85, 273–282. [Google Scholar] [PubMed]
- Ritter, H.; Schulz, G.E. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Plant Cell 2004, 16, 3426–3436. [Google Scholar] [PubMed]
- Hahlbrock, K.; Scheel, D. Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 347–369. [Google Scholar]
- Li, C.L.; Bai, Y.C.; Chen, H.; Zhao, H.X.; Shao, J.R.; Wu, Q. Cloning, characterization and functional analysis of a phenylalanine ammonia-lyase gene (FtPAL) from Fagopyrum tataricum Gaertn. Plant Mol. Biol. Rep. 2012, 30, 1172–1182. [Google Scholar]
- Chaman, M.E.; Copaja, S.V.; Argandoña, V.H. Relationships between salicylic acid content, phenylalanine ammonia-lyase (PAL) activity, and resistance of barley to aphid infestation. J. Agric. Food Chem. 2003, 51, 2227–2231. [Google Scholar]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [PubMed]
- Li, H.; Yu, Y.; Li, Z.Z.; Arkorful, E.; Yang, Y.Y.; Liu, X.Q.; Li, X.H.; Li, R.L. Benzothiadiazole and b-aminobutyricacid induce resistance to Ectropis obliqua in tea plants (Camellia sinensis (L.) O. Kuntz). Molecules 2018, 23, 1290. [Google Scholar]
- Hou, X.M.; Shao, F.J.; Ma, Y.M.; Lu, S.F. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: Genome-wide characterization, molecular cloning and expression analysis. Mol. Biol. Rep. 2013, 40, 4301–4310. [Google Scholar]
- Zhu, L.B.; Cui, W.J.; Fang, Y.Q.; Liu, Y.; Gao, X.X.; Zhou, Z.M. Cloning, expression and characterization of phenylalanine ammonia-lyase from Rhodotorula glutinis. Biotechnol. Lett. 2013, 35, 751–756. [Google Scholar]
- Hsieh, L.S.; Hsieh, Y.L.; Yeh, C.S.; Cheng, C.Y.; Yang, C.C.; Lee, P.D. Molecular characterization of a phenylalanine ammonia-lyase gene (BoPAL1) from Bambusa oldhamii. Mol. Biol. Rep. 2011, 38, 283–290. [Google Scholar]
- Mahesh, V.; Rakotomalala, J.J.; Le Gal, L.; Vigne, H.; de Kochko, A.; Hamon, S.; Noirot, M.; Campa, C. Isolation and genetic mapping of a Coffea canephora phenylalanine ammonia-lyase gene (CcPAL1) and its involvement in the accumulation of caffeoyl quinic acids. Plant Cell Rep. 2006, 25, 986–992. [Google Scholar] [PubMed]
- Gao, J.H.; Zhang, S.W.; Cai, F.; Zheng, X.J.; Lin, N.; Qin, X.B.; Ou, Y.C.; Gu, X.P.; Zhu, X.H.; Xu, Y.; et al. Characterization, and expression profile of a phenylalanine ammonia lyase gene from Jatropha curcas L. Mol. Biol. Rep. 2012, 39, 3443–3452. [Google Scholar] [PubMed]
- Dehghan, S.; Sadeghi, M.; Pöppel, A.; Fischer, R.; Lakes-Harlan, R.; Kavousi, H.R.; Vilcinskas, A.; Rahnamaeian, M. Differential inductions of phenylalanine ammonia-lyase and chalcone synthase during wounding, salicylic acid treatment, and salinity stress in safflower, Carthamus tinctorius. Biosci. Rep. 2014, 34, e114. [Google Scholar]
- Khakdan, F.; Alizadeh, H.; Ranjbar, M. Molecular cloning, functional characterization and expression of a drought inducible phenylalanine ammonia-lyase gene (ObPAL) from Ocimum basilicum L. Plant Physiol. Biochem. 2018, 130, 464–472. [Google Scholar] [PubMed]
- Calabrese, J.C.; Jordan, D.B.; Boodhoo, A.; Sariaslani, S.; Vannelli, T. Crystal structure of phenylalanine ammonia lyase: Multiple helix dipoles implicated in catalysis. Biochemistry 2004, 43, 11403–11416. [Google Scholar]
- Weitzel, C.; Petersen, M. Enzymes of phenylpropanoid metabolism in the important medicinal plant Melissa officinalis L. Planta 2010, 232, 731–742. [Google Scholar] [PubMed]
- Okada, T.; Mikage, M.; Sekita, S. Molecular characterization of the phenylalanine ammonia-lyase from Ephedra sinica. Biol. Pharm. Bull. 2008, 31, 2194–2199. [Google Scholar]
- Wanner, L.A.; Li, G.; Ware, D.; Somssich, I.E.; Davis, K.R. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1995, 27, 327–338. [Google Scholar]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar]
- Song, J.; Wang, Z.Z. Molecular cloning, expression and characterization of a phenylalanine ammonia-lyase gene (SmPAL1) from Salvia miltiorrhiza. Mol. Biol. Rep. 2008, 36, 939–952. [Google Scholar]
- Bassard, J.E.; Richert, L.; Geerinck, J.; Renault, H.; Duval, F.; Ullmann, P.; Schmitt, M.; Meyer, E.; Mutterer, J.; Boerjan, W. Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 2012, 24, 4465–4482. [Google Scholar] [PubMed]
- Esposito, D.; Chatterjee, D.K. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 2006, 17, 353–358. [Google Scholar] [PubMed]
- Li, S.J.; Zhang, B.Y.; Zhu, H.M.Y.; Zhu, T.H. Cloning and expression of the chitinase encoded by ChiKJ406136 from Streptomyces sampsonii (Millard & Burr) Waksman KJ40 and its antifungal effect. Forests 2018, 9, 699. [Google Scholar]
- Oneda, H.; Inouye, K. Refolding and recovery of recombinant human matrix metalloproteinase 7 (matrilysin) from inclusion bodies expressed by Escherichia coli. J. Biochem. 1999, 126, 905–911. [Google Scholar] [PubMed]
- Baneyx, F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 1999, 10, 411–421. [Google Scholar] [PubMed]
- Urban, A.; Ansmant, I.; Motorin, Y. Optimisation of expression and purification of the recombinant Yol066 (Rib2) protein from Saccharomyces cerevisiae. J. Chromatogr. B 2003, 786, 187–195. [Google Scholar]
- Ma, W.L.; Wu, M.; Wu, Y.; Ren, Z.M.; Zhong, Y. Cloning and characterisation of a phenylalanine ammonia-lyase gene from Rhus chinensis. Plant Cell Rep. 2013, 32, 1179–1190. [Google Scholar]
- Sarma, A.D.; Sharma, R. Purification and characterization of uv-b induced phenylalanine ammonia-lyase from rice seedlings. Phytochemistry 1999, 50, 729–737. [Google Scholar]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar]
- Logemann, E.; Parniske, M.; Hahlbrock, K. Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc. Natl. Acad. Sci. USA 1995, 92, 5905–5909. [Google Scholar]
- Rösler, J.; Krekel, F.; Amrhein, N.; Schmid, J. Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 1997, 113, 175–179. [Google Scholar] [PubMed]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [PubMed]
- Zhu, Q.; Dabi, T.; Beeche, A.; Yamamoto, R.; Lawton, M.A.; Lamb, C. Cloning and properties of a rice gene encoding phenylalanine ammonia-lyase. Plant Mol. Biol. 1995, 29, 535–550. [Google Scholar] [PubMed]
- Kumar, A.; Ellis, B.E. The phenylalanine ammonia-lyase gene family in raspberry. Structure, expression, and evolution. Plant Physiol. 2001, 127, 230–239. [Google Scholar] [PubMed]
- Lee, B.K.; Park, M.R.; Srinivas, B.; Chun, J.C.; Kwon, I.S.; Chung, I.M.; Yoo, N.H.; Choi, K.G.; Yun, S.J. Induction of phenylalanine ammonia-lyase gene expression by paraquat and stress-related hormones in Rehmannia glutinosa. Mol. Cells 2003, 16, 34–39. [Google Scholar] [PubMed]
- Appert, C.; Logemann, E.; Hahlbrock, K.; Schmid, J.; Amrhein, N. Structural and catalytic properties of the four phenylalanine ammonia-lyase isoenzymes from parsley (Petroselinum crispum Nym.). Eur. J. Biochem. 1994, 225, 491–499. [Google Scholar] [PubMed]
- Lu, B.B.; Du, Z.; Ding, R.X.; Zhang, L.; Yu, X.J.; Liu, C.H.; Chen, W.S. Cloning and characterization of a differentially expressed phenylalanine ammonialyase gene (IiPAL) after genome duplication from tetraploid Isatis indigotica Fort. J. Integr. Plant Biol. 2006, 48, 1439–1449. [Google Scholar]
- Pellegrini, L.; Rohfritsch, O.; Fritig, B.; Legrand, M. Phenylalanine ammonia-lyase in tobacco. Molecular cloning and gene expression during the hypersensitive reaction to tobacco mosaic virus and the response to a fungal elicitor. Plant Physiol. 1994, 106, 877–886. [Google Scholar]
- Liu, R.R.; Xu, S.H.; Li, J.L.; Hu, Y.L.; Lin, Z.P. Expression profile of a PAL gene from Astragalus membranaceus var. Mongholicus and its crucial role in flux into flavonoid biosynthesis. Plant Cell Rep. 2006, 25, 705–710. [Google Scholar]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar]
- Wojtasik, W.; Kulma, A.; Dymińska, L.; Hanuza, J.; Czemplik, M.; Szopa, J. Evaluation of the significance of cell wall polymers in flax infected with a pathogenic strain of Fusarium oxysporum. BMC Plant Biol. 2016, 16, 75. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Liu, L.; Yang, S.; Zeng, Q.; He, Z.; Liu, Y. Cloning, Characterization and Expression of the Phenylalanine Ammonia-Lyase Gene (PaPAL) from Spruce Picea asperata. Forests 2019, 10, 613. https://doi.org/10.3390/f10080613
Liu Y, Liu L, Yang S, Zeng Q, He Z, Liu Y. Cloning, Characterization and Expression of the Phenylalanine Ammonia-Lyase Gene (PaPAL) from Spruce Picea asperata. Forests. 2019; 10(8):613. https://doi.org/10.3390/f10080613
Chicago/Turabian StyleLiu, Yufeng, Lijuan Liu, Shuai Yang, Qian Zeng, Zhiran He, and Yinggao Liu. 2019. "Cloning, Characterization and Expression of the Phenylalanine Ammonia-Lyase Gene (PaPAL) from Spruce Picea asperata" Forests 10, no. 8: 613. https://doi.org/10.3390/f10080613
APA StyleLiu, Y., Liu, L., Yang, S., Zeng, Q., He, Z., & Liu, Y. (2019). Cloning, Characterization and Expression of the Phenylalanine Ammonia-Lyase Gene (PaPAL) from Spruce Picea asperata. Forests, 10(8), 613. https://doi.org/10.3390/f10080613




