Nematophagous Pleurotus Species Consume Some Nematode Species but Are Themselves Consumed by Others
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Culture
2.2. Nematode Culture
- Family RhabditidaeOscheius dolichura (Schneider) Sudhaus [LKC50], Oscheius myriophilus (Poinar) Sudhaus [DF5020], Oscheius tipulae (Lam and Webster) Sudhaus [LKC57], Caenorhabditis elegans (Maupas) Dougherty [N2], Mesorhabditis inarimensis (Meyl) Dougherty [LKC51], Poikilolaimus oxycercus (de Man) Sudhaus and Koch [LKC64], and Metarhabditis rainai (Carta and Osbrink) Sudhaus [LKC20]
- Family CephalobidaeZeldia punctata (Thorne) Thorne [PS1192], Acrobeloides varius Kim, Kim and Park [LKC52], Acrobeloides varius [PS1959], Acrobeloides varius [LKC27], and Acrobeloides sp. cf amurensis Truskova [PS1146]
- Family PanagrolaimidaePanagrolaimus artyukhovskii Blinova & Mishina [LKC44] and Panagrellus redivivus (Linnaeus) Thorne [PS1163]
- Family DiplogastridaePristionchus aerivorus (Cobb in Merrill & Ford) Chitwood [LKC54]To avoid redundancy, only one population of A. varius (LKC52) was used for the P. ostreatus assay. All three populations were tested with P. pulmonarius.
2.3. Pleurotus–Nematode Interaction Assay
2.4. Phylogenetic Analysis
2.5. Fungal-Feeding Ability
3. Results
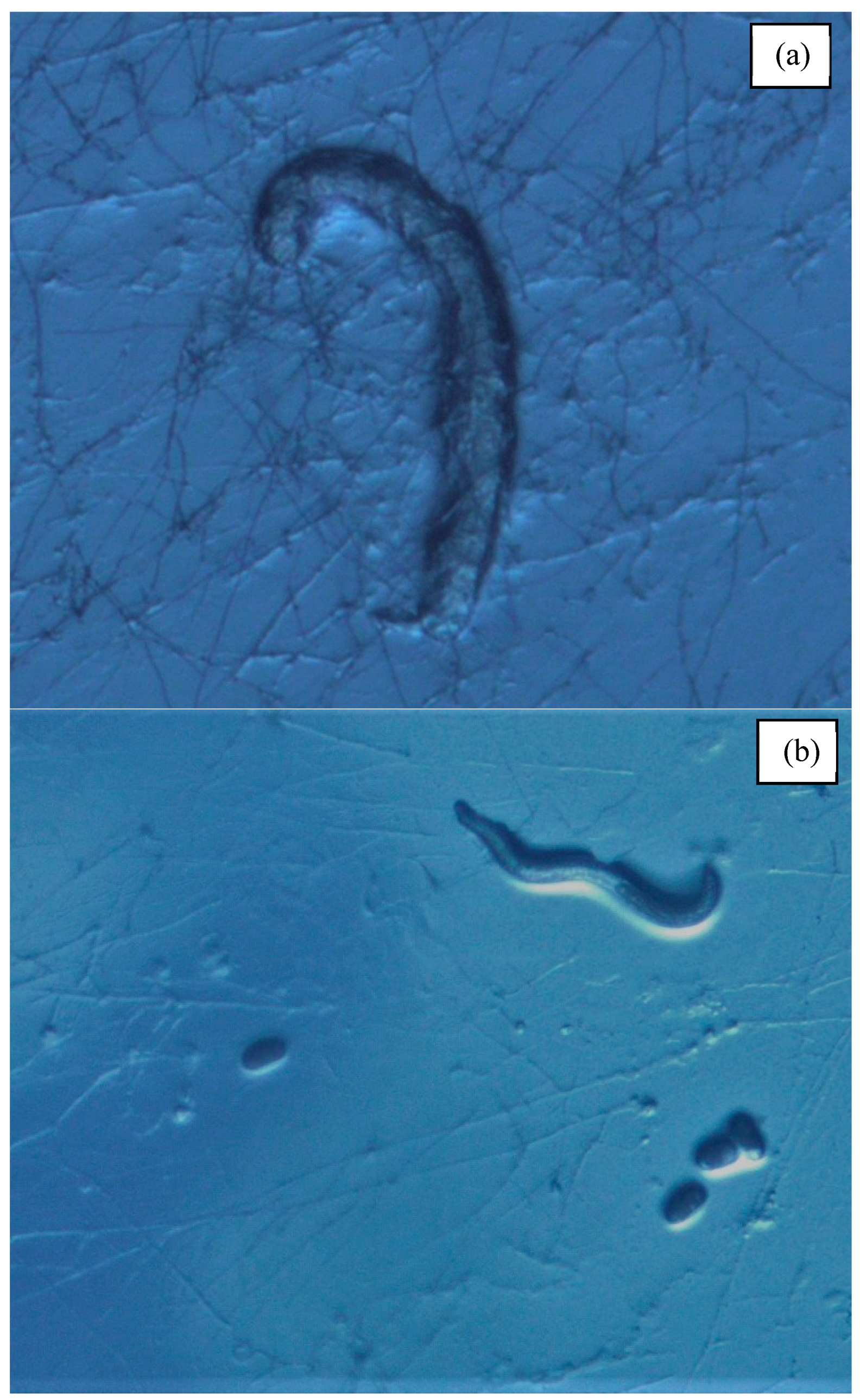
3.1. Pleurotus pulmonarius–Nematode Interaction Assay
3.2. Pleurotus ostreatus–Nematode Interaction Assay
3.3. Phylogenetic Analyses
3.4. Feeding Ability
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clarke, N.E.; Doi, S.A.R.; Wangdi, K.; Chen, Y.; Clements, A.C.A.; Nery, S.V. Efficacy of anthelminthic drugs and drug combinations against soil-transmitted helminths: A systematic review and network meta-analysis. Clin. Infect. Dis. 2019, 68, 96–105. [Google Scholar] [CrossRef]
- Stirling, G.R.; Stirling, A.M.; Walter, D.E. The Mesostigmatid Mite Protogamasellus mica, an Effective Predator of Free-Living and Plant-Parasitic Nematodes. J. Nematol. 2017, 49, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.-Z.; Hale, K.; Carta, L.; Platzer, E.; Wong, C.; Fang, S.-C.; Aroian, R.V. Bacillus thuringiensis crystal proteins that target nematodes. Proc. Acad. Sci. 2003, 100, 2760–2765. [Google Scholar] [CrossRef] [PubMed]
- Page, A.P.; Roberts, M.; Félix, M.-A.; Pickard, D.; Page, A.; Weir, W. The golden death bacillus Chryseobacterium nematophagum is a novel matrix digesting pathogen of nematodes. BMC Boil. 2019, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Nordbring-Hertz, B.; Jansson, H.B.; Tunlid, A. Nematophagous fungi. Enc. Life Sci. 2001. [Google Scholar] [CrossRef]
- Zhang, K.-Q.; Hyde, K.D. Nematode-Trapping Fungi; Springer: Dordrecht, The Netherlands, 2014. [Google Scholar]
- Thorn, R.G.; Moncalvo, J.-M.; Reddy, C.A.; Vilgalys, R. Phylogenetic Analyses and the Distribution of Nematophagy Support a Monophyletic Pleurotaceae within the Polyphyletic Pleurotoid-Lentinoid Fungi. Mycologia 2000, 92, 241. [Google Scholar] [CrossRef]
- Thorn, R.G.; Barron, G.L. Carnivorous mushrooms. Science 1984, 224, 76–78. [Google Scholar] [CrossRef]
- Soares, F.E.D.F.; Sufiate, B.L.; De Queiróz, J.H. Nematophagous fungi: Far beyond the endoparasite, predator and ovicidal groups. Agric. Resour. 2018, 52, 1–8. [Google Scholar] [CrossRef]
- Tzean, S.S. Nematophagous Resupinate Basidiomycetous Fungi. Phytopathology 1993, 83, 1015. [Google Scholar] [CrossRef]
- Okada, H.; Kadota, I. Host status of 10 fungal isolates for two nematode species, Filenchus misellus and Aphelenchus avenae. Soil Boil. Biochem. 2003, 35, 1601–1607. [Google Scholar] [CrossRef]
- Okada, H.; Harada, H.; Kadota, I. Fungal-feeding habits of six nematode isolates in the genus Filenchus. Soil Boil. Biochem. 2005, 37, 1113–1120. [Google Scholar] [CrossRef]
- Barron, G.L.; Thorn, R.G. Destruction of nematodes by species of Pleurotus. Can. J. Bot. 1987, 65, 774–778. [Google Scholar] [CrossRef]
- Okada, H.; Araki, M.; Tsukiboshi, T.; Harada, H. Characteristics of Tylencholaimus parvus (Nematoda: Dorylaimida) as a fungivorus nematode. Nematology 2005, 7, 843–849. [Google Scholar] [CrossRef]
- Thorn, R.G.; Tsuneda, A. Interactions between various wood-decay fungi and bacteria: antibiosis, attack, lysis, or inhibition. Rep. Tottori Mycol. Inst. 1992, 30, 13–20. [Google Scholar] [CrossRef]
- Larsen, M.; Nansen, P. Ability of the fungus Pleurotus pulmonarius to immobilize preparasitic nematode larvae. Res. Vet. Sci. 1991, 51, 246–249. [Google Scholar] [CrossRef]
- Hibbett, D.S.; Thorn, R.G. Nematode-Trapping in Pleurotus tuberregium. Mycologia 1994, 86, 696. [Google Scholar] [CrossRef]
- Kwok, O.C.H.; Plattner, R.; Weisleder, D.; Wicklow, D.T. A nematicidal toxin fromPleurotus ostreatus NRRL 3526. J. Chem. Ecol. 1992, 18, 127–136. [Google Scholar] [CrossRef]
- Stadler, M.; Mayer, A.; Anke, H.; Sterner, O. Fatty Acids and Other Compounds with Nematicidal Activity from Cultures of Basidiomycetes. Planta Med. 1994, 60, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Trudell, S.; Ammirati, J. Mushrooms of the Pacific Northwest; Timber Press: Portland, OR, USA, 2009; pp. 134–135. [Google Scholar]
- Stiernagle, T. Maintenance of C. elegans. In C. elegans: A Practical Approach; Hope, I.A., Ed.; Oxford University Press: New York, NY, USA, 1999; pp. 51–68. [Google Scholar]
- Carta, L.K.; Li, S. Improved 18S small subunit rDNA primers for problematic nematode amplification. J. Nematol. 2018, 50, 533–542. [Google Scholar] [CrossRef]
- Van Megen, H.; Elsen, S.V.D.; Holterman, M.; Karssen, G.; Mooyman, P.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 2009, 11, 927–950. [Google Scholar]
- Blaxter, M.; Koutsovoulos, K. The evolution of parasitism in Nematoda. Parasitology 2015, 142 (Suppl. S1), S26–S39. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F.; Hall, B. MrBayes: A Program for the Bayesian Inference of Phylogeny; Software Manual; University of Rochester: Rochester, NY, USA, 2000. [Google Scholar]
- Barron, G.L. Nematophagous Fungi: Endoparasites of Rhabditis terricola. Microb. Ecol. 1978, 4, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Carnegie, A.J.; Lidbetter, J.R. Rapidly expanding host range for Puccinia psidii sensu lato in Australia. Australas. Australas. Plant Pathol. 2012, 41, 13–29. [Google Scholar] [CrossRef]
- Marx, D.H. Tree host range and world distribution of the ectomycorrhizal fungus Pisolithus tinctorius. Can. J. Microbiol. 1977, 23, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Merckx, V.S.F.T.; Janssens, S.B.; Hynson, N.A.; Specht, C.D.; Bruns, T.D.; Smets, E.F. Mycoheterotrophic interactions are not limited to a narrow phylogenetic range of arbuscular mycorrhizal fungi. Mol. Ecol. 2012, 21, 1524–1532. [Google Scholar] [CrossRef]
- Procter, D.L.C. Fecundity, reproductive effort, age-specific reproductive tactics and intrinsic rate of natural increase of a High Arctic nematode belonging to the genus Chiloplacus. Ecography 1986, 9, 104–108. [Google Scholar] [CrossRef]
- Waller, P.; Faedo, M. The potential of nematophagous fungi to control the free-living stages of nematode parasites of sheep: screening studies. Vet. Parasitol. 1993, 49, 285–297. [Google Scholar] [CrossRef]
- Jansson, H.-B.; Jeyaprakash, A.; Zuckerman, B.M. Differential Adhesion and Infection of Nematodes by the Endoparasitic Fungus Meria coniospora (Deuteromycetes). Appl. Environ. Microbiol. 1985, 49, 552–555. [Google Scholar]
- Jansson, H.-B.; Nordbring-Hertz, B. Involvement of Sialic Acid in Nematode Chemotaxis and Infection by an Endoparasitic Nematophagous Fungus. Microbiology 1984, 130, 39–43. [Google Scholar] [CrossRef][Green Version]
- Dirksen, P.; Marsh, S.A.; Braker, I.; Heitland, N.; Wagner, S.; Nakad, R.; Mader, S.; Petersen, C.; Kowallik, V.; Rosenstiel, P.; et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Boil. 2016, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Köthe, M.; Antl, M.; Huber, B.; Stoecker, K.; Ebrecht, D.; Steinmetz, I.; Eberl, L. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 2003, 5, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Carta, L.K. Bacterial-Feeding Nematode Growth and Preference for Biocontrol Isolates of the Bacterium Burkholderia cepacia. J. Nematol. 2000, 32, 362–369. [Google Scholar] [PubMed]
- Tahseen, Q.; Clark, I.M. Attraction and preference of bacteriophagous and plant-parasitic nematodes towards different types of soil bacteria. J. Hist. 2014, 48, 1485–1502. [Google Scholar] [CrossRef]
- Ferris, H.; Bongers, T.; De Goede, R. A framework for soil food web diagnostics: extension of the nematode faunal analysis concept. Appl. Soil Ecol. 2001, 18, 13–29. [Google Scholar] [CrossRef]
- Kitchen, S.; Ratnappan, R.; Han, S.; Leasure, C.; Grill, E.; Iqbal, Z.; Granger, O.; O’Halloran, D.M.; Hawdon, J.M. Isolation and characterization of a naturally occurring multidrug-resistant strain of the canine hookworm, Ancylostoma caninum. Int. J. Parasitol. 2019, 49, 397–406. [Google Scholar] [CrossRef]
- Mamiya, Y.; Hiratsuka, M.; Murata, M. Ability of wood-decay fungi to prey on the pinewood nematode, Bursaphelenchus xylophilus (Steiner and Buhrer) Nickle. Jpn. J. Nematol. 2005, 35, 21–30. [Google Scholar] [CrossRef]
- Renker, C.; Alphei, J.; Buscot, F. Soil nematodes associated with the mammal pathogenic fungal genus Malassezia (Basidiomycota: Ustilaginomoycetes) in Central European forests. Biol. Fertil. Soils 2003, 37, 70–72. [Google Scholar]
- Platt, H.M. The Phylogenetic Systematics of Free-Living Nematodes; The Ray Society: London, UK, 1994. [Google Scholar]
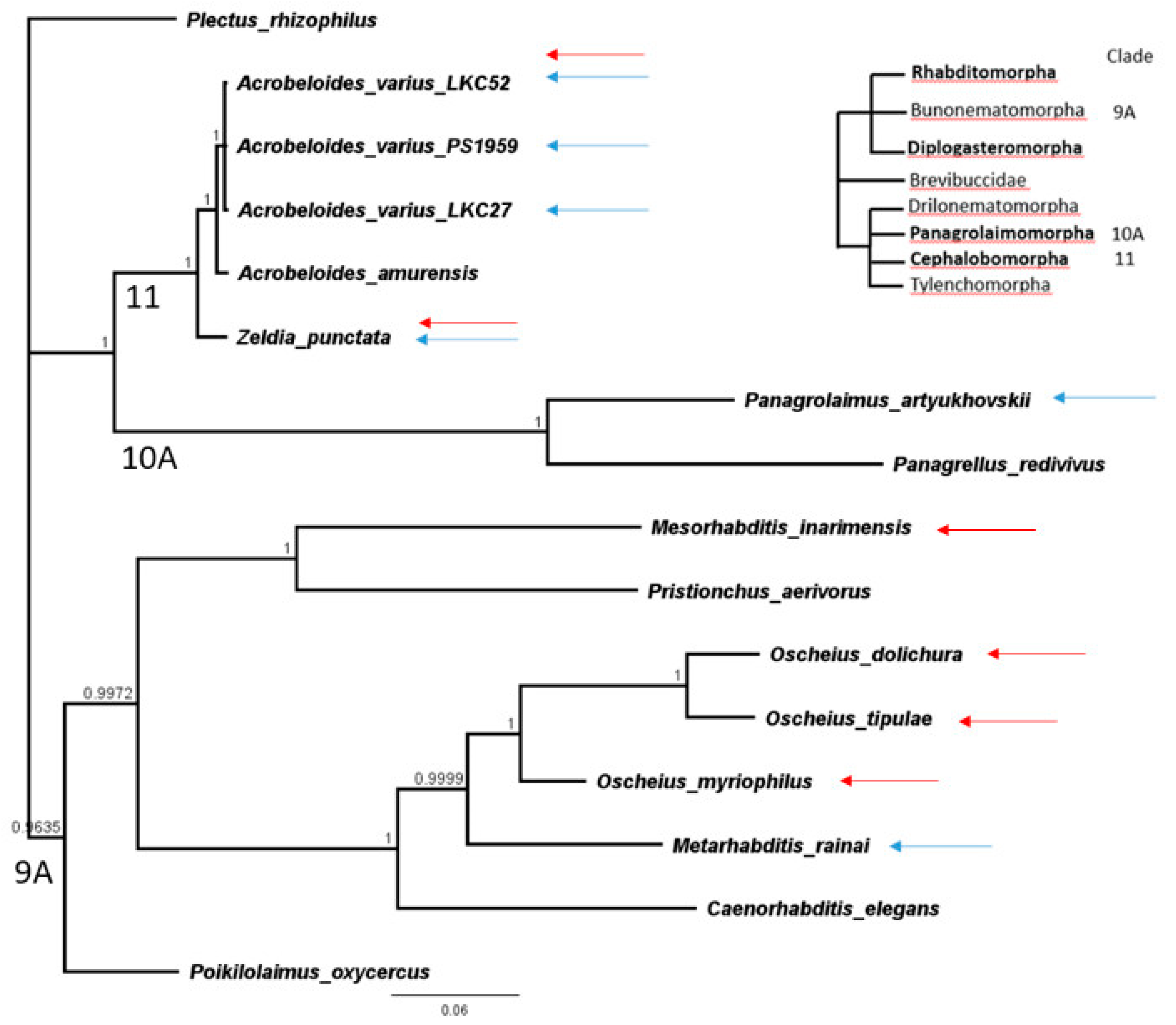
| Species | Culture Isolate | GenBank Accession/Isolate | Clade in van Megen et al., 2009 [23] |
|---|---|---|---|
| Oscheius dolichura | LKC50 | KP756940 JU72 | 9A Rhabditomorpha |
| Oscheius myriophilus | DF5020 | U81588 | 9A |
| Oscheius tipulae | LKC57 | CEW1 KP756939 | 9A |
| Caenorhabditis elegans | N2 | NR000054 | 9A |
| Mesorhabditis inarimensis | LKC51 | 90A3 * MK636575 | 9A |
| Poikilolaimus oxycercus | LKC64 | 101A3* MK636576 | 9A |
| Metarhabditis rainai | LKC20 | AF083008 PS1191 | 9A |
| Pristionchus aerivorus | LKC54 | 90C1 * MK636577 | 9A Diplogasteromorpha |
| Panagrolaimus artyukhovskii | LKC44 | 90E9 * MK636578 | 10A Panagrolaimomorpha |
| Panagrellus redivivus | PS1163 | AF083007 | 10A |
| Zeldia punctata | PS1192 | U61760 | 11 Cephalobomorpha |
| Acrobeloides amurensis | PS1146 | AF034391 | 11 |
| Acrobeloides varius ** | LKC27 | 94A6 * MK636581 | 11 |
| Acrobeloides varius | LKC52 | 100H3 * MK636579 | 11 |
| Acrobeloides varius ** | PS1959 | 104M16 * MK636580 | 11 |
| Plectus rhizophilus | PlecRhi1 | AY593928 | 6 (Outgroup) |
| Nematode Species | Pleurotus pulmonarius | Pleurotus ostreatus |
|---|---|---|
| Tylencholaimus parvus | S | S |
| Aphelenchus avenae | R | R |
| Species | Culture Isolate | Pleurotus pulmonarius | Pleurotus ostreatus |
|---|---|---|---|
| Oscheius dolichura | LKC50 | S | R |
| Oscheius myriophilus | DF5020 | S | R |
| Oscheius tipulae | LKC57 | S | R |
| Caenorhabditis elegans | N2 | S | S |
| Mesorhabditis inarimensis | LKC51 | S | R |
| Poikilolaimus oxycercus | LKC64 | S | S |
| Metarhabditis rainai | LKC20 | R | S |
| Pristionchus aerivorus | LKC54 | S | S |
| Panagrolaimus artyukhovskii | LKC44 | R | S |
| Panagrellus redivivus | PS1163 | S | S |
| Zeldia punctata | PS1192 | R | R |
| Acrobeloides amurensis | PS1146 | S | S |
| Acrobeloides varius ** | LKC27 | R | NA |
| Acrobeloides varius | LKC52 | R | R |
| Acrobeloides varius ** | PS1959 | R | NA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marlin, M.; Wolf, A.; Alomran, M.; Carta, L.; Newcombe, G. Nematophagous Pleurotus Species Consume Some Nematode Species but Are Themselves Consumed by Others. Forests 2019, 10, 404. https://doi.org/10.3390/f10050404
Marlin M, Wolf A, Alomran M, Carta L, Newcombe G. Nematophagous Pleurotus Species Consume Some Nematode Species but Are Themselves Consumed by Others. Forests. 2019; 10(5):404. https://doi.org/10.3390/f10050404
Chicago/Turabian StyleMarlin, Maria, Avery Wolf, Maryam Alomran, Lynn Carta, and George Newcombe. 2019. "Nematophagous Pleurotus Species Consume Some Nematode Species but Are Themselves Consumed by Others" Forests 10, no. 5: 404. https://doi.org/10.3390/f10050404
APA StyleMarlin, M., Wolf, A., Alomran, M., Carta, L., & Newcombe, G. (2019). Nematophagous Pleurotus Species Consume Some Nematode Species but Are Themselves Consumed by Others. Forests, 10(5), 404. https://doi.org/10.3390/f10050404






