Genetic Diversity and Differentiation of Relict Plant Liriodendron Populations Based on 29 Novel EST-SSR Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
2.2. EST-SSR Development and Genotyping
2.3. Analysis of Genetic Diversity and Genetic Relationship
3. Results
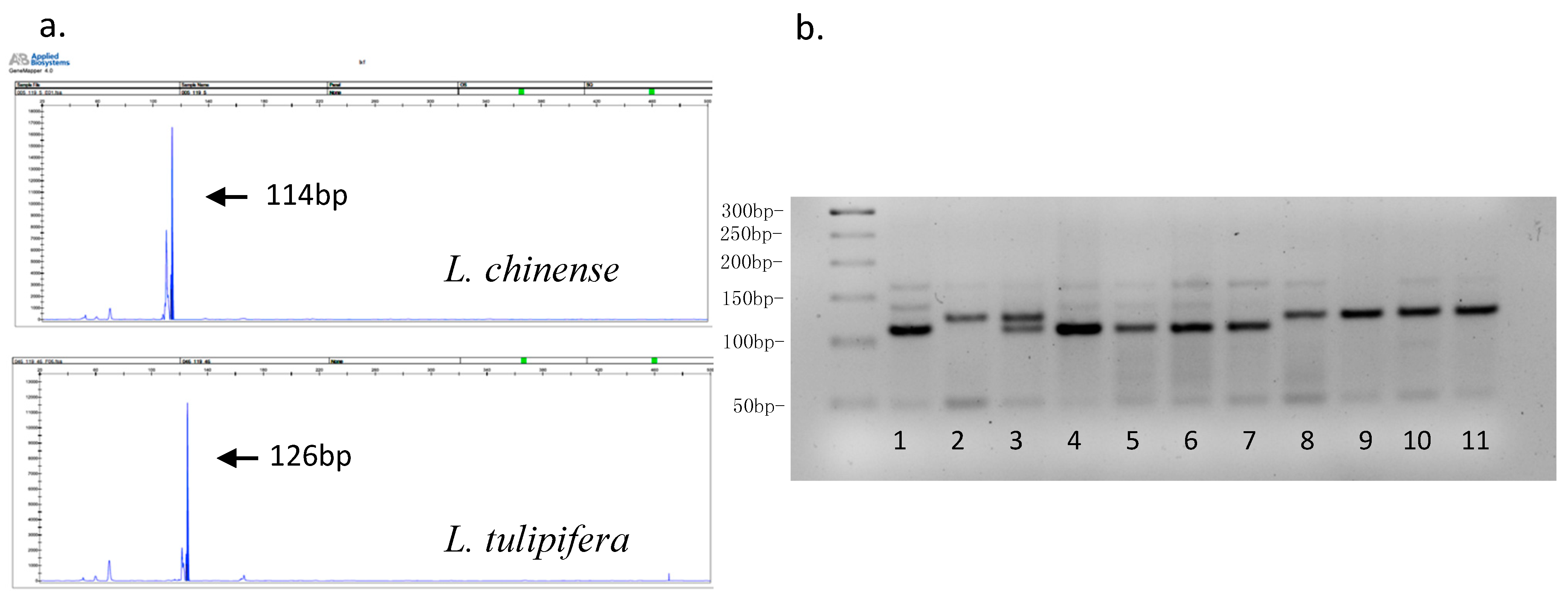
3.1. Microsatellite Development
3.2. EST-SSR Polymorphism and Genetic Diversity
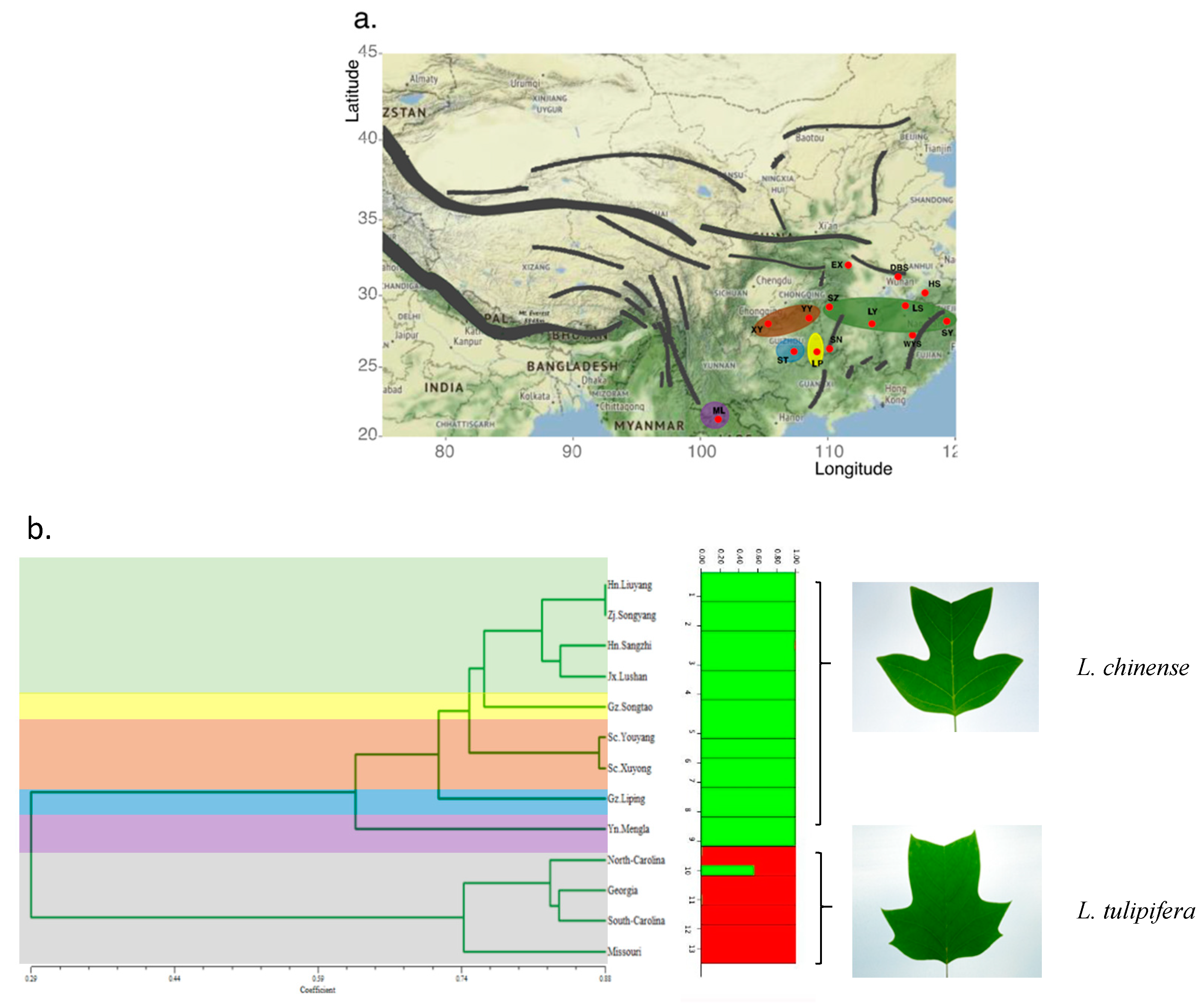
3.3. Population Genetic Structure of Liriodendron
3.4. Cluster Analysis Using EST-SSR Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| L. chinense | Liriodendron chinense |
| L. tulipifera | Liriodendron tulipifera |
| EST-SSR | Expressed Sequence Tag/Microsatellites or simple sequence repeats |
| MAS | Marker-assisted selection |
| CTAB | Cetyltrimethyl Ammonium Bromide |
| Na | Number of alleles |
| Ho | observed heterozygosity |
| He | expected heterozygosity |
| HWE | Hardy-Weinberg equilibrium text |
| I | Shannon index |
| PIC | Polymorphism Information Content |
References
- Parks, C.R.; Wendel, J.F. Molecular Divergence between Asian and North American Species of Liriodendron (Magnoliaceae) with Implications for Interpretation of Fossil Floras. Am. J. Bot. 1990, 77, 1243–1256. [Google Scholar] [CrossRef]
- Xiang, Q.-Y.; Soltis, D.E.; Soltis, P.S.; Manchester, S.R.; Crawford, D.J. Timing the Eastern Asian–Eastern North American Floristic Disjunction: Molecular Clock Corroborates Paleontological Estimates. Mol. Phylogenet. Evol. 2000, 17, 335. [Google Scholar] [CrossRef]
- Ming, H.R.; An, H.S. Geographical distribution of Liriodendron chinense in China and its significance. J. Plant Resour. Environ. 1995, 4, 1–6. [Google Scholar]
- Xiang, Q.-Y.; Soltis, D.E.; Soltis, P.S. The Eastern Asian and Eastern and Western North American Floristic Disjunction: Congruent Phylogenetic Patterns in Seven Diverse Genera. Mol. Phylogenet. Evol. 1998, 10, 178–190. [Google Scholar] [CrossRef]
- Zheng, W. Dendrology of China; China Forestry Press: Beijing, China, 1983; Volume 1. (In Chinese) [Google Scholar]
- Zhang, Y.-B.; Ma, K.-P. Geographic distribution patterns and status assessment of threatened plants in China. Biodivers. Conserv. 2008, 17, 1783–1798. [Google Scholar] [CrossRef]
- Fang, Y. Geographical distribution and spatial pattern of Liriodendron chinense. J. Nanjing For. Univ. Nat. Sci. Ed. 1994, 18, 13–18. (In Chinese) [Google Scholar]
- Hao, R.; He, S. Studies on Natural Population Dynamics and Dangerous Habitats of Liriodendron chinense. J. Plant Ecol. 1999, 23, 1. [Google Scholar] [CrossRef]
- Jin, X.U.; Wang, Z. Genetic variation of floral character and pollen viability of Liriodendron hybrid and its parents. J. Plant Resour. Environ. 2001, 10, 31–34. [Google Scholar]
- Williams, R.S.; Feist, W.C. Durability of yellow-poplar and sweetgum and service life of finishes after long-term exposure. For. Prod. J. 2004, 54, 96–101. [Google Scholar]
- Chen, J.-H.; Lin, S.-S.; Wang, W.-X.; Yuan, S.-T.; Shi, J.-S.; Jia, A.-Q. The extract, LXB-1, from the barks of Liriodendron × hybrid, induced apoptosis via Akt, JNK and ERK1/2 pathways in A549 lung cancer cells. Z. Für Nat. C 2015, 70, 305–311. [Google Scholar] [CrossRef]
- Chen, J.-H.; Yang, G.-X.; Ding, Q.; Xia, T.-S.; Shi, J.; Jia, A.-Q. In vitro tumor cytotoxic activities of extracts from three Liriodendron plants. Pak. J. Pharm. Sci. 2013, 26, 233–237. [Google Scholar]
- Moon, M.K.; Oh, H.M.; Kwon, B.-M.; Baek, N.-L.; Kim, S.-H.; Kim, J.S.; Kim, D.K. Farnesyl protein transferase and tumor cell growth inhibitory activities of lipiferolide isolated fromLiriodendron tulipifera. Arch. Pharmacal Res. 2007, 30, 299–302. [Google Scholar] [CrossRef]
- Gerloff, U.; Schlötterer, C.; Rassmann, K.; Rambold, I.; Hohmann, G.; Fruth, B.; Tautz, D. Amplification of hypervariable simple sequence repeats (microsatellites) from excremental DNA of wild living bonobos (Pan paniscus). Mol. Ecol. 1995, 4, 515–518. [Google Scholar] [CrossRef]
- Vasemägi, A.; Nilsson, J.; Primmer, C.R. Expressed Sequence Tag-Linked Microsatellites as a Source of Gene-Associated Polymorphisms for Detecting Signatures of Divergent Selection in Atlantic Salmon (Salmo salar L.). Mol. Boil. Evol. 2005, 22, 1067–1076. [Google Scholar]
- Reeve, A. Genetic diversity and differentiation of the Critically Endangered Hispaniolan palm Coccothrinax jimenezii M.M. Mejía & M.M. García based on novel SSR markers). Biochem. Syst. Ecol. 2016, 66, 216–223. [Google Scholar]
- Gutiérrez-Ozuna, R.; Hamilton, M.B. Identification and characterization of microsatellite loci in the tuliptree, Liriodendron tulipifera (Magnoliaceae). Appl. Sci. 2017, 5, 1700032. [Google Scholar] [CrossRef]
- Xu, M.; Liu, X.; Wang, J.W.; Teng, S.Y.; Shi, J.Q.; Li, Y.Y.; Huang, M.R. Transcriptome sequencing and development of novel genic SSR markers for Dendrobium officinale. Mol. Breed. 2017, 37, 18. [Google Scholar]
- Xu, M.; Sun, Y.; Li, H. EST-SSRs development and paternity analysis for Liriodendron spp. New For. 2010, 40, 361–382. [Google Scholar] [CrossRef]
- Zhang, H.L.; Li, H.G.; Xu, M.; Feng, Y.H. Identification of Liriodendron tulipifera, Liriodendron chinense and hybrid Liriodendron using species-specific SSR markers. Sci. Silvae Sin. 2010, 46, 36–39. [Google Scholar]
- Yang, A.-H.; Zhang, J.-J.; Tian, H.; Yao, X.-H. Characterization of 39 novel EST-SSR markers for Liriodendron tulipifera and cross-species amplification in L. chinense (Magnoliaceae). Am. J. Bot. 2012, 99, e460–e464. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Zhang, J.; Ye, Q.; Huang, H. Characterization of 14 novel microsatellite loci in the endangered Liriodendron chinense (Magnoliaceae) and cross-species amplification in closely related taxa. Conserv. Genet. 2008, 9, 483–485. [Google Scholar] [CrossRef]
- Kobayashi, N. A simple and efficient DNA extraction method for plants, especially woody plants. Plant Tissue Cult Biotech 1998, 4, 76–80. [Google Scholar]
- Lalitha, S. Primer Premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Bo, J.; Hai-Bin, X.U.; Yang, X.U.; Long, X.F.; Chen, J.H.; Shi, J.S. Optimization of Genomic-SSR Reaction System in Liriodendron. For. Res. 2013, 26, 506–510. [Google Scholar]
- Zimmermann, J.; Voss, H.; Erfle, H.; Rupp, T.; Dietrich, T.; Hewitt, N.A.; Schwager, C.; Stegemann, J.; Ansorge, W. Direct sequencing of PCR products using magnetic beads and fluorescein-12-Dutp. Methods Mol. Cell. Biol. 1992, 3, 114–115. [Google Scholar]
- Yeh, F.; Yang, R.; Boyle, T.; Ye, Z.; Mao, J.; Yeh, C.; Timothy, B.; Mao, X. POPGENE version 1.32; the User-Friendly Shareware for Population Genetic Analysis; Center for International Forestry Research, University of Alberta: Edmonoto, AB, Canada, 1997. [Google Scholar]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, version 2.2, 2.1; Department of Ecoloy and Evolution, State University of New York: New York, NY, USA, 2005. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J.J.M.E. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2010, 14, 2611–2620. [Google Scholar] [CrossRef]
- Metzgar, D.; Bytof, J.; Wills, C. Selection Against Frameshift Mutations Limits Microsatellite Expansion in Coding DNA. Genome Res. 2000, 10, 72–80. [Google Scholar]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Chen, J. Studies on the somatic embryogenesis of liriodendron hybrids (L. chinense × L. tulipifera). Sci. Silvae Sin. 2003, 39, 49–53. [Google Scholar]
- Zhang, X.; Carlson, A.; Tian, Z.; Staton, M.; E Schlarbaum, S.; E Carlson, J.; Liang, H. Genetic characterization of Liriodendron seed orchards with EST-SSR markers. J. Sci. Mol. Breed. 2015, 4, 1. [Google Scholar] [CrossRef]
- Lei, M.; Wang, Q.; Wu, Z.-J.; López-Pujol, J.; Li, D.-Z.; Zhang, Z.-Y. Molecular phylogeography of Fagus engleriana (Fagaceae) in subtropical China: Limited admixture among multiple refugia. Tree Genet. Genomes 2012, 8, 1203–1212. [Google Scholar] [CrossRef]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat Plants 2019, 5, 18–25. [Google Scholar] [CrossRef]
- Grohme, M.A.; Soler, R.F.; Wink, M.; Frohme, M. Microsatellite marker discovery using single molecule real-time circular consensus sequencing on the Pacific Biosciences RS. BioTechniques 2013, 55, 253–256. [Google Scholar] [CrossRef]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef]
- Wright, S.I.; Andolfatto, P. The Impact of Natural Selection on the Genome: Emerging Patterns in Drosophila and Arabidopsis. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 193–213. [Google Scholar] [CrossRef]
- Zhu, X.; Ma, J.; Yao, Q.; Hao, R.; He, S. Allozyme verification on the population differentiation of Liriodendron chinense (Hemsl.) Sarg. J. Plant Resour. Environ. 1995, 4, 9–14. [Google Scholar]
- Zaouali, Y.; Chograni, H.; Trimech, R.; Boussaid, M. Genetic diversity and population structure among Rosmarinus officinalis L. (Lamiaceae) varieties: Var. typicus Batt. and var. troglodytorum Maire. based on multiple traits. Ind. Crop. Prod. 2012, 38, 166–176. [Google Scholar] [CrossRef]
- Wen, J. Evolution of eastern asian and eastern north american disjunct distributions in flowering plants. Annu. Rev. Ecol. Syst. 1999, 30, 421–455. [Google Scholar] [CrossRef]


| Locus | Primer Sequences (5′–3′) | Repeat Motif | Product Size | BLAST Top Hit Description (Organism) | BLAST Top Hit Accession No. | E-Value | |
|---|---|---|---|---|---|---|---|
| ESSR8 | F | ACAGAATCCTCGCTCGCA | (CCTCTC) 4 | 398 | NO hit | none | none |
| R | ACGGTAACCAAACCCCCA | ||||||
| ESSR12 | F | ATTCCTCGCACCCTAAACCTAGCC | (GACTCC) 4 | 407 | PREDICTED: Nelumbo nucifera carbon catabolite repressor protein 4 homolog 2-like (LOC104597619), mRNA | XM_010259268.2 | 4 × 10−56 |
| R | CGATCTCTCCCTCTCTTTCACTCC | ||||||
| ESSR26 | F | GGTTGTGCCACCTCTCCCTCC | (TAA) 8 | 207 | NO hit | none | none |
| R | TGCATTTCCCCAAATTCCTGA | ||||||
| ESSR27 | F | GTGGTGGGTTTGATTTTCTTT | (CCA) 9 | 209 | NO hit | none | none |
| R | TGATCGTGGATGCTACCTGAT | ||||||
| ESSR77 | F | GCAACTTACGCAACAACACG | (GT) 10 | 368 | NO hit | none | none |
| R | CACTGCACAAAGGCCATCTA | ||||||
| ESSR92 | F | TTTACTAATTTGTCCGCAAT | (AGC) 7 | 314 | NO hit | none | none |
| R | CCTTCTTCTCCTTCTCTCCT | ||||||
| ESSR118 | F | CGCCCCCAGCTTCTATATCCT | (CA) 10 | 289 | NO hit | none | none |
| R | TTCTTCCTTTCAAGTCCTCCA | ||||||
| ESSR131 | F | TTGGATTTCCATATCTGGGA | (CTTT) 5 | 419 | PREDICTED: Asparagus officinalis uncharacterized LOC109826068 (LOC109826068), mRNA | XM_020393028.1 | 4 × 10−56 |
| R | ACAATAACTTTGGTGTGCCT | ||||||
| ESSR10 | F | AATTCCTCGCACCCTAAACCTAGCC | (GACTCC) 4 | 409 | PREDICTED: Nelumbo nucifera carbon catabolite repressor protein 4 homolog 2-like (LOC104597619), mRNA | XM_010259268.2 | 4 × 10−56 |
| R | TCGATCTCTCCCTCTCTTTCACTCC | ||||||
| ESSR39 | F | CCACCTCTCGCAAAATCTCC | (AAGGA) 4 | 262 | NO hit | none | none |
| R | CCTGCAACGTCTCCCTCCTT | ||||||
| ESSR51 | F | GGGAATGAGGGATCAAACCT | (GCA) 10 | 363 | PREDICTED: Nelumbo nucifera uncharacterized LOC104596120 (LOC104596120), transcript variant X2, mRNA | XM_010257158.2 | 5 × 10−21 |
| R | ATGCACTCCTTCACACGGAA | ||||||
| ESSR54 | F | CCTCCCAAAAATCAAACAGA | (TGT) 7 | 368 | NO hit | none | none |
| R | AGCCAGAGTAGTCCAAGCAC | ||||||
| ESSR72 | F | GACCATCCTTCACATTACAACA | (AGA) 10 | 256 | NO hit | none | none |
| R | AGAACAGAACCTCGCAACCACT | ||||||
| ESSR82 | F | GGGAAAAAATGAGAATACCAAG | (AAAG) 5 | 350 | NO hit | none | none |
| R | CAGTAGAAACCATCTGAAAGCA | ||||||
| ESSR87 | F | CTATCTATTCATCTCCCCCG | (GA) 12 | 313 | NO hit | none | none |
| R | ATGTCGTCCAACACACCTTC | ||||||
| ESSR90 | F | AAGAGAGATAAAGAGAGGGA | (TCCCCT) 4 | 446 | PREDICTED: Nelumbo nucifera GDP-L-galactose phosphorylase 1-like (LOC104609968), mRNA | XM_010276420.2 | 2 × 10−19 |
| R | TTAGAAACTACGGTAGGAAC | ||||||
| ESSR93 | F | CACAAAAAAGTCACGCCAATC | (AAAG) 5 | 391 | PREDICTED: Vitis vinifera protein FAM133A (LOC100265155), transcript variant X3, mRNA | XM_010662230.2 | 3 × 10−68 |
| R | TTTCACAACAGTCCATCCATC | ||||||
| ESSR96 | F | TTCCCCACACACCCCGTCAA | (GCT) 7 | 486 | NO hit | none | none |
| R | CGAATCCCCCCATCATCCCT | ||||||
| ESSR97 | F | CTTTCTGTATGGGAGTGGGT | (CTT) 7 | 432 | PREDICTED: Gossypium raimondii DEAD-box ATP-dependent RNA helicase 13 (LOC105800653), mRNA | XM_012631890.1 | 3 × 10−47 |
| R | TGACGAGTCGGAGTATGGTT | ||||||
| ESSR107 | F | CATATACGCATTTTTTCGCCC | (CTTT) 5 | 485 | NO hit | none | none |
| R | ACCTCACCTACATTCGGACCG | ||||||
| ESSR110 | F | CCTTGTCTTGGTTTCTGTTCT | (CTT) 8 | 253 | NO hit | none | none |
| R | GTTTATTTTTGTGGAGGGGCT | ||||||
| ESSR116 | F | CTGGATATCTACGAGTCACGACT | (TC) 10 | 280 | NO hit | none | none |
| R | AACAAAAGGACATAAAAGACCAT | ||||||
| ESSR119 | F | TACCATCCGTTAGTGCTTCCGCTCCTG | (TCCTTC) 4 | 126 | PREDICTED: Nelumbo nucifera TIP41-like protein (LOC104611680), mRNA | XM_010278853.2 | 2 × 10−43 |
| R | CTTAGAATCGGGCATTCCACGCATCCA | ||||||
| ESSR124 | F | AGGAAAAAAAGGGTTTGAAGCGGATGA | (GAA) 7 | 480 | NO hit | none | none |
| R | AATGAGGAGCCATGTACTTCGCATTGC | ||||||
| ESSR127 | F | TGTCATAACAGTTCCTACCAAAAGCAG | (TGC) 7 | 463 | NO hit | none | none |
| R | TGTATTAGAGACGGAGTAGAAGATGCG | ||||||
| ESSR132 | F | AATGGGCTAGAGAATCCAAAAAACT | (CACCAG) 4 | 281 | PREDICTED: Elaeis guineensis dnaJ homolog subfamily B member 13-like (LOC105043518), transcript variant X2, mRNA | XM_010921102.2 | 4 × 10−56 |
| R | CCCGATAAAAACCCTAATAACAAAA | ||||||
| ESSR133 | F | CGCTCGTCTCCTTCCATTGCTCT | (AGA) 7 | 387 | NO hit | none | none |
| R | GCTGTGTTCTGGGGTGTGTTTGC | ||||||
| ESSR149 | F | AGCACTTTTCTGTCTCTCTCC | (CAGGAG) 4 | 319 | NO hit | none | none |
| R | CCAAGCTGTTCACTCTTTCAC | ||||||
| ESSR197 | F | GGGCTGTGATTAGAAAGAGAGAGG | (TTTC) 6 | 234 | Spirodela polyrhiza strain 9509 chromosome 2 sequence | CP019094.1 | 5 × 10−4 |
| L. chinense (Hemsl.) Sarg. (n = 35) | L. tulipifera L. (n = 13) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Na | Ne | I | Ho | He | Nei | PIC | Na | Ne | I | Ho | He | Nei | PIC |
| ESSR8 | 5 | 2.786 | 1.151 | 1 | 0.651 | 0.641 | 0.576 | 7 | 5.143 | 1.772 | 1 | 0.841 | 0.81 | 0.779 |
| ESSR12 | 4 | 2.396 | 0.998 | 1 | 0.592 | 0.583 | 0.494 | 4 | 2.396 | 0.998 | 1 | 0.610 | 0.58 | 0.494 |
| ESSR26 | 3 | 2.820 | 1.067 | 0 | 0.655 | 0.645 | 0.571 | 2 | 1.800 | 0.637 | 0 | 0.464 | 0.44 | 0.346 |
| ESSR27 | 5 | 3.568 | 1.378 | 0 | 0.731 | 0.720 | 0.671 | 3 | 1.946 | 0.824 | 0 | 0.507 | 0.49 | 0.424 |
| ESSR77 | 17 | 13.462 | 2.712 | 1 | 0.939 | 0.926 | 0.921 | 7 | 4.173 | 1.639 | 1 | 0.791 | 0.76 | 0.728 |
| ESSR92 | 4 | 2.409 | 0.987 | 0.143 | 0.593 | 0.585 | 0.498 | 3 | 2.153 | 0.829 | 0.923 | 0.557 | 0.54 | 0.428 |
| ESSR118 | 13 | 8.942 | 2.321 | 0.486 | 0.901 | 0.888 | 0.878 | 7 | 4.881 | 1.745 | 0.167 | 0.830 | 0.80 | 0.768 |
| SSR131 | 7 | 3.379 | 1.440 | 1 | 0.714 | 0.704 | 0.656 | 2 | 2.000 | 0.693 | 1 | 0.520 | 0.50 | 0.375 |
| ESSR10 | 4 | 2.649 | 1.103 | 1 | 0.632 | 0.622 | 0.547 | 6 | 2.748 | 1.229 | 1 | 0.662 | 0.64 | 0.568 |
| ESSR39 | 4 | 2.503 | 1.045 | 1 | 0.609 | 0.600 | 0.518 | 2 | 2.000 | 0.693 | 1 | 0.520 | 0.50 | 0.375 |
| ESSR51 | 7 | 2.418 | 1.133 | 0.882 | 0.595 | 0.587 | 0.516 | 4 | 2.504 | 1.054 | 1 | 0.627 | 0.60 | 0.524 |
| ESSR54 | 8 | 5.518 | 1.845 | 1 | 0.831 | 0.819 | 0.795 | 3 | 2.253 | 0.898 | 0.923 | 0.579 | 0.56 | 0.477 |
| ESSR72 | 10 | 3.796 | 1.666 | 1 | 0.748 | 0.737 | 0.699 | 5 | 4.390 | 1.530 | 1 | 0.803 | 0.77 | 0.734 |
| ESSR82 | 9 | 7.595 | 2.094 | 1 | 0.883 | 0.868 | 0.854 | 4 | 3.200 | 1.256 | 1 | 0.717 | 0.69 | 0.630 |
| ESSR87 | 7 | 4.111 | 1.625 | 1 | 0.768 | 0.757 | 0.721 | 5 | 4.225 | 1.487 | 0.923 | 0.794 | 0.76 | 0.722 |
| ESSR90 | 5 | 4.562 | 1.560 | 0.371 | 0.792 | 0.781 | 0.745 | 4 | 2.683 | 1.119 | 0.308 | 0.652 | 0.63 | 0.556 |
| ESSR93 | 4 | 2.288 | 0.995 | 0.4 | 0.571 | 0.563 | 0.505 | 2 | 1.899 | 0.666 | 0.615 | 0.492 | 0.47 | 0.361 |
| ESSR96 | 6 | 2.955 | 1.292 | 1 | 0.671 | 0.662 | 0.606 | 2 | 2.000 | 0.693 | 1 | 0.520 | 0.50 | 0.375 |
| ESSR97 | 3 | 2.110 | 0.845 | 0.114 | 0.534 | 0.526 | 0.436 | 2 | 1.899 | 0.666 | 0.462 | 0.492 | 0.47 | 0.361 |
| ESSR107 | 5 | 2.934 | 1.243 | 0.441 | 0.669 | 0.659 | 0.605 | 5 | 4.299 | 1.520 | 0.25 | 0.801 | 0.77 | 0.729 |
| ESSR110 | 11 | 8.892 | 2.269 | 0.882 | 0.901 | 0.888 | 0.877 | 7 | 6.857 | 1.936 | 1 | 0.891 | 0.85 | 0.836 |
| ESSR116 | 2 | 2.000 | 0.693 | 1 | 0.507 | 0.500 | 0.375 | 6 | 3.714 | 1.483 | 1 | 0.760 | 0.73 | 0.687 |
| ESSR119 | 2 | 2.000 | 0.693 | 1 | 0.507 | 0.500 | 0.375 | 2 | 2.000 | 0.693 | 1 | 0.520 | 0.50 | 0.375 |
| ESSR124 | 3 | 2.080 | 0.837 | 0 | 0.527 | 0.519 | 0.431 | 4 | 3.130 | 1.237 | 0.333 | 0.710 | 0.68 | 0.622 |
| ESSR127 | 4 | 2.802 | 1.156 | 0.941 | 0.653 | 0.643 | 0.578 | 3 | 2.315 | 0.911 | 0.923 | 0.591 | 0.57 | 0.472 |
| ESSR132 | 8 | 4.867 | 1.838 | 1 | 0.806 | 0.795 | 0.774 | 7 | 5.828 | 1.838 | 1 | 0.862 | 0.83 | 0.806 |
| ESSR133 | 8 | 6.806 | 1.977 | 1 | 0.865 | 0.853 | 0.835 | 4 | 2.361 | 0.980 | 1 | 0.601 | 0.58 | 0.485 |
| ESSR149 | 2 | 1.059 | 0.130 | 0 | 0.056 | 0.056 | 0.054 | 4 | 2.683 | 1.119 | 0 | 0.652 | 0.63 | 0.556 |
| ESSR197 | 8 | 3.977 | 1.673 | 0.514 | 0.759 | 0.749 | 0.721 | 4 | 2.195 | 0.935 | 0.154 | 0.566 | 0.54 | 0.462 |
| mean | 6 | 4.058 | 1.371 | 0.696 | 0.678 | 0.668 | 0.615 | 4.1379 | 3.092 | 1.141 | 0.724 | 0.653 | 0.63 | 0.554 |
| St.Dev | 3 | 2.723 | 0.568 | 0.398 | 0.174 | 0.172 | 1.7672 | 1.343 | 0.406 | 0.385 | 0.131 | 0.13 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, X.; Weng, Y.; Liu, S.; Hao, Z.; Sheng, Y.; Guan, L.; Shi, J.; Chen, J. Genetic Diversity and Differentiation of Relict Plant Liriodendron Populations Based on 29 Novel EST-SSR Markers. Forests 2019, 10, 334. https://doi.org/10.3390/f10040334
Long X, Weng Y, Liu S, Hao Z, Sheng Y, Guan L, Shi J, Chen J. Genetic Diversity and Differentiation of Relict Plant Liriodendron Populations Based on 29 Novel EST-SSR Markers. Forests. 2019; 10(4):334. https://doi.org/10.3390/f10040334
Chicago/Turabian StyleLong, Xiaofei, Yuhao Weng, Siqin Liu, Zhaodong Hao, Yu Sheng, Lanhua Guan, Jisen Shi, and Jinhui Chen. 2019. "Genetic Diversity and Differentiation of Relict Plant Liriodendron Populations Based on 29 Novel EST-SSR Markers" Forests 10, no. 4: 334. https://doi.org/10.3390/f10040334
APA StyleLong, X., Weng, Y., Liu, S., Hao, Z., Sheng, Y., Guan, L., Shi, J., & Chen, J. (2019). Genetic Diversity and Differentiation of Relict Plant Liriodendron Populations Based on 29 Novel EST-SSR Markers. Forests, 10(4), 334. https://doi.org/10.3390/f10040334





