Abstract
Surviving relict populations of species that were more widespread in ancient times can teach us a lot, such as evolution and genetic differentiation. One such relict plant is Liriodendron, of which populations remain in China (L. chinense (Hemsl.) Sarg.) and the USA (L. tulipifera L.). Studying the genetic structure of these populations would give insight into the genetic differentiation and the breeding strategy. In this work, we developed and characterized 29 novel simple sequence repeat (SSR) markers based on expressed sequence tags (ESTs) from hybrid Liriodendron (Liriodendron chinense × tulipifera) callus. In total, 29 SSRs with perfect primer-designed were used to assess genetic diversity and differentiation. The set of polymorphic EST-SSR loci was identified in 48 Liriodendron individuals, represented by 35 individuals sampled from 14 provenances of L. chinense and 13 individuals sampled from 5 provenances of L. tulipifera. Our results indicated that L. chinense populations possess slightly higher genetic diversity than L. tulipifera populations. Based on genetic distances, 48 Liriodendron individuals clustered into three groups (the eastern China L. chinense, the western China L. chinense and L. tulipifera), although the STRUCTURE analysis of the Liriodendron populations revealed just two clear genetic clusters (L. chinense and L. tulipifera). Among these 29 novel markers, ESSR119 showed an obvious species-specific characteristic which can be very useful in marker-assisted selection (MAS). In general, all these EST-SSR markers may have agronomic potential and constitute a basis for future studies on the identification, innovation, and even preservation of Liriodendron germplasms.
1. Introduction
Liriodendron is a genus of the Magnoliaceae family, which is comprised of a pair of sister species with a standard intercontinental disjunction distribution [1]. Both these populations suffered through extinctions caused by Late Tertiary climate oscillations and the Pleistocene glaciations [2]. Liriodendron tulipifera L. (L. tulipifera) is widely distributed in eastern North America broadleaf forests, while Liriodendron chinense (Hemsl.) Sarg. (L. chinense) inhabits central-western and southern China, usually in the mountains at elevations from 450 to 1800 meters [3]. Bio-geographers have regarded Liriodendron as an ideal natural resource for research into the disjunctive distribution of flowering plants between eastern Asia and eastern North America [2,4]. Generally speaking, the leaves of L. chinense are Chinese jacket-shaped with three lobes and both sepals and petals are green. L. tulipifera has goosefoot-shaped, five-lobed leaves and yellow-green sepals and petals [5]. However, L. chinense is now an endangered species due to changes in its ecosystem and geographical surroundings, as well as due to its biological characteristics, such as a low germination rate and limited seed production [3,6]. To make matters worse, the data showed that there might be less than 10 trees in over 70% of the natural population of L. chinense, especially in eastern part of China. Even more serious is the lack of up-to-date and detailed investigation reports [7,8]. Hybrid Liriodendron (L. chinense × L. tulipifera) was first cultivated in 1963 and has been reported to show heterosis, such as a distinct growth advantage and good adaptability [9]. Liriodendron wood is used in a diverse range of products, such as furniture and farming equipment [10]. Many studies have shown that extracts from Liriodendron leave exhibit potent cytotoxic effects on tumors cell lines [11,12] and inhibitory activity towards farnesyl protein transferase (FPTase) and tumors cell growth [13]. Despite the significant value of Liriodendron and the endangered status of L. chinense, information on their population genetic structure and geographic variation are limited. Microsatellites or simple sequence repeats (SSRs) consist of tandem arrays of short nucleotide motifs, which are randomly dispersed throughout eukaryotic genomes [14]. SSRs offer several advantages for use as genetic markers, such as high reproducibility, a high degree of polymorphism, co-dominant inheritance, and relatively high abundance across the entire genome [15]. These excellent characteristics make SSRs powerful high-resolution tools for the study of population genetics, genetic variation, and marker-assisted selection (MAS) in plants [16,17,18]. However, there are only a few SSR markers available which have been tested for polymorphism in Liriodendron [19,20,21,22]. Compared to other species, genomic resources for Liriodendron remain severely limited. In this study, 29 novel EST-SSR markers were isolated and characterized and subsequently used for preliminary population genetic structure analyses.
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
Young leaves were collected from 48 individuals representing fourteen provenances of L. chinense and five provenances of L. tulipifera, respectively (Figure 1 and Table S1). All of them were collected from the major planting areas of the species and now preserved at the Huzhua Mountain Forestry Station in Hubei Province (31°04′ N, 112°54′ E). These 48 samples were individually ground into fine powder with liquid nitrogen, and total genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method [23]. The quality and concentration of the DNA were determined by electrophoresis on 1% agarose gels. All DNA samples were subsequently diluted to 50 ng/µL, which was the working concentration for PCR.

Figure 1.
(a) Geographic distribution of the origin of 14 Liriodendron chinense (Hemsl.) Sarg. (L. chinense) populations sampled in this study; (b) Geographic distribution of the origin of five Liriodendron tulipifera L. (L. tulipifera) populations sampled in this study; (c) The UPGMA (unweight pair-group method with arithmetic means) dendrogram generated cluster analysis based on the genetic diversity of eight different provinces L. chinense samples. The L. tuilipifera plants were taken as a whole part. The L. chinense plants in this study were grouped into three clusters (blue, Eastern China; Orange, Western China; purple, Yunan Province). Bootstrap replicates = 1000. Note: LY: Liuyang; SN: Suining; YY: Youyang; SY: Songyang; LP: Liping; HS: Huangshan; SZ: Sangzhi; LS: Lushan; XY: Xuyong; EX: Exi region; ST: Songtao; ML: Mengla; WYS: Wuyishan; DBS: Dabieshan; NC: North Carolina; LA: Louisiana; GE: Georgia; SC: South Carolina; MO: Missouri. Hunan (LY, SN, SZ); Jiangxi (LS, WYS); Zhejiang (SY); Anhui (HS, DBS); Hubei (Exi); Sichuan (YY, XY); Guizhou (ST, LP); Yunan (ML); USA (NC, LA, GE, SC, MO).
2.2. EST-SSR Development and Genotyping
In the previous research, an RNA-Seq experiment was conducted using Illumina HiSeq 2000. Expressed sequence tags (ESTs) were sequenced from hybrid Liriodendron (L. chinense × L. tulipifera) callus and screened for the presence of SSRs using microsatellite identification tool (MISA) (http//www.pgrc.ipk-gatersleben.de/misa). Four hundred and eighteen unique sequences with at least one SSR were yielded with the criteria of 20, 9, 6, 5, 4, and 3 repeat units for mono-, di-, tri-, tetra-, penta-, and hexanucleotide motifs, respectively. Amplification primers which were ranging from 20 to 25 base pairs, and with moderate GC content flanking the SSR region, were designed using Primer5 [24] (http://frodo.wi.mit.edu/primer5/). Microsatellite polymorphisms were initially assessed within four individuals of L. chinense and four individuals of L. tulipifera from different provenances. PCR amplification was performed in a 10 µL reaction solution, containing 75 ng genomic DNA, 1.0 μL 10× PCR Buffer, 1.25 μl 2.5 mM·L−1 MgCl, 0.2 μL 10 mM·L−1 dNTP, 0.25 μL 10 μM·L−1 each of forward and reverse primers, and 0.5 units of Taq polymerase (Takara Biotechnology, Dalian, China). A touchdown PCR protocol was applied for all primers and performed on the SimpliAmp™ Thermal Cycler (Applied Biosystems, Foster City, CA, USA). Samples were incubated by Touchdown PCR at 95 °C for 5 min, followed by 15 touchdown cycles, first at 94 °C for 15 s, 15 s at annealing temperature (61 °C in the first cycle and decreased 1 °C every second cycle down to 47 °C), and 30 s at 72 °C. Next, the samples were subjected to 15 cycles at 94 °C for 15 s, then at 52 °C for 15 s, and finally at 72 °C for 30 s [25]. Fragments resulting from PCR amplifications were separated by electrophoresis on 8% polyacrylamide denaturing gels and visualized with silver nitrate staining. For further verification of the accuracy of novel EST-SSR polymorphisms, PCR amplification was performed with Fluorescein-12-dUTP (Thermo Scientific, Waltham, MA, USA), which can be enzymatically incorporated into the PCR product with Taq DNA polymerase [26]. Then, we performed capillary electrophoresis using an ABI3730xl DNA Automatic Analyzer with a GeneScan-500LIZ size standard (Applied Biosystems by life technology). Based on these data, allele sizes were determined using PeakScanner (v1.0) software (Thermo Scientific, Waltham, MA, USA).
2.3. Analysis of Genetic Diversity and Genetic Relationship
The number of alleles (Na), observed heterozygosity (Ho), expected heterozygosity (He), Hardy-Weinberg equilibrium text (HWE), Shannon information index (I), F-statistics (Fst), and genetic distance (Nei’s distance) were calculated using POPGEN (v1.32) [27]. Polymorphism information content (PIC) was derived according to the formula [28]:
where n is the number of alleles at one locus; pi and pj are the frequencies of the ith and jth alleles at one locus.
Clustering analysis is conducted by utilizing software NTSYS-pc (v2.2) [29], and the clustering figure should be established with the method of UPGMA (unweight pair-group method with arithmetic means). Population structure was determined using the model-based program, STRUCTURE (v2.3.2) [26]. To identify the number of populations (K) capturing the major structure in the data, we used a burn-in period of 100,000 Markov Chain Monte Carlo iterations and 100,000 runs, with an admixture model following Hardy-Weinberg equilibrium, and correlated allele frequencies and independent loci for each run. Twenty independent runs were performed for each simulated value of K, ranging from 2 to 12. The true K value was determined using both an estimate of the posterior probability of the data for a given K and the Evanno ΔK [30,31].
3. Results
3.1. Microsatellite Development
A total of 82,933 new ESTs were identified, using hybrid Liriodendron (L. chinense × L. tulipifera) callus as sequencing material. Four hundred and eighteen contigs with at least one SSR were identified using criteria demanding the presence of at least 20, 9, 6, 5, 4, and 3 repeat units for mono-, di-, tri-, tetra-, penta-, and hexanucleotide motifs, respectively. Perfect trinucleotide motifs in the present study were found to be the most abundant, with a frequency of 25% of the total, followed by hexanucleotide motifs (24.8%; Figure S1), which may be attributed to lower tolerance for frame-shift mutation within coding regions [32]. Mononucleotide and pentanucleotide motifs occurred with a much lower frequency (Figure S1). Two hundred and ten SSR loci with available primer-designed sites were selected in this study. We designed primers of 20 to 25 base pairs in length with moderate GC content, flanking the SSR region. Overall, 29 of the 210 primer pairs amplified clear, stable, and polymorphic products on eight individuals, including four L. chinense plants (from Sangzhi, Hunan Prov.; Youyang, Sichuan Prov.; Xuyong, Sichuan Prov.; Lushan, Jiangxi Prov.) and four L. tulipifera plants (from North Carolina, USA; Georgia, USA; South Carolina, USA; Missouri, USA). To further verify the authenticity of novel EST-SSR polymorphisms after polyacrylamide gel electrophoresis (data not shown), we selected 48 Liriodendron plants to perform PCR amplification then capillary electrophoresis. Thirty-five individuals belonged to 14 different L. chinense provenances, distributed over eight Chinese provinces and the remaining 13 individuals belonged to five different L. tulipifera provenances (Figure 1a,b and Table S1).
To gain more insight into the putative functions of the 29 unique SSR-associated genes, we searched their sequences against the NCBI databases (nr/nt, refseq_rna, refseq_genomes, est, and refseq_genomic) using the BLASTN program, with a threshold E-value of 1.00 × 10−5, resulting in 10 sequences showing significant similarities to previously annotated genes (Table 1).

Table 1.
Characteristics of 29 microsatellite primers developed for L. chinense and L. tulipifera.
3.2. EST-SSR Polymorphism and Genetic Diversity
Among the tested 35 L. chinense plants, the number of alleles (Na) per locus ranged from 2 to 17, with an average of 6 alleles, while the Na per locus ranged from 2 to 7 (mean = 4.137) among the 13 L. tulipifera plants. The observed heterozygosity (Ho) within these two species both ranged from 0.000 to 1.000 with the average Ho being 0.696 within L. chinense and 0.724 within L. tulipifera. He (expected heterozygosity) ranged from 0.056 to 0.939 (mean = 0.678) within L. tulipifera and from 0.464 to 0.862 (mean = 0.653) within L. tulipifera. Further, the PIC values in L. chinense trees ranged from 0.054 to 0.921 with an average of 0.615. Similar observations were made in L. tulipifera with PIC values changing from 0.361 to 0.836 and a lower average PIC value of 0.554 (Table 2). Based on Shannon’s information index (I) analyzed by SSR markers, the highest genetic diversity was present in population Songyang, Zhejiang Prov. (I = 1.0789) (Table S5), followed by populations Liuyang, Hunan Prov. and Youyang Sichuan Prov., with a high level of diversity at the species level (I = 0.9855 and 0.9543). Population North Carolina, USA had the highest genetic diversity among L. tulipifera. Populations in Sangzhi, Hunan Prov. and Lushan, Jiangxi Prov. showed the lowest genetic diversity index (I = 0.4541).

Table 2.
Results of primer screening in L. chinense and cross-species amplification in L. tulipifera.
Both L. chinense and L. tulipifera groups showed a relatively high population differentiation index (Fst) of more than 0.15 [33], which means the genus has a rich genetic diversity. However, L. chinense having a higher population differentiation index than L. tulipifera is likely due to the former having been sampled across 14 provenances while the latter only had five provenances sampled (Fst = 0.2958 vs. 0.2214; Table S2). 20%–30% genetic variation exists among Liriodendron provenances, and about 70% genetic variation exists within provenance, which accounts for a greater part of the whole genetic variation, there out, the variation of genetic diversity, of these 17 Liriodendron provenances mainly exist in populations. Gene flow (Nm) mainly reflects the communication frequency of gene among most individuals within the population and populations [34]. Moreover, the gene flow of these L. chinense provenances is 0.59521 (Nm of L. tulipifera provenances = 0.8790). When gene flow (Nm) >1, gene flow can prevent genetic difference among populations effectively caused by genetic drift, and thus we can find that specific gene flow exists among them, but the level is relatively low.
The combination of multiple SSR markers can not only evaluate the population differentiation in general but also which with specific polymorphism give a compelling basis for marker-assisted selection in the early breeding stage, making the breeding cycle shortened. We used PeakScanner software (ThermoFisher Scientific) to analyze our capillary electrophoresis data, which detected a total of two alleles across both L. chinense and L. tulipifera trees for the ESSR119 marker. All L. chinense individuals showed one main peak at a length of 114 bp, while in all L. tulipifera individuals the ESSR119 peak located at 126 bp (Figure 2a). Gel screening also showed similar results. PCR amplification products from all L. chinense individuals showed the same size fragment located at 110–120 bp, while those from all L. tulipifera individuals showed the fragment longer than 120 bp (Figure 2b). In 2006, we also conducted a cross experiment. Pollen from L. tulipifera (South Carolina) was pollinated on the stigma of L. chinense (Lushan) with removing stamens and then treated flowers (L. chinense) protected by bagging. After that, F1 plants were successfully obtained. In this condition, hybrid Liriodendron individuals consistently were heterozygous for both ESSR119 alleles, one being from the L. chinense parent and the other being from the L. tulipifera parent. ESSR131 was composed of five different alleles in L. chinense individuals, while in L. tulipifera only one allele was detected, which located at 422 bp. ESSR96 presented similar characteristics in that alleles showed more consistency in L. tulipifera individuals (only 194/200 bp) than in L. chinense individuals showed six kinds of polymorphic forms (206/212 bp, 212/218 bp, 216/218 bp, 222/228 bp, 212/216 bp, 212/222 bp). By contrast, ESSR116 showed consistency only in L. chinense individuals (Figure S2). Although these two species separated 10–16 million years ago [31], they are quite similar morphologically and reproductively compatible [1,35], with hybrids showing a heterosis effect [36]. It is difficult to distinguish this pair of Liriodendron species in the early stages of growth [20]. However, the EST-SSR markers presented in this study, especially species-specific ESSR119 should facilitate this process through marker-assisted selection.
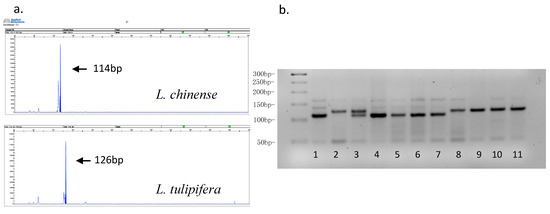
Figure 2.
PCR products amplified by expressed sequence tag-simple sequence repeat (EST-SSR) marker (ESSR119) by two methods. (a) Peakscanner analysis showed one main peak located at 114 bp in one of the L. chinense individuals and one main peak located at 126 bp in one of the L. tulipifera individuals. (b) Representative gel showing amplification profiles of ESSR119 marker and its fragment length polymorphism among four individuals of L. chinense and four individuals of L. tulipifera. The amplifications are resolved in 3% agarose gel along with 50 bp DNA size standard. Note: lane 1–3 are standard samples. 1, L. chinense (114 bp); 2, L. tulipifera (126 bp); 3, Hybrid Liriodendron (L. chinense (Lushan) X L. tulipifera (South Carolina)) (114 and 126bp); 4–7, four plants of L. chinense come from Sangzhi, Hunan Prov.; Youyang, Sichuan Prov.; Xuyong, Sichuan Prov.; Lushan, Jiangxi Prov., respectively. They all showed one main 114 bp band; 8–11, four individuals of L. tulipifera come from North Carolina, USA; Georgia, USA; South Carolina, USA; Missouri, USA, respectively. They all showed one main 126 bp band.
3.3. Population Genetic Structure of Liriodendron
The population structure was analyzed using a Bayesian approach on 19 provenances implemented in the STRUCTURE software. Following the method of Evanno, the K values were plotted and had a peak of 2 (Figure S3). So, the maximum K occurred at K = 2 for the EST-SSR markers. Nineteen provenances (48 individuals) could be divided into two main groups, one consists of all L. chinense populations with one lobe on each side of a blade, and other consists of left L. tulipifera populations with two pairs of lobes on a blade (Figure 3b and Figure S3). After then, we also analyzed the genetic structure of 13 provenances with a sample size greater than three individuals. K was tested from two to twelve with 20 independent replications. The ΔK values computed for these populations indicated a strong signal for K = 2 (Table S6), which is the same as the previously estimated populations. This result implied that 13 provenances under study were grouped into two clusters (Figure 3b and Figure S4). Due to long-term geographical isolation between those two Liriodendron species, we assumed that geographic difference should mainly contribute to the current genetic structure of populations.

Figure 3.
The UPGMA dendrogram based on Nei’s (1978) genetic distance. Bootstrap replicates = 1000. Nine L. chinense populations and four L. tulipifera populations were analyzed with a population sample size of more than three individuals. The L. chinense plants grouped into five clusters (green, Eastern China; yellow, Songtao, Guizhou Province; Orange, Sichuan Province; blue, Liping, Guizhou Province; purple, Yunan Province.). The dark lines represent the movement of the major mountain chains. The information comes from https://zh.wikipedia.org/zh-cn/, and the mountain chains chart are redrawn based on http://m.sohu.com/n/484766869/?wp=2. The population structure based on 13 provenances at K = 2 as determined using the ΔK method of Evanno et al.
3.4. Cluster Analysis Using EST-SSR Markers
The tested SSR markers gave us some information about the genetic identity and genetic distance, which can accurately assess the similarity among Liriodendron populations. The genetic identity (GI) of these 19 provenances changes from 0.1487 to 0.9567 (Table S3b), the average genetic identity is 0.5302, and the genetic distance (D) varies from 0.0443 to 1.9057, and the average genetic relationship is 0.7527, hence, it can be deduced that these 19 Liriodendron provenances have significant genetic variation, and remote genetic relationship. Among which, the maximal genetic identity of the Exi region and Youyang is 0.9567, which shows that the genetic relationship of these two populations is closer than other populations. However, the minimum genetic relationship of Mengla and Louisiana (USA) is 0.1478, which indicates that the genetic relationship of these two populations is farther than other populations. Moreover, the genetic distance of these two populations is 1.9057 (Table S3b), which was the highest among all comparisons. Furthermore, many loci (93% among L. chinense and 68.97% among L. tulipifera) significantly deviated from Hardy–Weinberg proportions in all independent sample groups originating from a single provenance, possibly due to insufficient sample size. So, in the preliminary view of genetic distance, we took the provinces of China as units and individuals from USA as a whole. A UPGMA dendrogram dependent on Nei’s genetic distance (Table S3a) showed that the 35 L. chinense plants collected from eight different provinces were grouped into three distinct clusters (Figure 3). In the first cluster, all members belonged to eastern China (Hunan, Jiangxi, Anhui, and Zhejiang). The second cluster consisted of plants from Hubei, Sichuan, and Guizhou, all located in western China. The third cluster contained plants coming exclusively from Yunan, which has been described as one of the main refugia for plants that suffered the quaternary glacial period [37]. Moreover, Nei’s genetic distance of Yunan cluster was the farthest from populations of L. tulipifera (1.3302; Table S3a). After we removed population samples of insufficient size (numbering less than three individuals), 13 populations were left, comprising nine L. chinense populations and four L. tulipilfera populations. Using this data, we constructed a new and robust UPGMA dendrogram. Nine L. chinense populations grouped into five main clusters (Figure 3). In the first cluster, all populations were located in eastern China (Liuyang, Songyang, Sangzhi, Lushan). The second cluster contained only one population from Songtao, Guizhou Provinces. The third cluster contained two populations mainly distributed in Sichuan Province, over western China. Plants from Liping, belonging to another Guizhou Province population, were taken as the fourth cluster. What is more, the population was originating from Mengla, Yunnan Province was still an independent branch from other L. chinense populations, similar to its position in the previous UPGMA dendrogram (Figure 3).
4. Discussion
Many genomic resources have been developed for L. tulipifera, even whole genome sequencing of L. chinense has been completed [36,38]. However, only a few L. tulipifera SSR markers have been tested for their degree of polymorphism by polyacrylamide denaturing gels [19,21]. Compared to other species, a sufficient collection of polymorphic and informative SSR markers within Liriodendron is still lacking. A large number of EST-SSRs were developed, which will allow a better understanding of the genetic diversity and facilitate the application in breeding programs. Traditional cultivar identification and classification depended on morphological characters like leaf blade, flower color and so on [5], but the accuracy was often affected by environmental factors. The molecular markers developed from our study were efficient alternatives to morphological identification, especially for hybrids, which will lay a foundation for Liriodendron breeding in the future. Previous studies did report the presence of a handful of species-specific SSR markers used to genotype L. tulipifera, L. chinense, and hybrid Liriodendron. One such reported marker is LT008 (Figure S5), which is a species-specific primer pair. With the LT008, a 190 bp fragment from L. chinense, 180 bp fragment from L. tulipifera, and a 190 bp and a 180 bp fragment from hybrid Liriodendron were respectively amplified [20]. Compared to LT008, ESSR119 shows some advantages for species-specific identification. ESSR119 contains a tetranucleotide motif, making it easier and more precise to genotype using a 3% agarose gel with a 12 bp size difference (Figure S5). Nowadays, Next-Generation-Sequencing technologies are constantly developing. Third generation sequencing platform has the advantage of longer reading length, including SMRT (single-molecule real time) sequencing, single-molecule nanopore DNA sequencing, and others. They are being considered as effective methods for developing SSR markers and other molecular markers such as SNP [39,40].
Based on the study of the whole genome sequence of Liriodendron, the whole-genome single nucleotide polymorphism (SNP) analysis and structure analysis showed that 20 Liriodendron accessions formed three distinct phylogenetic groups [39]. In our study, we also used similar samples except for one L. tulipifera accession, Tennessee (USA). However, just based on transcriptome data, we could classify this population into two clusters by ΔK values. This difference indicates that the variation from transcriptome is more conservative than that from genome, and the SNP may provide more genetic information that suffered from the natural selection [41].
We find that there are north-south direction mountain chains between the eastern L. chinense populations (Liuyang, Songyang, Dabieshan and Lushan) and the western ones (Songtao, Youyang, Xuyong, Liping and Mengla) (Figure 3), therefore, based on combination with UPGMA dendrogram results and geographic information, there may be a correlation between the trend of clustering of different populations and the distribution of mountain chains. Furthermore, the L. chinense populations in the Mengla, Songtao, and Liping areas cluster into relatively independent branches and which in Mengla areas are relatively independent to L. tulipifera groups, even to L. chinense (Nei’s distance = 1.3302, Table S3b). Since the genetic identity (GI) of Exi region and Sichuan Province populations (Youyang and Xuyong) are all more than 0.9, we made a further analysis based on the combination of them. The result indicates there is probably high geographic variation among the populations of western China (Yunan-Guizhou region, Sichuan Province, and Exi region). Previous studies [38,42] have shown that the L. chinense populations in western China have higher genetic diversity. However, the genetic diversity of L. chinense in this area as shown by our EST-SSR markers is relatively lower than others. Analysis based on trend lines of He and the proportion of polymorphic loci showed populations in Yunan regions has the lowest genetic diversity (Figure 4). Also, the results of Shannon’s index and Nei’s expected heterozygosity also indicated that the genetic diversity of the western China L. chinense populations was lower than which of the eastern China ones (1.1214 and 1.2021 vs. 1.2764, Table S4). Compared to previous studies, the selected populations in this paper cover larger distribution regions of L. chinense and represent the typical groups of the existing populations. Therefore, we hold the view that there is still a particular controversy to evaluate the genetic diversity of the eastern and western populations based on a single kind of molecular marker. In future studies, the number of samples should be expanded as much as possible under funds taken into account, so that relatively more reliable results could be obtained through combining multiple markers. On the other hand, there are many mountains in the Yunnan–Guizhou region which are natural barriers hindering further communication between population, making them distributed like many small isolated islands. Most populations there are small [3,43], limiting the flow of genes (Nm = 0.7797) from one population to another, so the geographical variation is very high in this area, meanwhile, low-level genetic variation of L. chinense is prone to be influenced by habitat destruction, as the potentials of adaptation to environmental changes of the species [43]. In addition, from Figure 4, we also found that the genetic diversity of populations is higher in the middle and lower reaches of the Yangtze River (Liuyang; Sangzhi; Lushang; Songyang), which may be closely related to the direction of mountains and rivers which benefits communication between different populations (Figure 3). Overall, based on information provided by SSR, such as Nei’s genetic information (GI), F-statistics (Tables S2 and S5), we found that the genetic diversity of L. chinense is higher than that of L. tulipifera, which is consistent with the results obtained by Nucleotide diversity analysis reported in the paper about genome of Liriodendron [38]. This may be related to numbers of suitable shelters in East Asia [44]. However, we also need to consider the reproduction system. Considering that the population number and size of L. chinense is decreasing gradually, the inbreeding probability in the population increases greatly, which will inevitably increase the homozygous probability of harmful genes, thus possibly aggravate inbreeding depression, affecting diversity and increasing the degree of endangerment. Due to the lack of up-to-date data on the population survival status of L. chinense in natural state, our research conclusions still have some limitations.
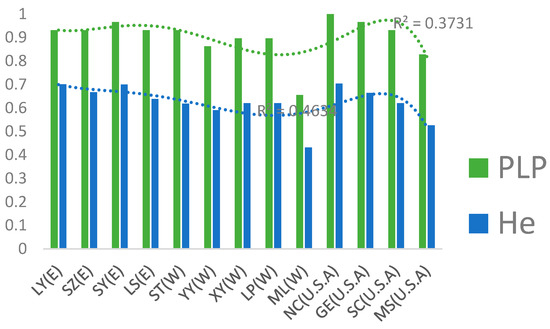
Figure 4.
Genetic diversity and trendline of nine L. chinense provenances and four L. tulipifera provenances. The genetic diversity of the western China provenances was lowest compared to the others. E in the bracket represents the eastern China population and W indicates the Western China population. The conjunction curve is a sextuple polynomial. He: expected heterozygosity; PLP: the proportion of polymorphic loci at the 5% level.
5. Conclusions
In conclusion, the novel 29 EST-SSRs developed in this study will be useful tools for future applications, such as prediction of genetic gain, Tree breeding based on molecular markers, and molecular phytogeography. The results of this study revealed a higher level of genetic differentiation among populations of L. chinense than those of L. tulipifera. Based on genetic distances, L. chinense individuals clustered into two main groups (the eastern China L. chinense and the western China L. chinense). According to such a clustering result, the genetic diversity of provenance in eastern China is higher than that in western China. Such information can be useful in the protection of this endangered species. The most urgent measure is to preserve existing populations and individuals, since small and isolated populations with high possibility of low-level genetic diversity are more vulnerable to the change of climate or habitat. Therefore, whether in the protective process or the utilization of L. chinense, we must pay attention to maximize the genetic diversity of L. chinense, endangered species. Furthermore, in-depth study of inbreeding depression and loss of genetic diversity can provide a basis for the protection of genetic resources of L. chinense. It should not be neglected that we must take necessary measures to conduct a detailed and comprehensive survival status investigation of L. chinense.
Further studies are needed to expand the samples as much as possible. For a more consolidated conclusion, genetic diversity of provenance was further assessed by combining multiple genetic markers in consideration of cost. There is no doubt that these novel EST-SSR markers will be helpful for future research on cultivar identification, population structure, and QTL analysis for Liriodendron. Also, the analysis of genetic diversity is a prerequisite for its exploration and utilization.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/10/4/334/s1. Figure S1: (a) Frequencies of the various 418 EST-SSR motifs; (b) PCR amplification results separated by electrophoresis on 8% polyacrylamide denaturing gels and visualized with silver nitrate staining based on random eight Liriodendron individuals. Figure S2: PCR products amplified by EST-SSR marker (ESSR116). Figure S3: Structure clustering results of 14 L. chinense and 5 L. tulipifera provenances based on EST-SSR markers. Figure S4: The population structure based on 13 provenances whose sample greater than three individuals at K = 2 as determined using both an estimate of the posterior probability of the data for a given K and the Evanno ΔK. Figure S5: PCR products amplified by two EST-SSR markers (a. ESSR119; b. LT008.) within the same set of samples. Table S1: Provenances information for the L. chinense and L. tulipifera samples used in this study. Table S2: Fixation index and F-statistics in L. chinense and L. tulipifera. Table S3: (a) Nei’s (1978) unbiased Measures of Genetic Identity and Genetic distances between L. chinense trees from eight provinces and L. tulipifera trees from the USA; (b) Nei’s (1978) Unbiased Measures of Genetic Identity and Genetic distance based on 19 populations provenances. Table S4: Genetic diversity analysis in three different L. chinense populations (Eastern China region, Sichuan Provinceand Exi region, Yunan-Guizhou region) and L. tulipifera populations. Table S5: Shannon’s information index (I) analysed by SSR markers. Table S6: The true K value was determined using both an estimate of the posterior.
Author Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. conceptualization, J.C. and X.L.; methodology, Z.H. and X.L.; software, Z.H.; validation, X.L. and Y.W.; formal analysis, Z.H.; investigation, S.L. and Y.S.; resources, L.G.; data curation, S.L and X.L.; writing—original draft preparation, X.L.; writing—review and editing, X.L., J.Shi and J.C; visualization, Y.S.; supervision, J.C. and J.Shi; project administration, J.Shi; funding acquisition, J.C.
Funding
This research was supported by Key research and development plan of Jiangsu Province (BE2017376), Foundation of Jiangsu forestry bureau (LYKJ[2017]42), the Nature Science Foundation of China (31770715), the Qinglan project of Jiangsu province, and Priority Academic Program Development of Jiangsu Higher Education Institutions. The funders played no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Acknowledgments
We would like to thank Sheng Zhu at College of biology and the environment, Nanjing Forestry University (NJFU), for valuable advices in improving the paper.
Conflicts of Interest
The authors declare that they have no competing interest.
Abbreviations
| L. chinense | Liriodendron chinense |
| L. tulipifera | Liriodendron tulipifera |
| EST-SSR | Expressed Sequence Tag/Microsatellites or simple sequence repeats |
| MAS | Marker-assisted selection |
| CTAB | Cetyltrimethyl Ammonium Bromide |
| Na | Number of alleles |
| Ho | observed heterozygosity |
| He | expected heterozygosity |
| HWE | Hardy-Weinberg equilibrium text |
| I | Shannon index |
| PIC | Polymorphism Information Content |
References
- Parks, C.R.; Wendel, J.F. Molecular Divergence between Asian and North American Species of Liriodendron (Magnoliaceae) with Implications for Interpretation of Fossil Floras. Am. J. Bot. 1990, 77, 1243–1256. [Google Scholar] [CrossRef]
- Xiang, Q.-Y.; Soltis, D.E.; Soltis, P.S.; Manchester, S.R.; Crawford, D.J. Timing the Eastern Asian–Eastern North American Floristic Disjunction: Molecular Clock Corroborates Paleontological Estimates. Mol. Phylogenet. Evol. 2000, 17, 335. [Google Scholar] [CrossRef]
- Ming, H.R.; An, H.S. Geographical distribution of Liriodendron chinense in China and its significance. J. Plant Resour. Environ. 1995, 4, 1–6. [Google Scholar]
- Xiang, Q.-Y.; Soltis, D.E.; Soltis, P.S. The Eastern Asian and Eastern and Western North American Floristic Disjunction: Congruent Phylogenetic Patterns in Seven Diverse Genera. Mol. Phylogenet. Evol. 1998, 10, 178–190. [Google Scholar] [CrossRef]
- Zheng, W. Dendrology of China; China Forestry Press: Beijing, China, 1983; Volume 1. (In Chinese) [Google Scholar]
- Zhang, Y.-B.; Ma, K.-P. Geographic distribution patterns and status assessment of threatened plants in China. Biodivers. Conserv. 2008, 17, 1783–1798. [Google Scholar] [CrossRef]
- Fang, Y. Geographical distribution and spatial pattern of Liriodendron chinense. J. Nanjing For. Univ. Nat. Sci. Ed. 1994, 18, 13–18. (In Chinese) [Google Scholar]
- Hao, R.; He, S. Studies on Natural Population Dynamics and Dangerous Habitats of Liriodendron chinense. J. Plant Ecol. 1999, 23, 1. [Google Scholar] [CrossRef]
- Jin, X.U.; Wang, Z. Genetic variation of floral character and pollen viability of Liriodendron hybrid and its parents. J. Plant Resour. Environ. 2001, 10, 31–34. [Google Scholar]
- Williams, R.S.; Feist, W.C. Durability of yellow-poplar and sweetgum and service life of finishes after long-term exposure. For. Prod. J. 2004, 54, 96–101. [Google Scholar]
- Chen, J.-H.; Lin, S.-S.; Wang, W.-X.; Yuan, S.-T.; Shi, J.-S.; Jia, A.-Q. The extract, LXB-1, from the barks of Liriodendron × hybrid, induced apoptosis via Akt, JNK and ERK1/2 pathways in A549 lung cancer cells. Z. Für Nat. C 2015, 70, 305–311. [Google Scholar] [CrossRef]
- Chen, J.-H.; Yang, G.-X.; Ding, Q.; Xia, T.-S.; Shi, J.; Jia, A.-Q. In vitro tumor cytotoxic activities of extracts from three Liriodendron plants. Pak. J. Pharm. Sci. 2013, 26, 233–237. [Google Scholar]
- Moon, M.K.; Oh, H.M.; Kwon, B.-M.; Baek, N.-L.; Kim, S.-H.; Kim, J.S.; Kim, D.K. Farnesyl protein transferase and tumor cell growth inhibitory activities of lipiferolide isolated fromLiriodendron tulipifera. Arch. Pharmacal Res. 2007, 30, 299–302. [Google Scholar] [CrossRef]
- Gerloff, U.; Schlötterer, C.; Rassmann, K.; Rambold, I.; Hohmann, G.; Fruth, B.; Tautz, D. Amplification of hypervariable simple sequence repeats (microsatellites) from excremental DNA of wild living bonobos (Pan paniscus). Mol. Ecol. 1995, 4, 515–518. [Google Scholar] [CrossRef]
- Vasemägi, A.; Nilsson, J.; Primmer, C.R. Expressed Sequence Tag-Linked Microsatellites as a Source of Gene-Associated Polymorphisms for Detecting Signatures of Divergent Selection in Atlantic Salmon (Salmo salar L.). Mol. Boil. Evol. 2005, 22, 1067–1076. [Google Scholar]
- Reeve, A. Genetic diversity and differentiation of the Critically Endangered Hispaniolan palm Coccothrinax jimenezii M.M. Mejía & M.M. García based on novel SSR markers). Biochem. Syst. Ecol. 2016, 66, 216–223. [Google Scholar]
- Gutiérrez-Ozuna, R.; Hamilton, M.B. Identification and characterization of microsatellite loci in the tuliptree, Liriodendron tulipifera (Magnoliaceae). Appl. Sci. 2017, 5, 1700032. [Google Scholar] [CrossRef]
- Xu, M.; Liu, X.; Wang, J.W.; Teng, S.Y.; Shi, J.Q.; Li, Y.Y.; Huang, M.R. Transcriptome sequencing and development of novel genic SSR markers for Dendrobium officinale. Mol. Breed. 2017, 37, 18. [Google Scholar]
- Xu, M.; Sun, Y.; Li, H. EST-SSRs development and paternity analysis for Liriodendron spp. New For. 2010, 40, 361–382. [Google Scholar] [CrossRef]
- Zhang, H.L.; Li, H.G.; Xu, M.; Feng, Y.H. Identification of Liriodendron tulipifera, Liriodendron chinense and hybrid Liriodendron using species-specific SSR markers. Sci. Silvae Sin. 2010, 46, 36–39. [Google Scholar]
- Yang, A.-H.; Zhang, J.-J.; Tian, H.; Yao, X.-H. Characterization of 39 novel EST-SSR markers for Liriodendron tulipifera and cross-species amplification in L. chinense (Magnoliaceae). Am. J. Bot. 2012, 99, e460–e464. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Zhang, J.; Ye, Q.; Huang, H. Characterization of 14 novel microsatellite loci in the endangered Liriodendron chinense (Magnoliaceae) and cross-species amplification in closely related taxa. Conserv. Genet. 2008, 9, 483–485. [Google Scholar] [CrossRef]
- Kobayashi, N. A simple and efficient DNA extraction method for plants, especially woody plants. Plant Tissue Cult Biotech 1998, 4, 76–80. [Google Scholar]
- Lalitha, S. Primer Premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Bo, J.; Hai-Bin, X.U.; Yang, X.U.; Long, X.F.; Chen, J.H.; Shi, J.S. Optimization of Genomic-SSR Reaction System in Liriodendron. For. Res. 2013, 26, 506–510. [Google Scholar]
- Zimmermann, J.; Voss, H.; Erfle, H.; Rupp, T.; Dietrich, T.; Hewitt, N.A.; Schwager, C.; Stegemann, J.; Ansorge, W. Direct sequencing of PCR products using magnetic beads and fluorescein-12-Dutp. Methods Mol. Cell. Biol. 1992, 3, 114–115. [Google Scholar]
- Yeh, F.; Yang, R.; Boyle, T.; Ye, Z.; Mao, J.; Yeh, C.; Timothy, B.; Mao, X. POPGENE version 1.32; the User-Friendly Shareware for Population Genetic Analysis; Center for International Forestry Research, University of Alberta: Edmonoto, AB, Canada, 1997. [Google Scholar]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, version 2.2, 2.1; Department of Ecoloy and Evolution, State University of New York: New York, NY, USA, 2005. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J.J.M.E. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2010, 14, 2611–2620. [Google Scholar] [CrossRef]
- Metzgar, D.; Bytof, J.; Wills, C. Selection Against Frameshift Mutations Limits Microsatellite Expansion in Coding DNA. Genome Res. 2000, 10, 72–80. [Google Scholar]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Chen, J. Studies on the somatic embryogenesis of liriodendron hybrids (L. chinense × L. tulipifera). Sci. Silvae Sin. 2003, 39, 49–53. [Google Scholar]
- Zhang, X.; Carlson, A.; Tian, Z.; Staton, M.; E Schlarbaum, S.; E Carlson, J.; Liang, H. Genetic characterization of Liriodendron seed orchards with EST-SSR markers. J. Sci. Mol. Breed. 2015, 4, 1. [Google Scholar] [CrossRef]
- Lei, M.; Wang, Q.; Wu, Z.-J.; López-Pujol, J.; Li, D.-Z.; Zhang, Z.-Y. Molecular phylogeography of Fagus engleriana (Fagaceae) in subtropical China: Limited admixture among multiple refugia. Tree Genet. Genomes 2012, 8, 1203–1212. [Google Scholar] [CrossRef]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat Plants 2019, 5, 18–25. [Google Scholar] [CrossRef]
- Grohme, M.A.; Soler, R.F.; Wink, M.; Frohme, M. Microsatellite marker discovery using single molecule real-time circular consensus sequencing on the Pacific Biosciences RS. BioTechniques 2013, 55, 253–256. [Google Scholar] [CrossRef]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef]
- Wright, S.I.; Andolfatto, P. The Impact of Natural Selection on the Genome: Emerging Patterns in Drosophila and Arabidopsis. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 193–213. [Google Scholar] [CrossRef]
- Zhu, X.; Ma, J.; Yao, Q.; Hao, R.; He, S. Allozyme verification on the population differentiation of Liriodendron chinense (Hemsl.) Sarg. J. Plant Resour. Environ. 1995, 4, 9–14. [Google Scholar]
- Zaouali, Y.; Chograni, H.; Trimech, R.; Boussaid, M. Genetic diversity and population structure among Rosmarinus officinalis L. (Lamiaceae) varieties: Var. typicus Batt. and var. troglodytorum Maire. based on multiple traits. Ind. Crop. Prod. 2012, 38, 166–176. [Google Scholar] [CrossRef]
- Wen, J. Evolution of eastern asian and eastern north american disjunct distributions in flowering plants. Annu. Rev. Ecol. Syst. 1999, 30, 421–455. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).