Species Diversity Associated with Foundation Species in Temperate and Tropical Forests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Forest Dynamics Plots in the Americas
2.2. Identifying Candidate Foundation Species
2.3. Metrics of Forest Structure and Species Diversity
2.4. Codispersion Analysis
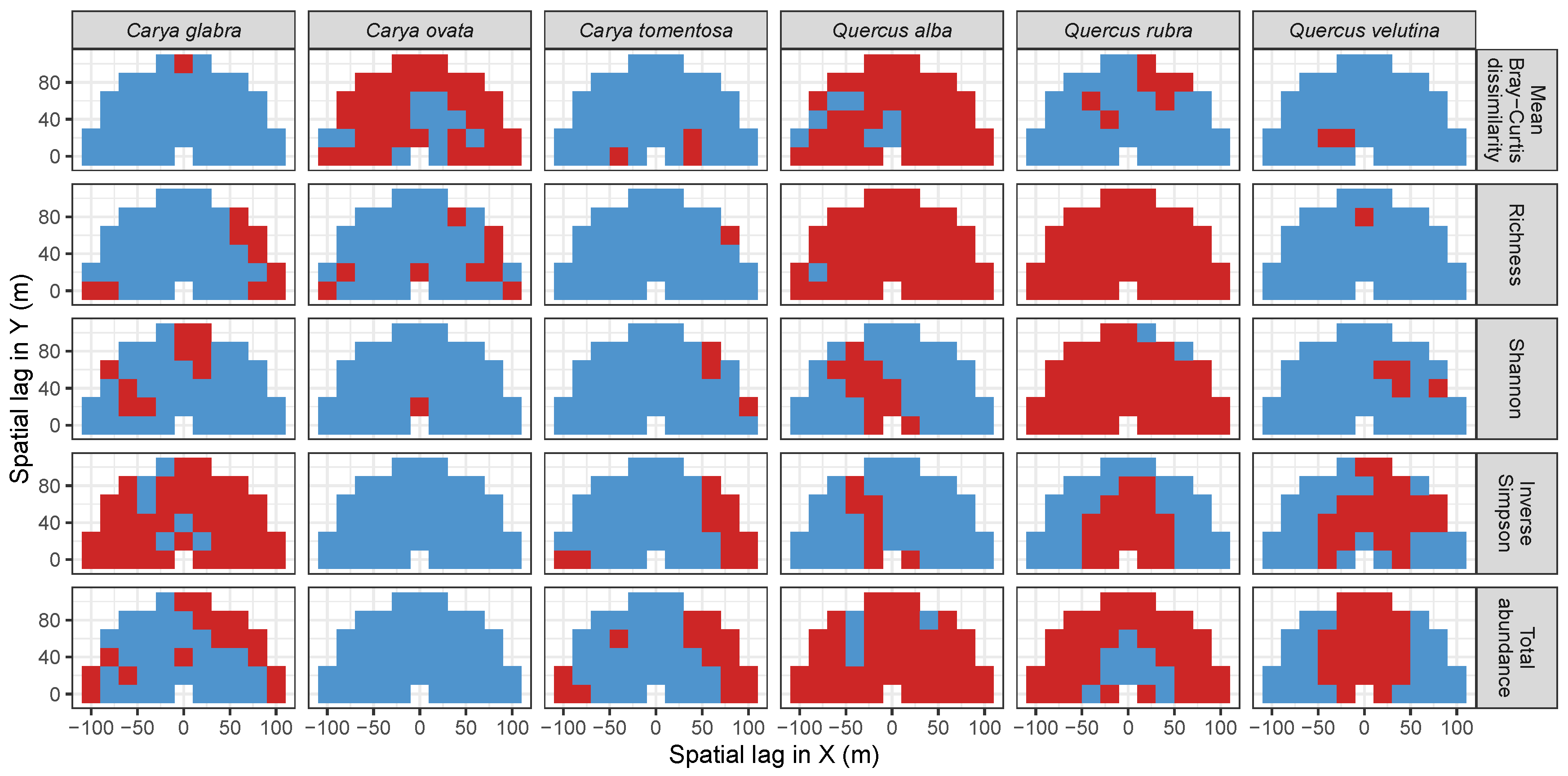
2.5. Significance Testing with Null Model Analysis
2.6. Data and Code Availability
3. Results
3.1. Forest Structure and Species Diversity
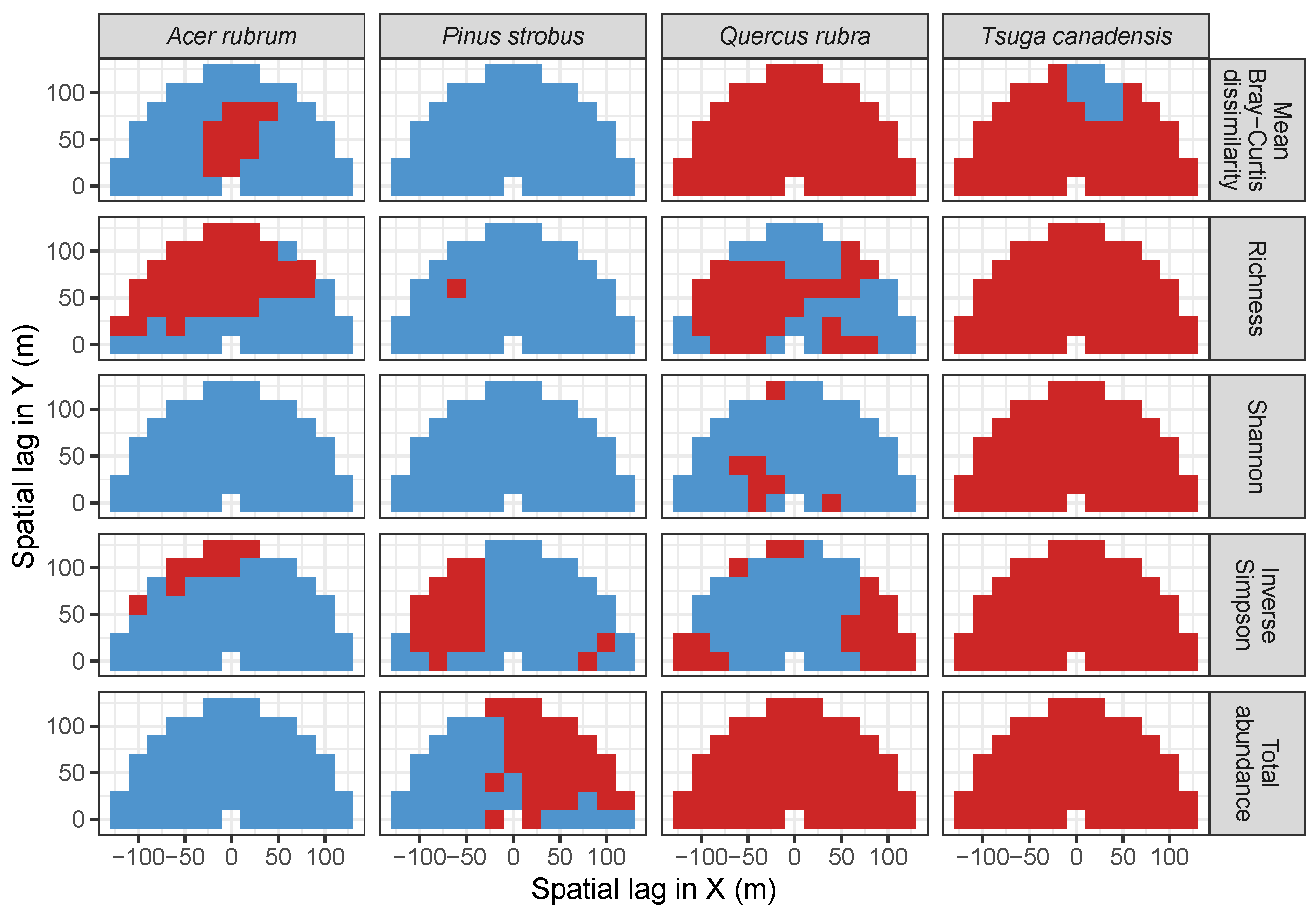
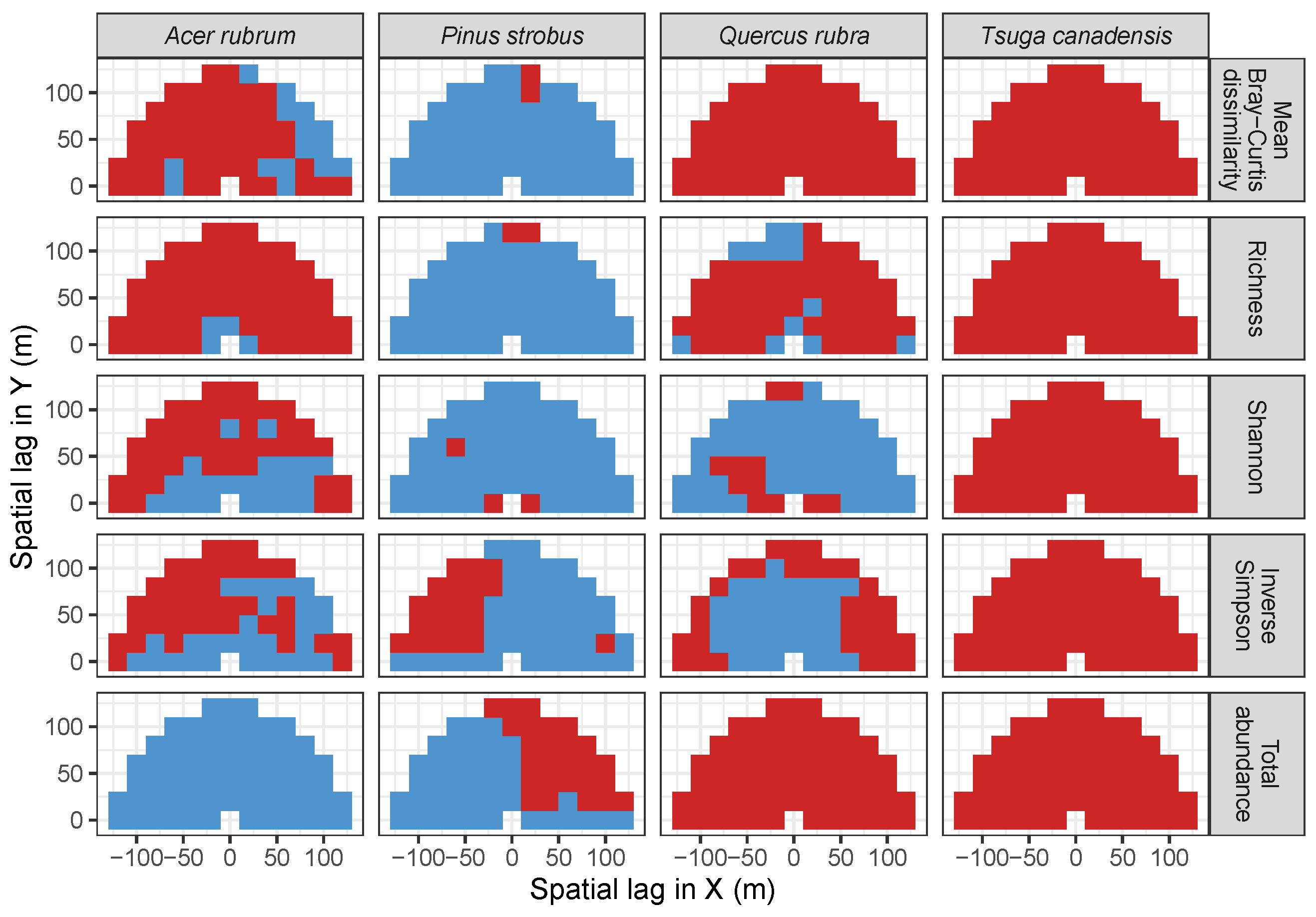
3.2. Codispersion Between Focal Species and Associated-species Diversity at Harvard Forest
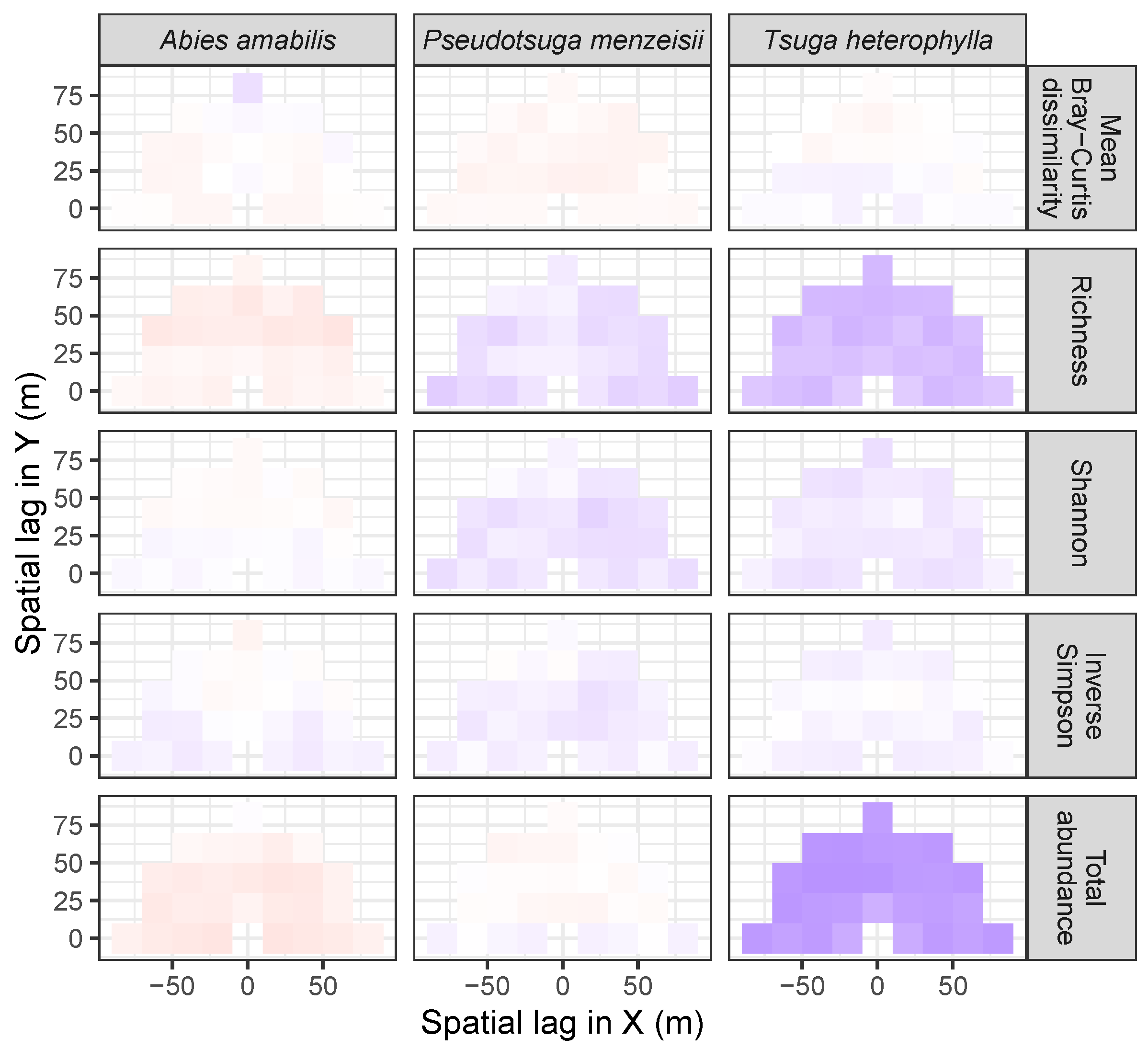
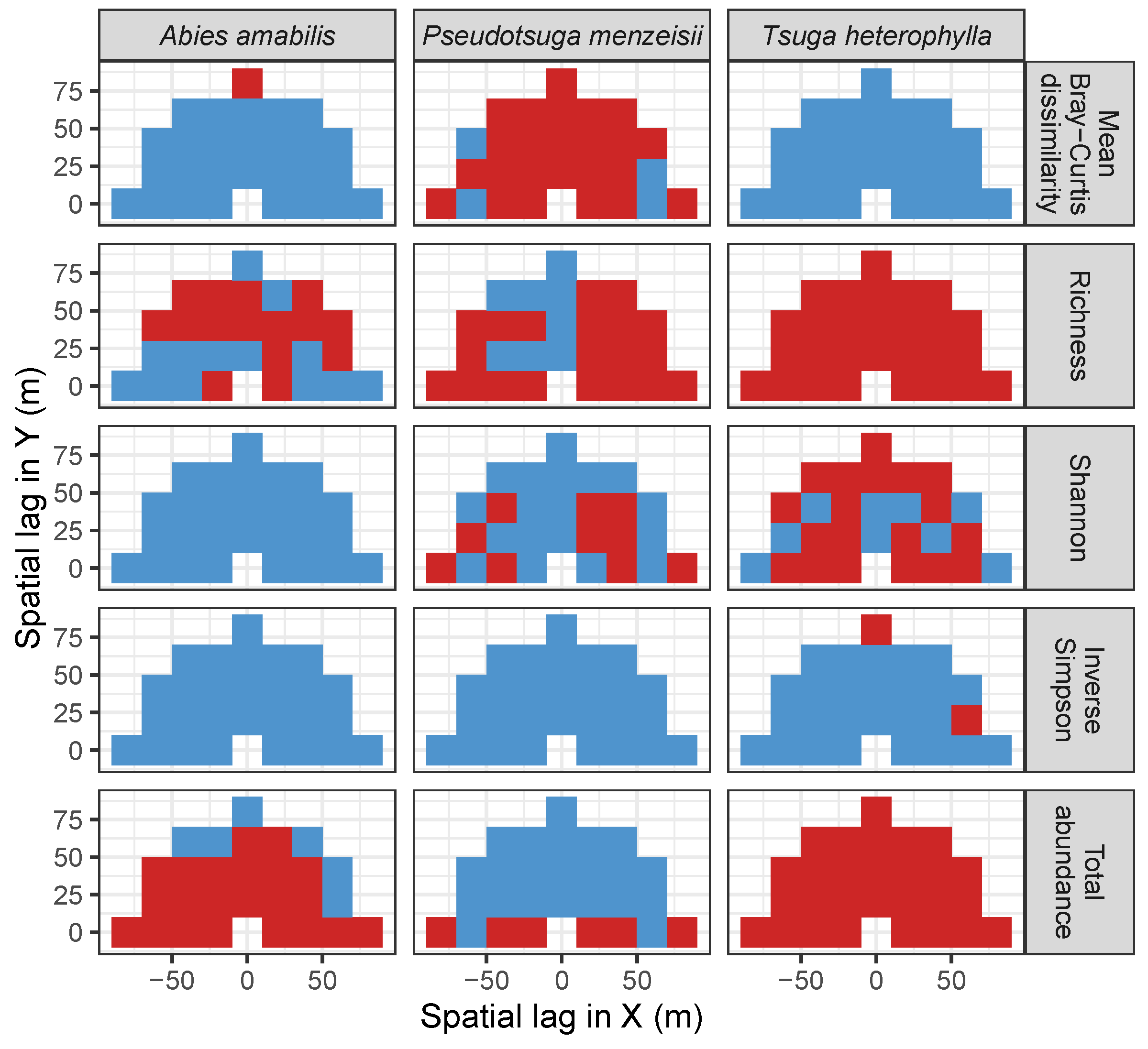
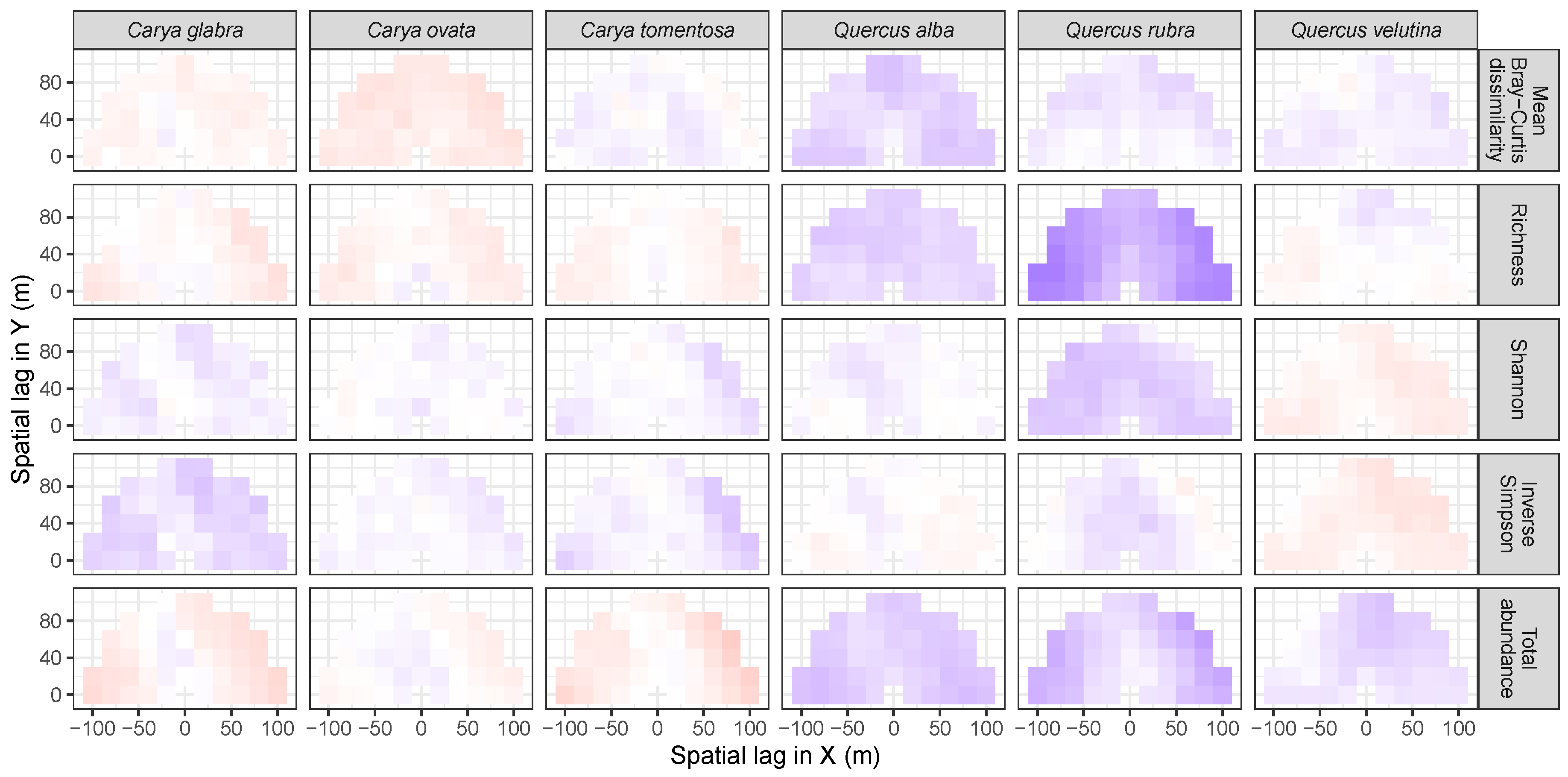
3.3. Codispersion Between Focal Tree Species and Associated-Species Diversity at the Other Five Sites
3.3.1. Trees at temperate sites
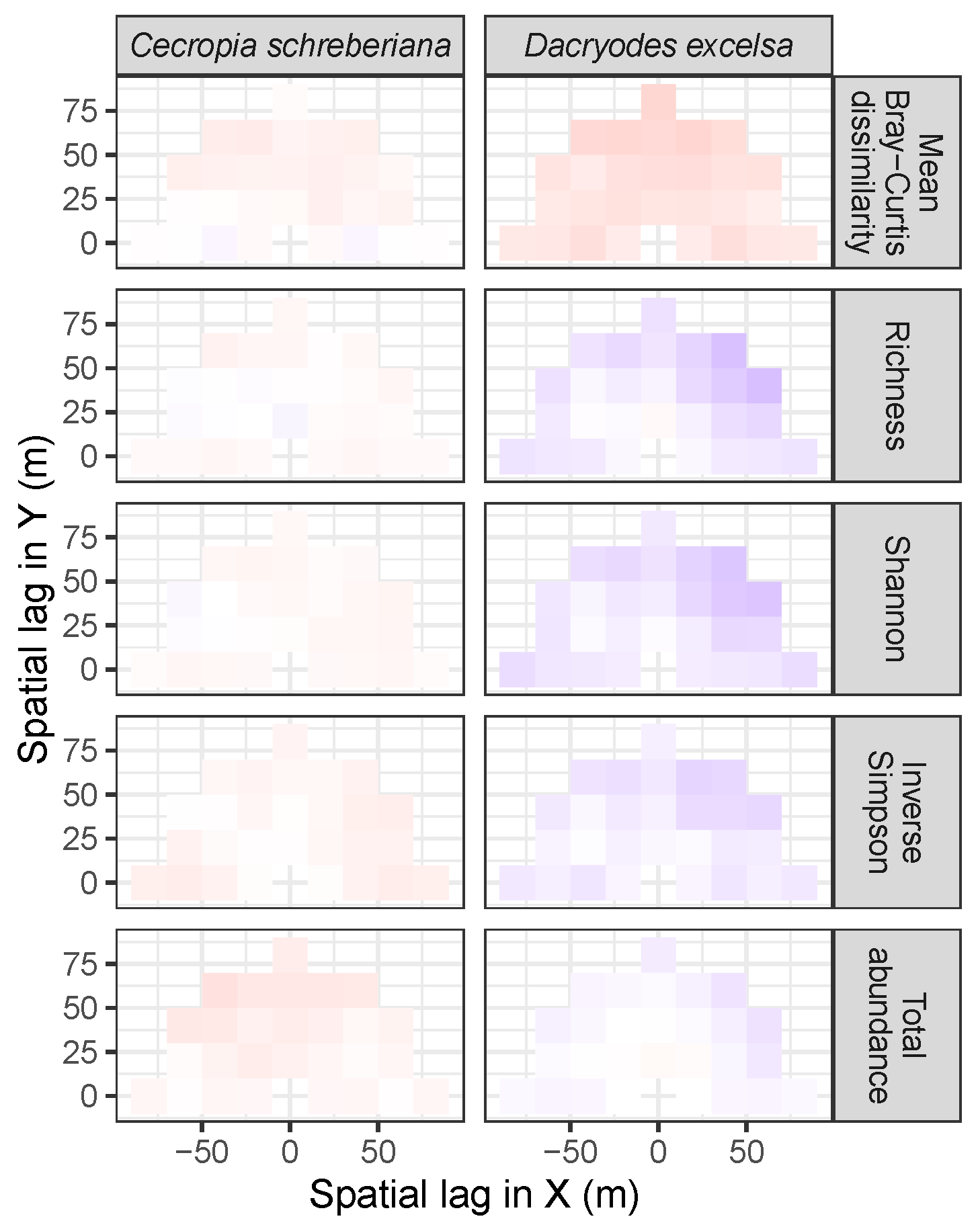
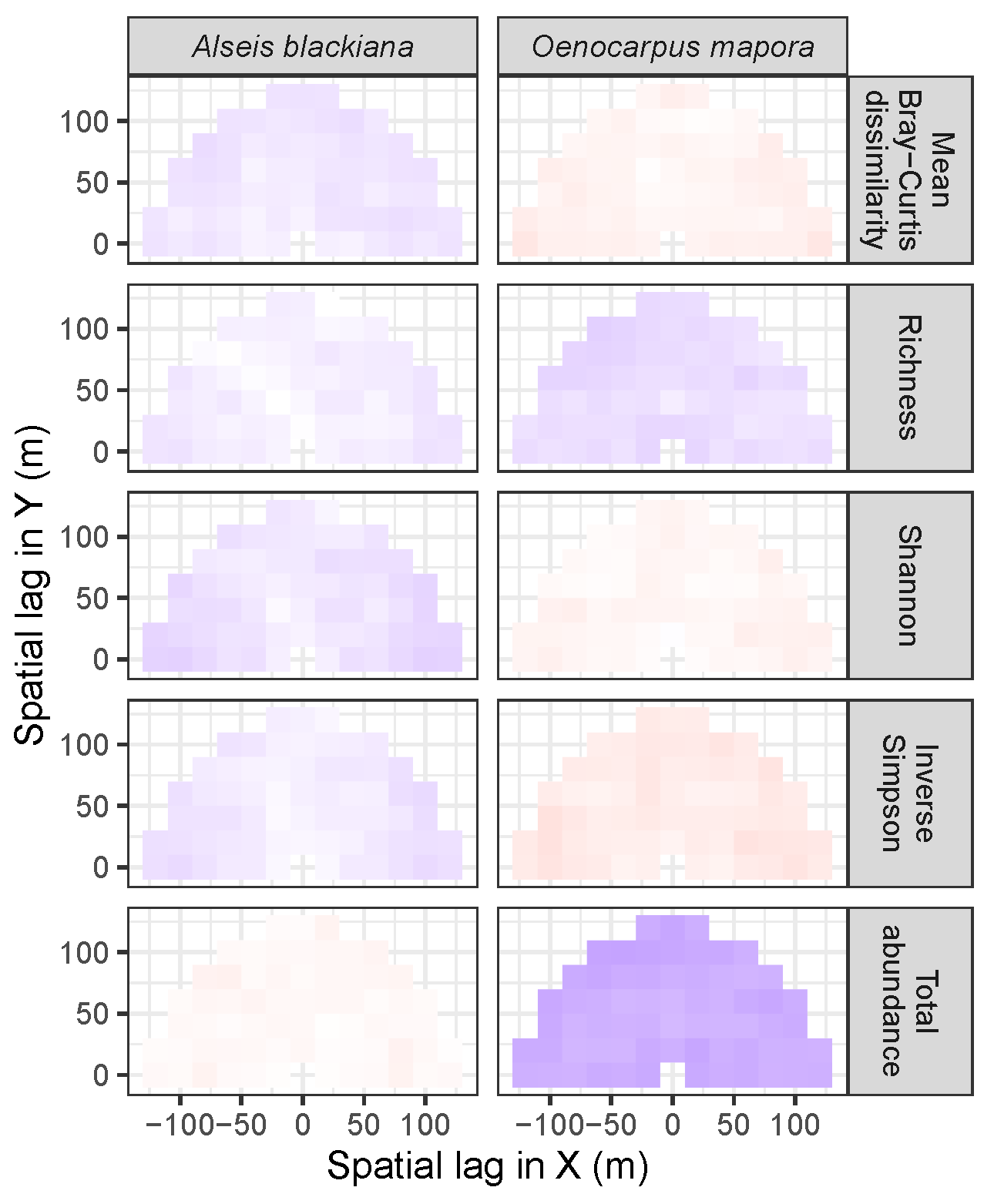
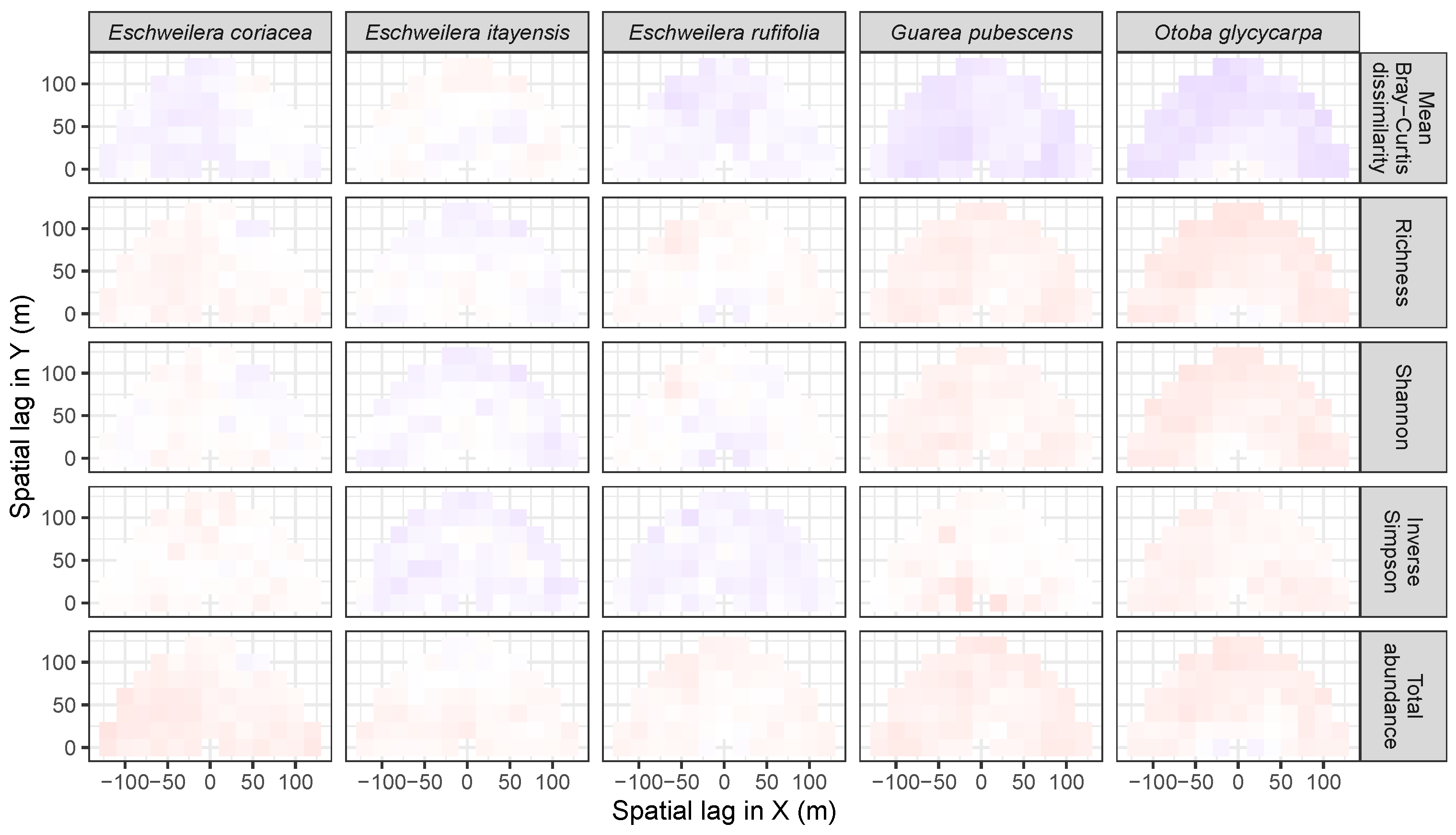
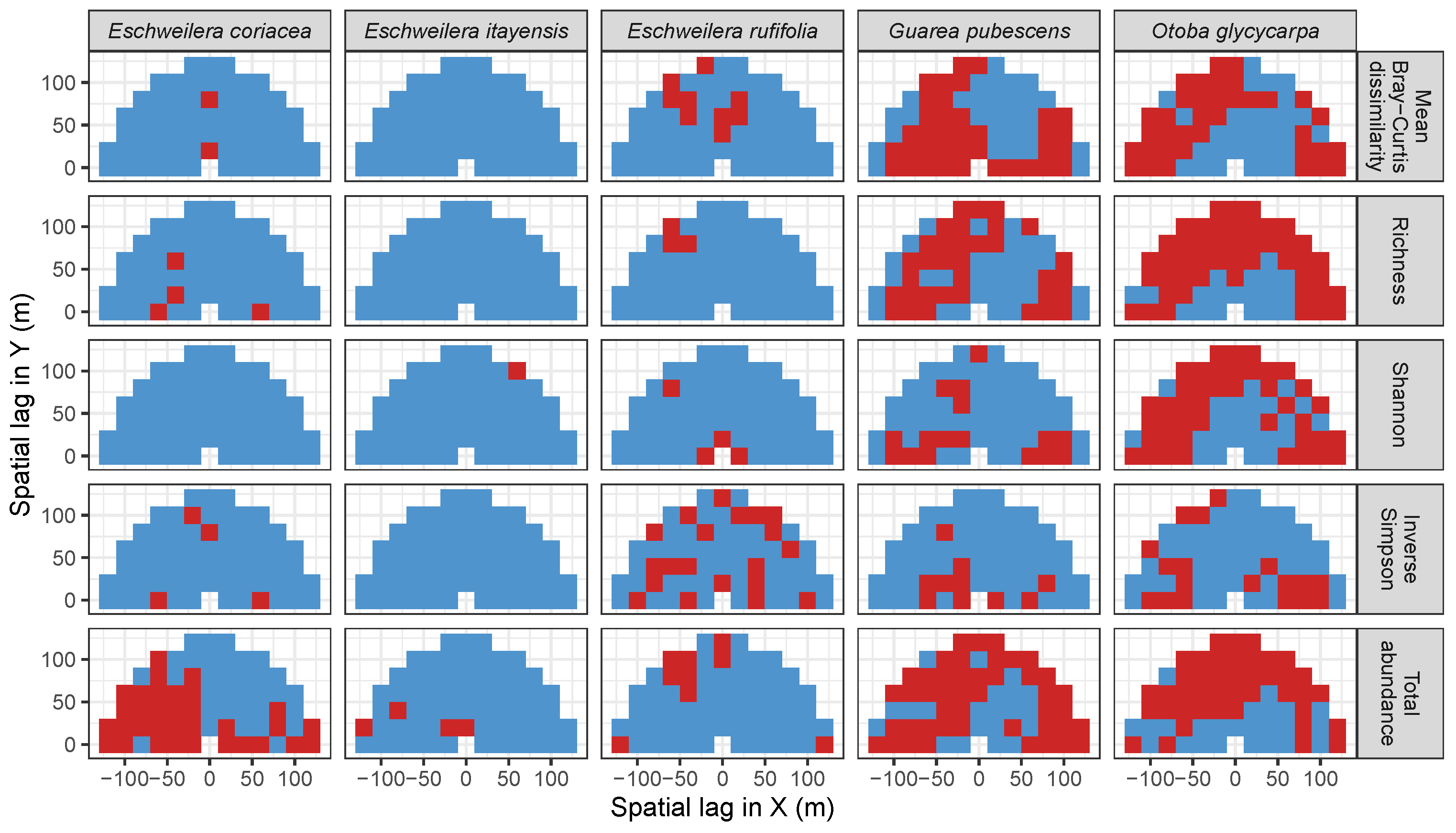
3.3.2. Trees at Tropical Sites
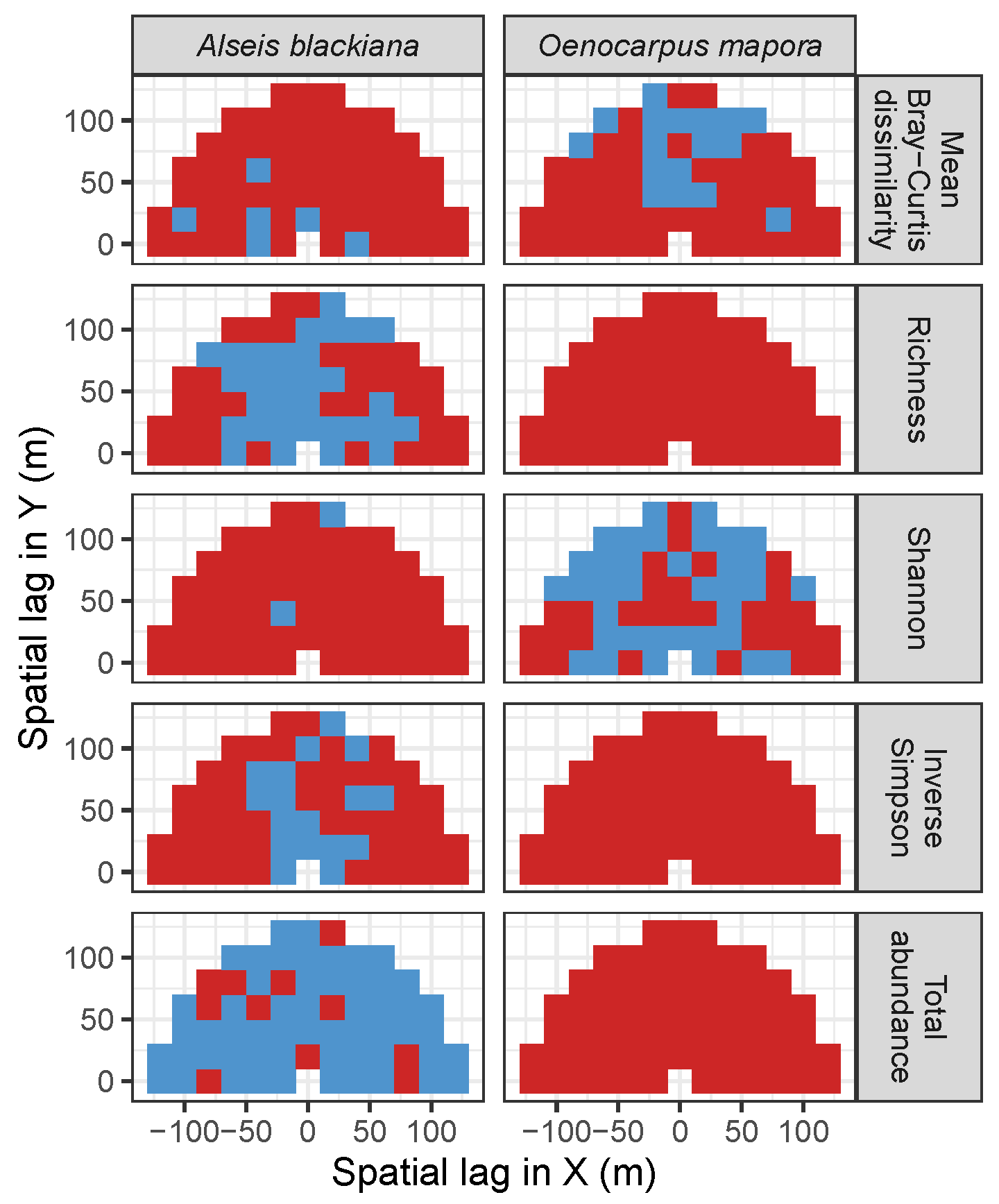
3.4. Codispersion Between Focal Understory Species and Associated-species Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Supplemental Tables and Figures
| Site | Species | Diversity Metric | Min | Median | Mean (SD) | Max |
|---|---|---|---|---|---|---|
| WR | Pseudotsuga menzeisii | Bray-Curtis | 0.05 | 0.11 | 0.11 (0.02) | 0.15 |
| Richness | −0.21 | −0.14 | −0.13 (0.05) | −0.06 | ||
| Shannon | −0.15 | −0.07 | −0.07 (0.04) | 0.02 | ||
| Simpson | −0.08 | −0.01 | 0 (0.04) | 0.08 | ||
| Abundance | −0.11 | −0.03 | −0.04 (0.04) | 0.03 | ||
| Tsuga heterophylla | Bray−Curtis | −0.04 | 0.01 | 0.01 (0.03) | 0.06 | |
| Richness | −0.31 | −0.27 | −0.27 (0.03) | −0.21 | ||
| Shannon | −0.13 | −0.09 | −0.09 (0.03) | −0.03 | ||
| Simpson | −0.09 | −0.04 | −0.04 (0.03) | 0.03 | ||
| Abundance | −0.47 | −0.44 | −0.43 (0.03) | −0.35 | ||
| Abies amabilis | Bray−Curtis | −0.14 | 0.03 | 0.02 (0.04) | 0.07 | |
| Richness | 0.04 | 0.09 | 0.1 (0.04) | 0.17 | ||
| Shannon | −0.02 | 0.01 | 0.02 (0.03) | 0.08 | ||
| Simpson | −0.06 | 0.01 | 0 (0.03) | 0.06 | ||
| Abundance | 0.03 | 0.13 | 0.13 (0.04) | 0.19 | ||
| HF | Acer rubrum | Bray−Curtis | 0.01 | 0.06 | 0.07 (0.03) | 0.13 |
| Richness | −0.01 | 0.11 | 0.11 (0.04) | 0.19 | ||
| Shannon | −0.05 | 0.05 | 0.05 (0.03) | 0.11 | ||
| Simpson | −0.04 | 0.07 | 0.07 (0.04) | 0.14 | ||
| Abundance | −0.06 | −0.01 | 0 (0.02) | 0.05 | ||
| Pinus strobus | Bray−Curtis | −0.1 | −0.04 | −0.04 (0.04) | 0.07 | |
| Richness | −0.09 | −0.01 | 0 (0.05) | 0.09 | ||
| Shannon | −0.06 | 0.03 | 0.02 (0.04) | 0.09 | ||
| Simpson | −0.03 | 0.07 | 0.07 (0.05) | 0.17 | ||
| Abundance | −0.21 | −0.09 | −0.11 (0.06) | −0.02 | ||
| Quercus rubra | Bray−Curtis | −0.28 | −0.21 | −0.2 (0.05) | −0.07 | |
| Richness | −0.2 | −0.11 | −0.12 (0.05) | 0 | ||
| Shannon | −0.11 | 0 | 0 (0.06) | 0.12 | ||
| Simpson | −0.05 | 0.09 | 0.09 (0.08) | 0.24 | ||
| Abundance | −0.4 | −0.28 | −0.27 (0.07) | −0.1 | ||
| Tsuga canadensis | Bray−Curtis | 0.12 | 0.21 | 0.21 (0.05) | 0.28 | |
| Richness | −0.64 | −0.49 | −0.49 (0.08) | −0.29 | ||
| Shannon | −0.46 | −0.34 | −0.34 (0.07) | −0.17 | ||
| Simpson | −0.26 | −0.2 | −0.2 (0.04) | −0.07 | ||
| Abundance | −0.45 | −0.33 | −0.34 (0.06) | −0.24 | ||
| TY | Quercus alba | Bray−Curtis | −0.27 | −0.18 | −0.18 (0.05) | −0.04 |
| Richness | −0.24 | −0.18 | −0.18 (0.03) | −0.12 | ||
| Shannon | −0.16 | −0.08 | −0.08 (0.04) | 0.01 | ||
| Simpson | −0.13 | −0.03 | −0.03 (0.05) | 0.05 | ||
| Abundance | −0.26 | −0.18 | −0.19 (0.05) | −0.09 | ||
| Quercus rubra | Bray−Curtis | −0.18 | −0.08 | −0.09 (0.05) | 0 | |
| Richness | −0.55 | −0.39 | −0.39 (0.1) | −0.21 | ||
| Shannon | −0.29 | −0.23 | −0.22 (0.05) | −0.09 | ||
| Simpson | −0.2 | −0.1 | −0.09 (0.06) | 0.07 | ||
| Abundance | −0.42 | −0.21 | −0.22 (0.1) | −0.05 | ||
| Quercus velutina | Bray−Curtis | −0.16 | −0.08 | −0.07 (0.04) | 0.06 | |
| Richness | −0.14 | 0 | −0.01 (0.06) | 0.1 | ||
| Shannon | 0.02 | 0.1 | 0.1 (0.04) | 0.17 | ||
| Simpson | −0.01 | 0.13 | 0.13 (0.05) | 0.22 | ||
| Abundance | −0.28 | −0.14 | −0.15 (0.07) | −0.01 | ||
| Carya glabra | Bray−Curtis | −0.06 | 0.06 | 0.05 (0.04) | 0.12 | |
| Richness | −0.05 | 0.05 | 0.06 (0.07) | 0.22 | ||
| Shannon | −0.16 | −0.06 | −0.07 (0.04) | 0.04 | ||
| Simpson | −0.26 | −0.18 | −0.16 (0.05) | 0 | ||
| Abundance | −0.09 | 0.11 | 0.09 (0.1) | 0.25 | ||
| Carya ovata | Bray−Curtis | 0.04 | 0.15 | 0.14 (0.04) | 0.22 | |
| Richness | −0.1 | 0.08 | 0.07 (0.06) | 0.17 | ||
| Shannon | −0.13 | −0.02 | −0.02 (0.04) | 0.07 | ||
| Simpson | −0.13 | −0.05 | −0.06 (0.03) | 0.03 | ||
| Abundance | −0.07 | 0.03 | 0.03 (0.05) | 0.13 | ||
| Carya tomentosa | Bray−Curtis | −0.12 | −0.04 | −0.04 (0.05) | 0.08 | |
| Richness | −0.05 | 0.05 | 0.05 (0.05) | 0.18 | ||
| Shannon | −0.19 | −0.04 | −0.06 (0.05) | 0.04 | ||
| Simpson | −0.26 | −0.07 | −0.09 (0.08) | 0.02 | ||
| Abundance | −0.03 | 0.12 | 0.12 (0.1) | 0.35 |
| Site | Species | Diversity Metric | Min | Median | Mean (SD) | Max |
|---|---|---|---|---|---|---|
| Richness | −0.04 | 0.04 | 0.03 (0.03) | 0.09 | ||
| Shannon | −0.03 | 0.05 | 0.04 (0.03) | 0.07 | ||
| Simpson | 0.01 | 0.07 | 0.06 (0.04) | 0.12 | ||
| Abundance | 0.01 | 0.08 | 0.09 (0.05) | 0.2 | ||
| Dacryodes excelsa | Bray−Curtis | 0.12 | 0.19 | 0.19 (0.05) | 0.28 | |
| Richness | −0.28 | −0.11 | −0.11 (0.07) | 0.03 | ||
| Shannon | −0.24 | −0.1 | −0.12 (0.06) | −0.02 | ||
| Simpson | −0.18 | −0.09 | −0.09 (0.05) | −0.01 | ||
| Abundance | −0.12 | −0.03 | −0.03 (0.04) | 0.04 | ||
| BCI | Alseis blackiana | Bray−Curtis | −0.15 | −0.11 | −0.1 (0.02) | −0.05 |
| Richness | −0.12 | −0.07 | −0.07 (0.03) | 0.01 | ||
| Shannon | −0.19 | −0.12 | −0.12 (0.04) | −0.03 | ||
| Simpson | −0.16 | −0.09 | −0.09 (0.03) | −0.02 | ||
| Abundance | 0.01 | 0.05 | 0.05 (0.02) | 0.1 | ||
| Oenocarpus mapora | Bray−Curtis | 0.01 | 0.08 | 0.08 (0.03) | 0.17 | |
| Richness | −0.2 | −0.14 | −0.14 (0.02) | −0.08 | ||
| Shannon | −0.01 | 0.06 | 0.06 (0.03) | 0.11 | ||
| Simpson | 0.08 | 0.13 | 0.14 (0.03) | 0.2 | ||
| Abundance | −0.39 | −0.34 | −0.34 (0.02) | −0.3 | ||
| AM | Eschweilera coriacea | Bray−Curtis | −0.09 | −0.05 | −0.04 (0.03) | 0.06 |
| Richness | −0.07 | 0.05 | 0.04 (0.03) | 0.09 | ||
| Shannon | −0.06 | 0.01 | 0.01 (0.03) | 0.08 | ||
| Simpson | −0.02 | 0.02 | 0.03 (0.03) | 0.11 | ||
| Abundance | −0.04 | 0.09 | 0.09 (0.04) | 0.18 | ||
| Eschweilera itayensis | Bray−Curtis | −0.06 | 0.02 | 0.02 (0.03) | 0.09 | |
| Richness | −0.09 | −0.02 | −0.01 (0.03) | 0.06 | ||
| Shannon | −0.11 | −0.03 | −0.03 (0.03) | 0.03 | ||
| Simpson | −0.1 | −0.05 | −0.04 (0.03) | 0.04 | ||
| Abundance | −0.02 | 0.04 | 0.04 (0.03) | 0.12 | ||
| Eschweilera rufifolia | Bray−Curtis | −0.12 | −0.04 | −0.05 (0.03) | 0.01 | |
| Richness | −0.06 | 0.03 | 0.03 (0.04) | 0.13 | ||
| Shannon | −0.09 | 0.01 | 0 (0.04) | 0.13 | ||
| Simpson | −0.12 | −0.05 | −0.05 (0.02) | 0.02 | ||
| Abundance | −0.02 | 0.05 | 0.05 (0.03) | 0.12 | ||
| Guarea pubescens | Bray−Curtis | −0.14 | −0.09 | −0.09 (0.03) | −0.04 | |
| Richness | −0.01 | 0.09 | 0.09 (0.03) | 0.14 | ||
| Shannon | 0 | 0.08 | 0.08 (0.03) | 0.15 | ||
| Simpson | −0.01 | 0.03 | 0.04 (0.04) | 0.18 | ||
| Abundance | 0.01 | 0.1 | 0.1 (0.03) | 0.17 | ||
| Otoba glycarpa | Bray−Curtis | −0.16 | −0.11 | −0.1 (0.04) | 0.04 | |
| Richness | −0.02 | 0.13 | 0.11 (0.05) | 0.19 | ||
| Shannon | 0 | 0.11 | 0.11 (0.04) | 0.17 | ||
| Simpson | 0 | 0.07 | 0.07 (0.03) | 0.13 | ||
| Abundance | −0.05 | 0.1 | 0.09 (0.05) | 0.17 |
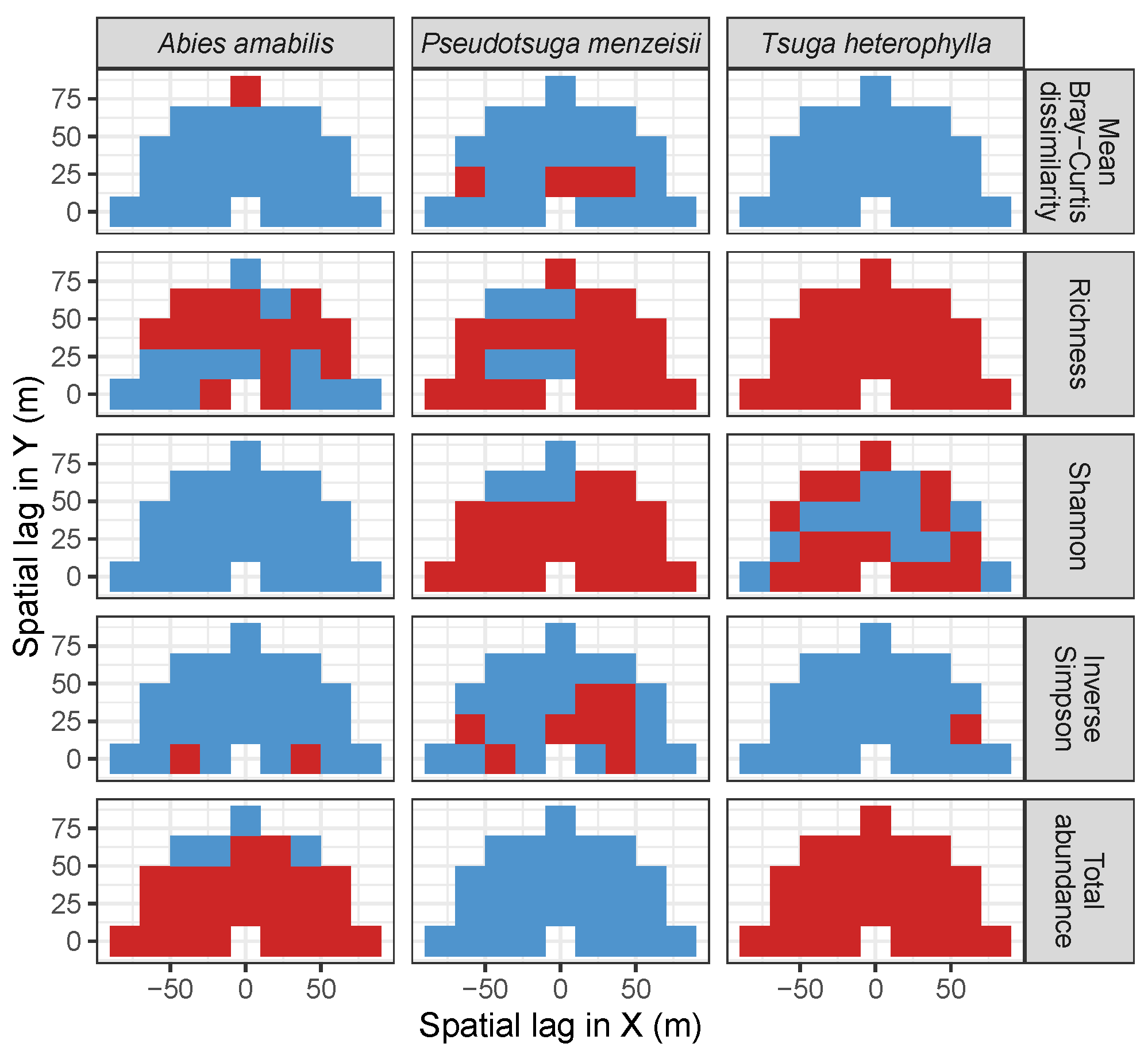
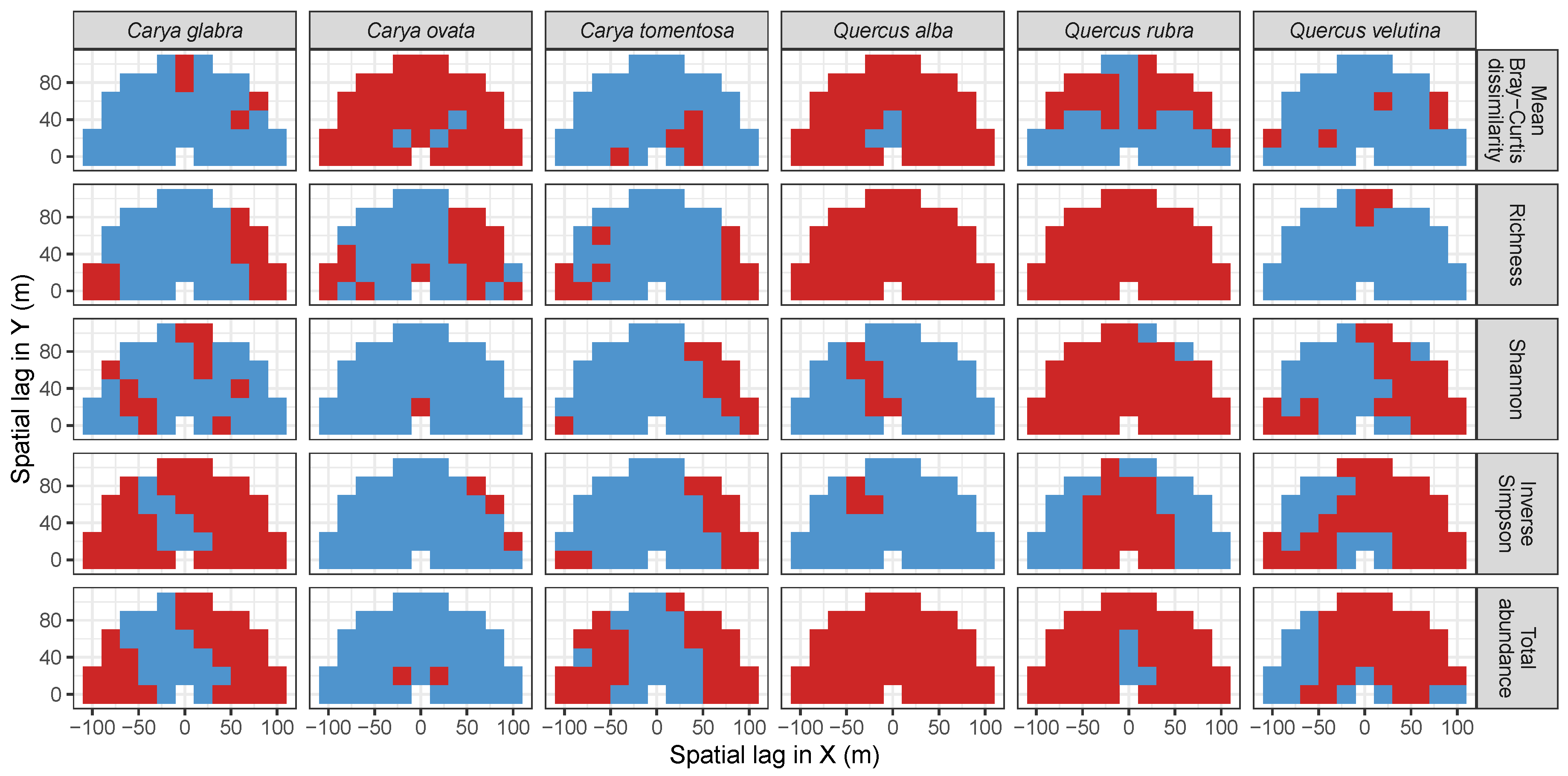
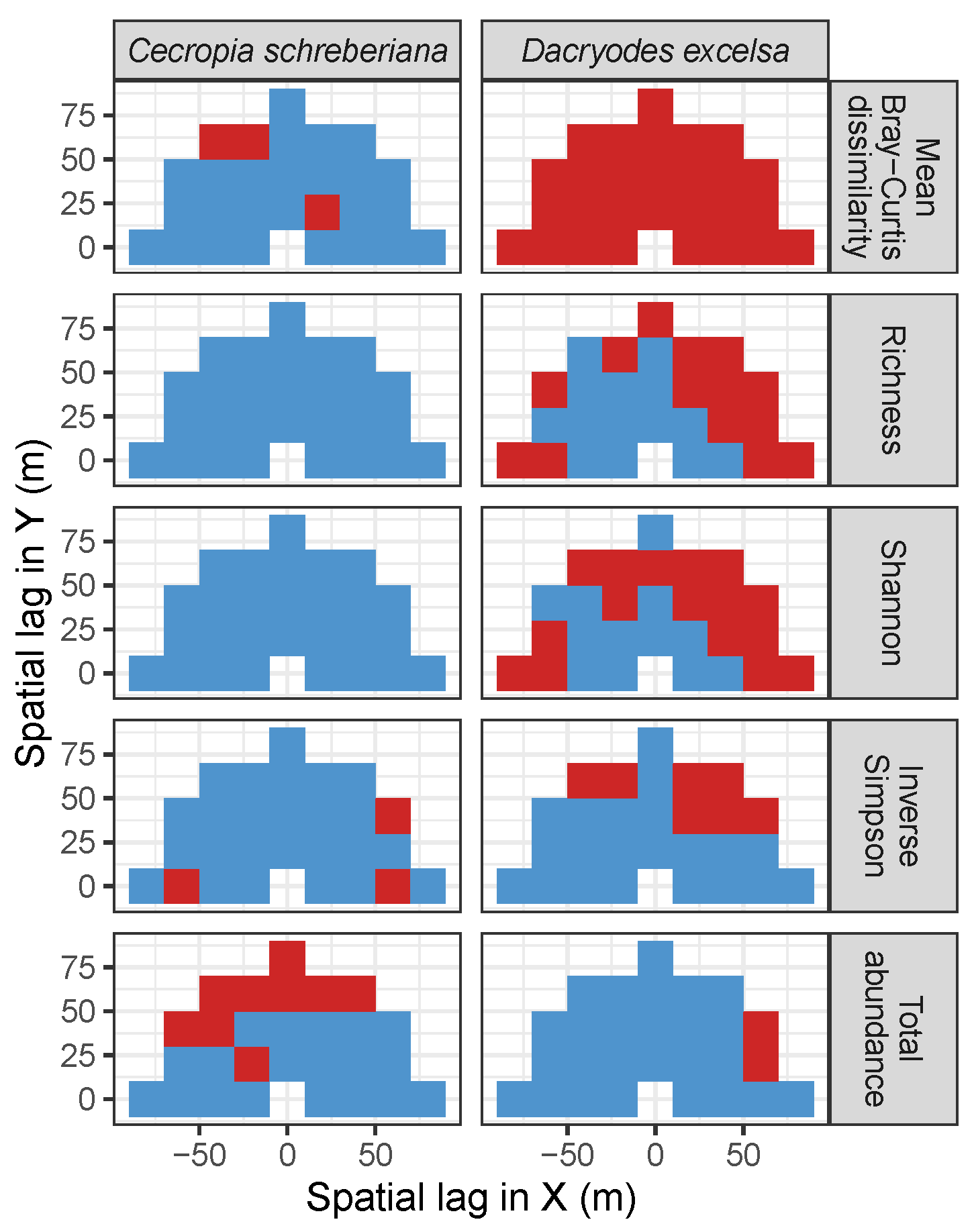
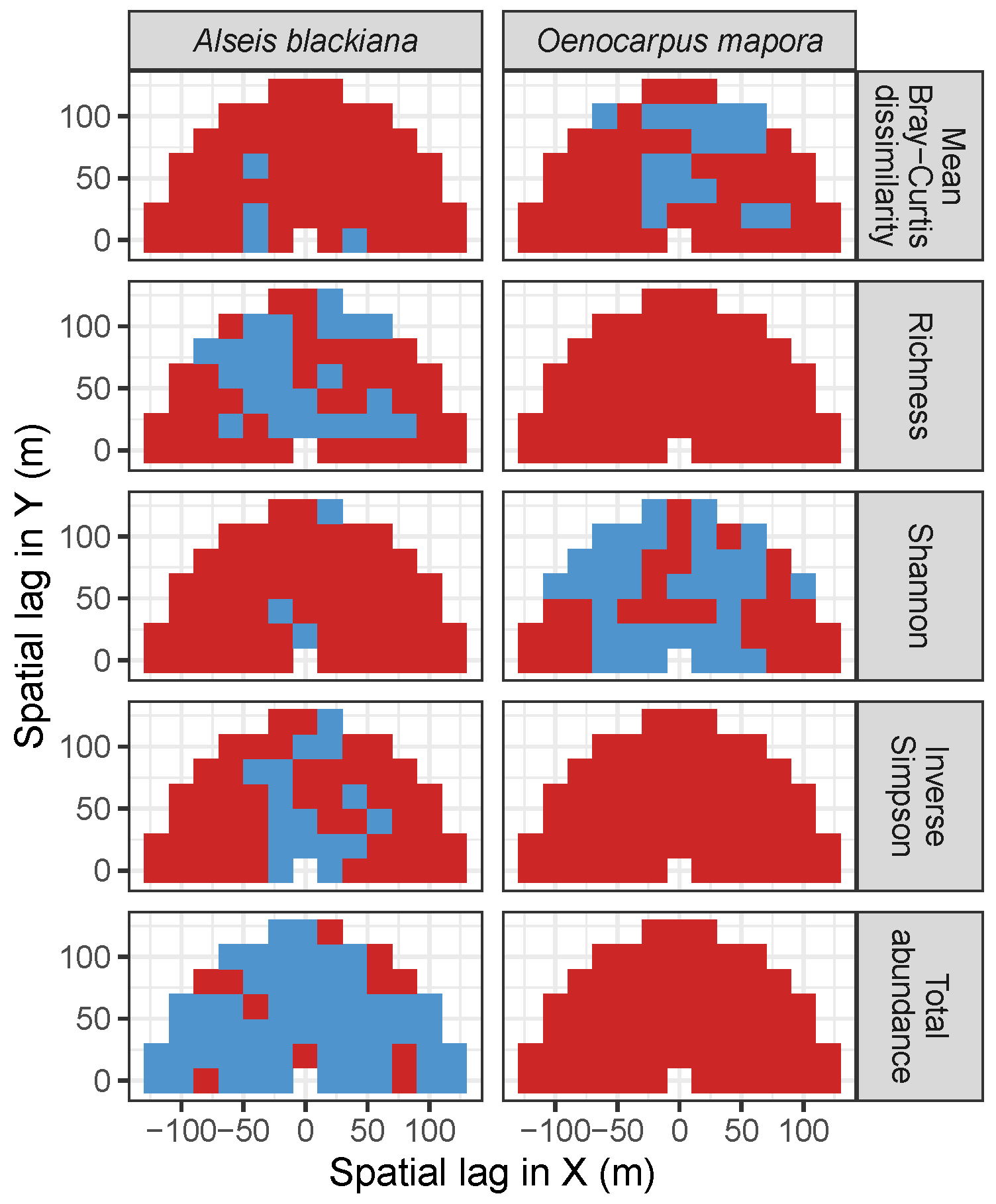
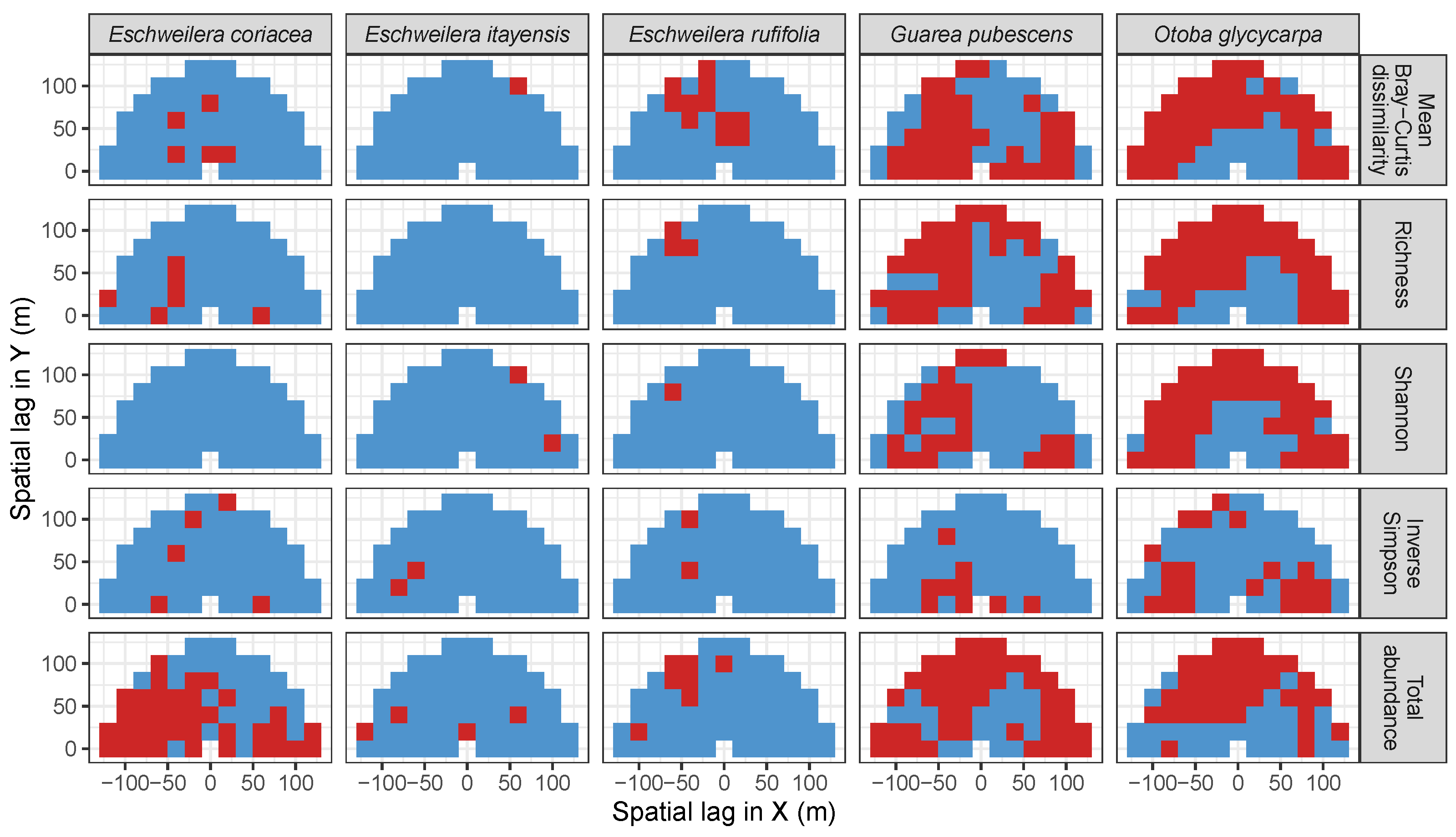
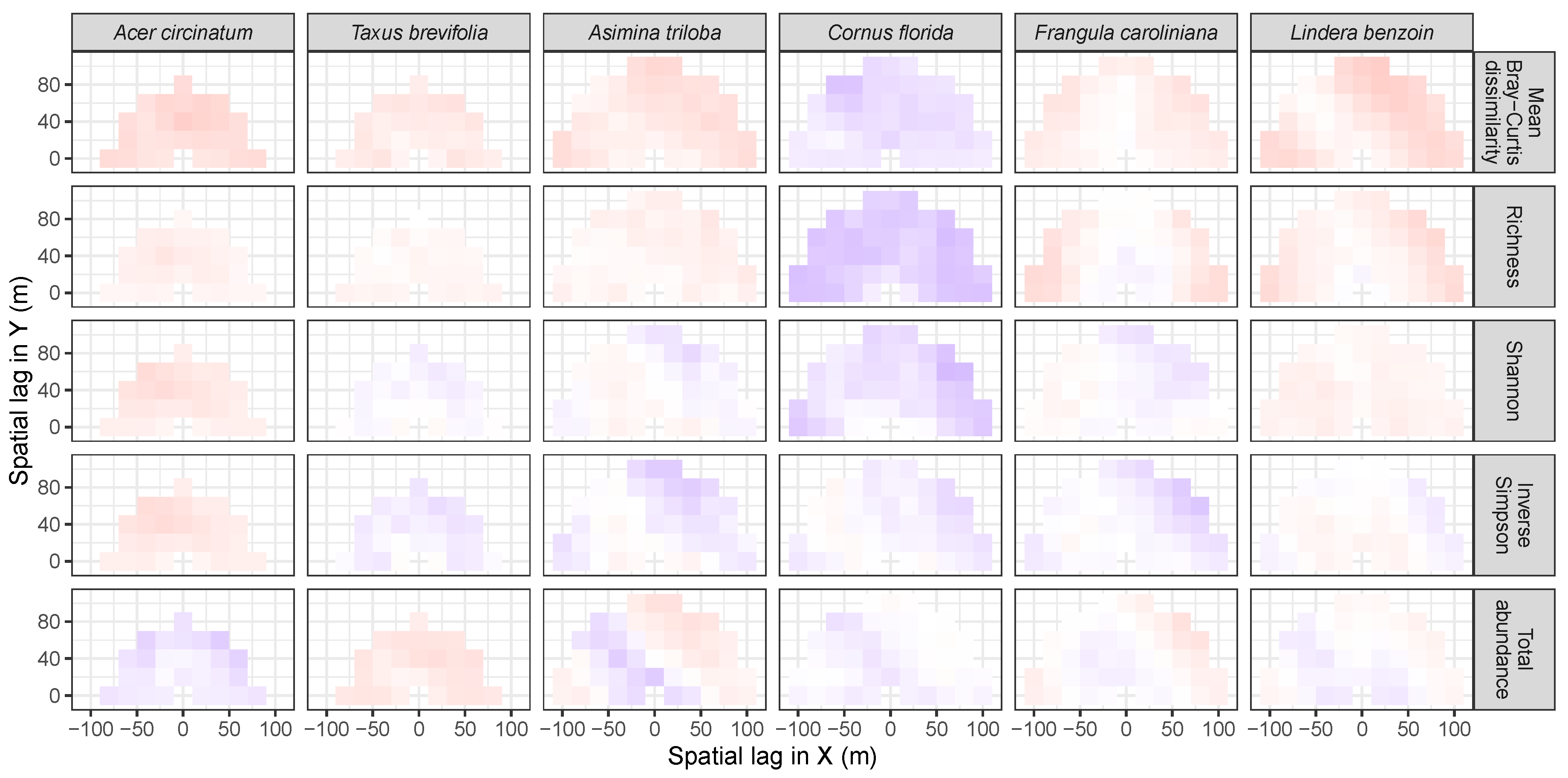
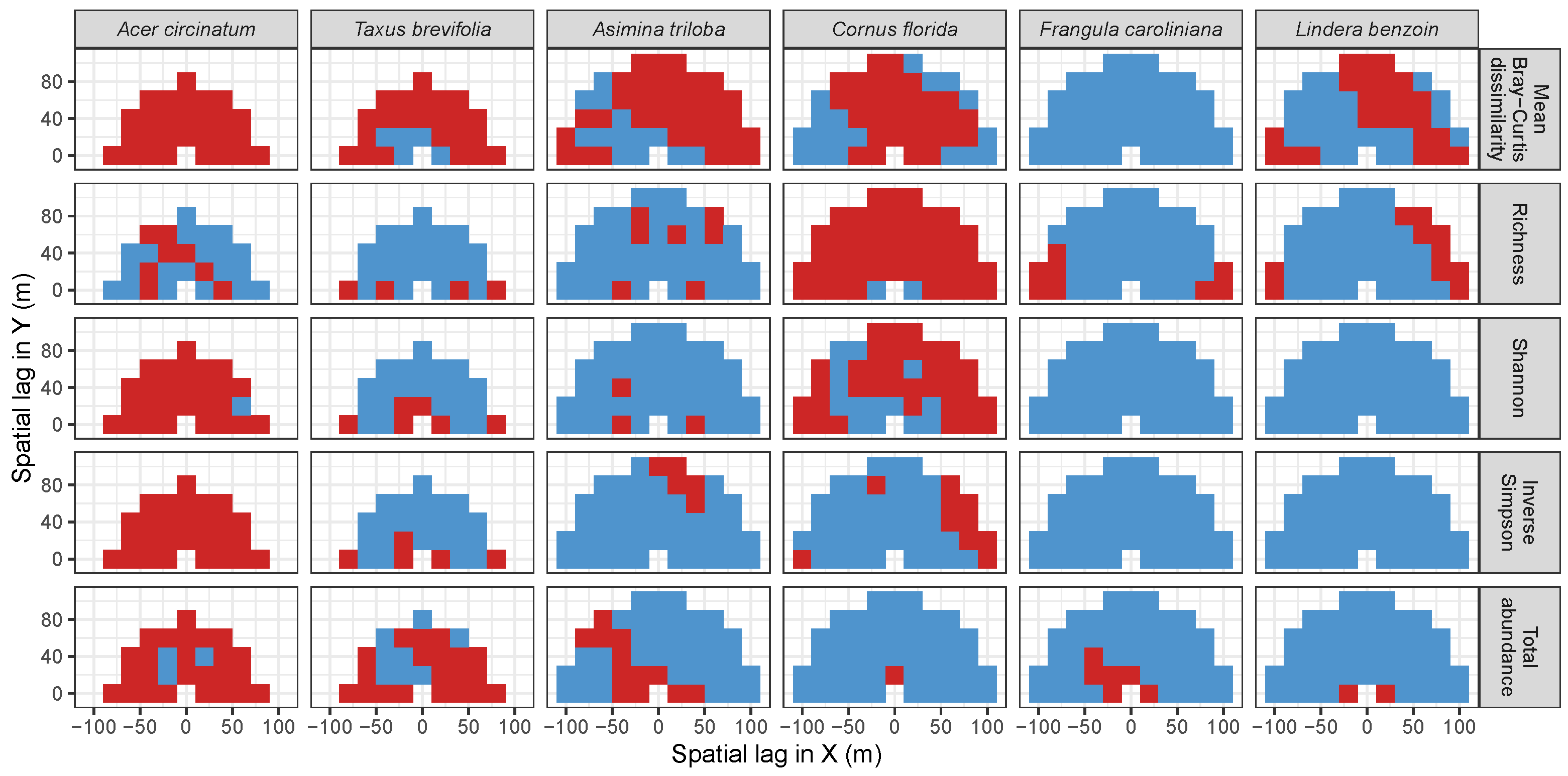
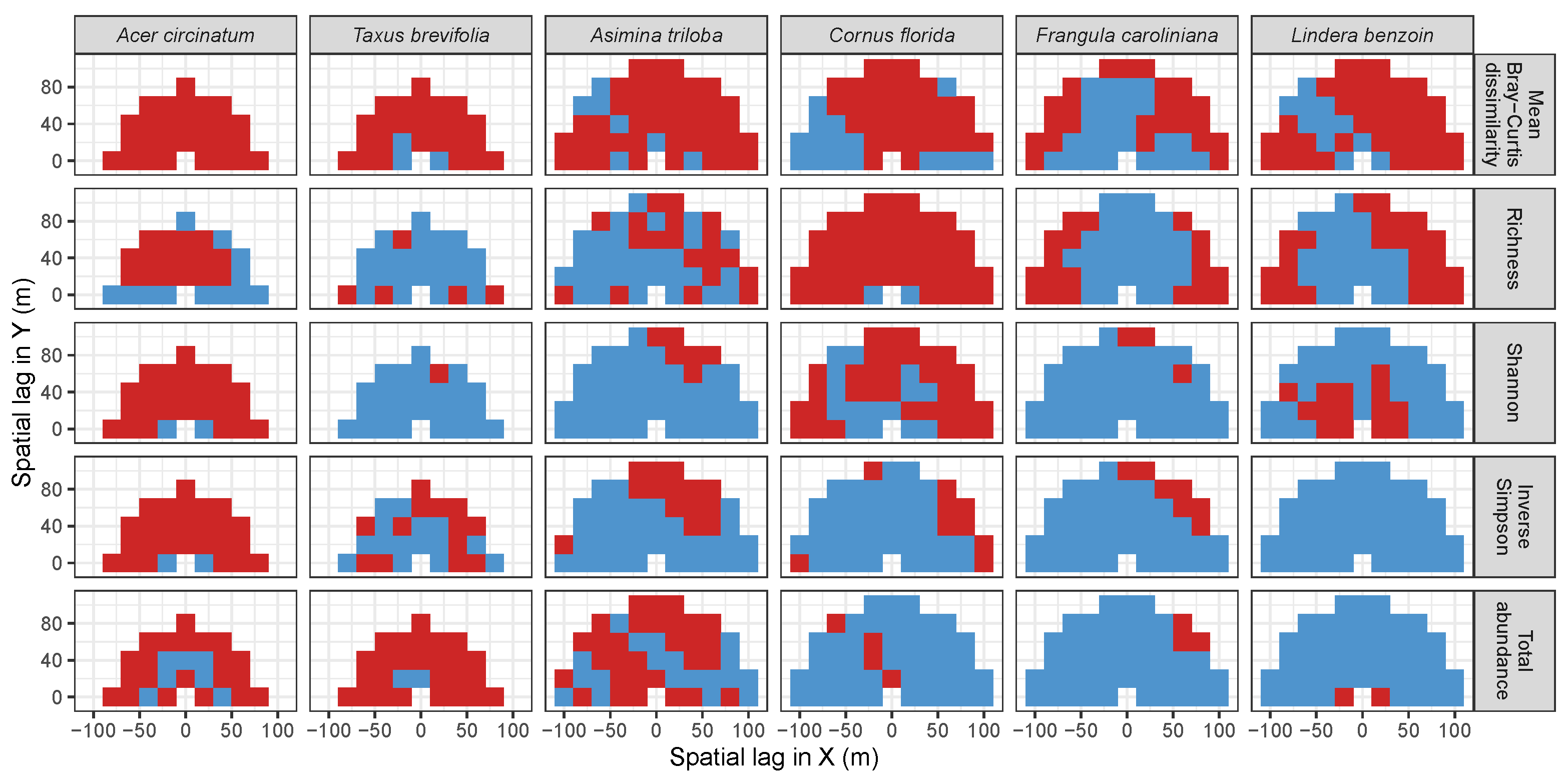
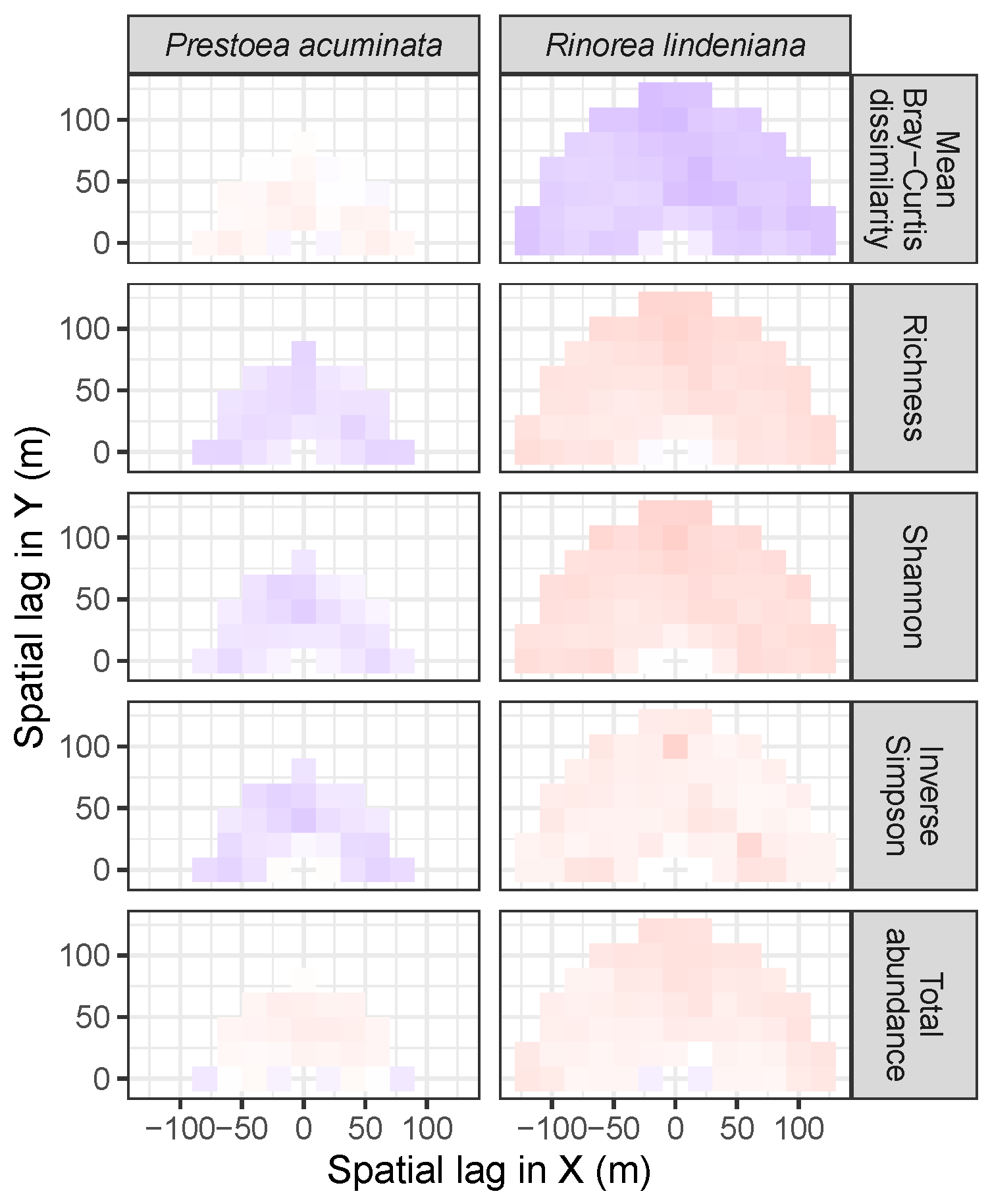
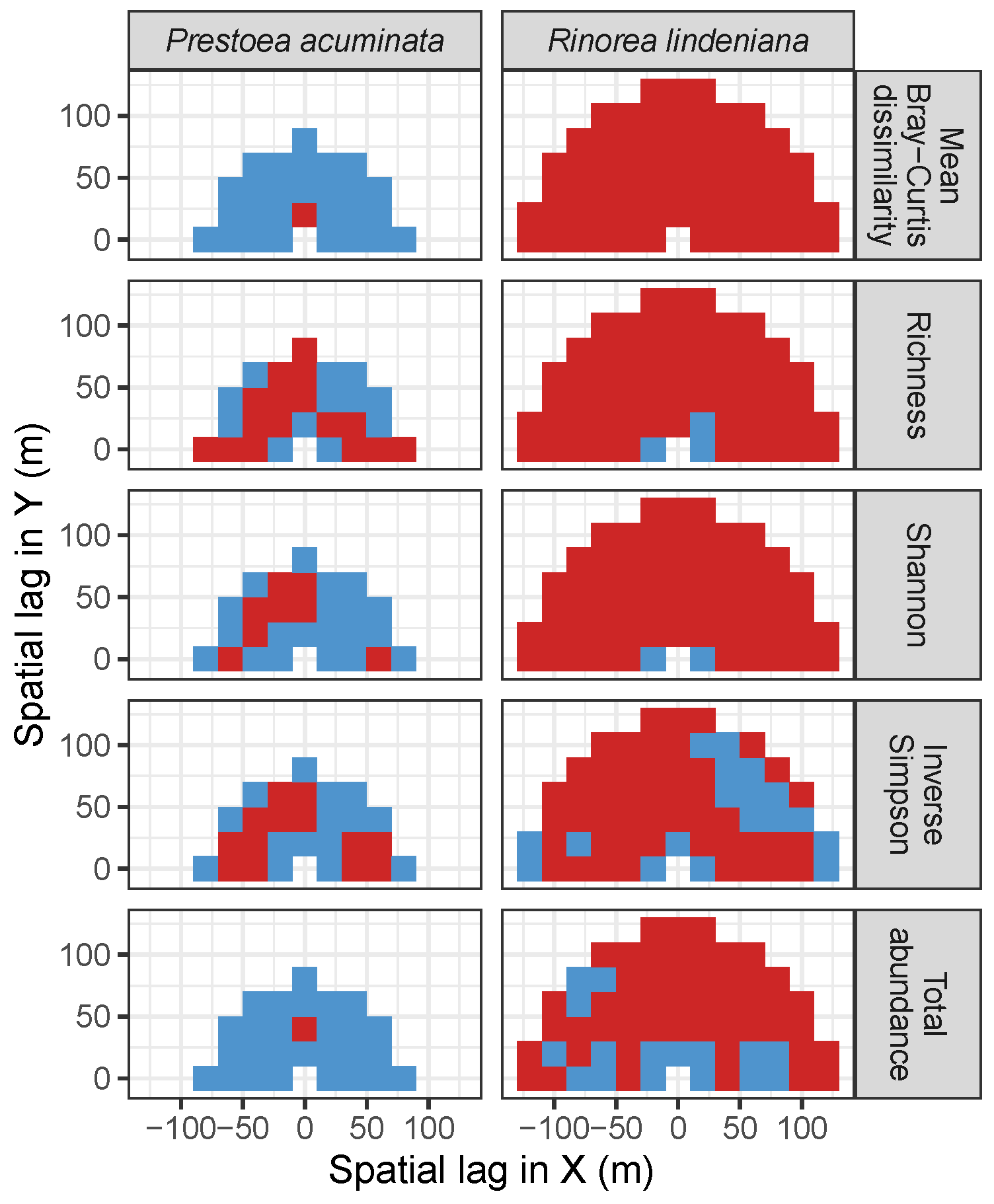
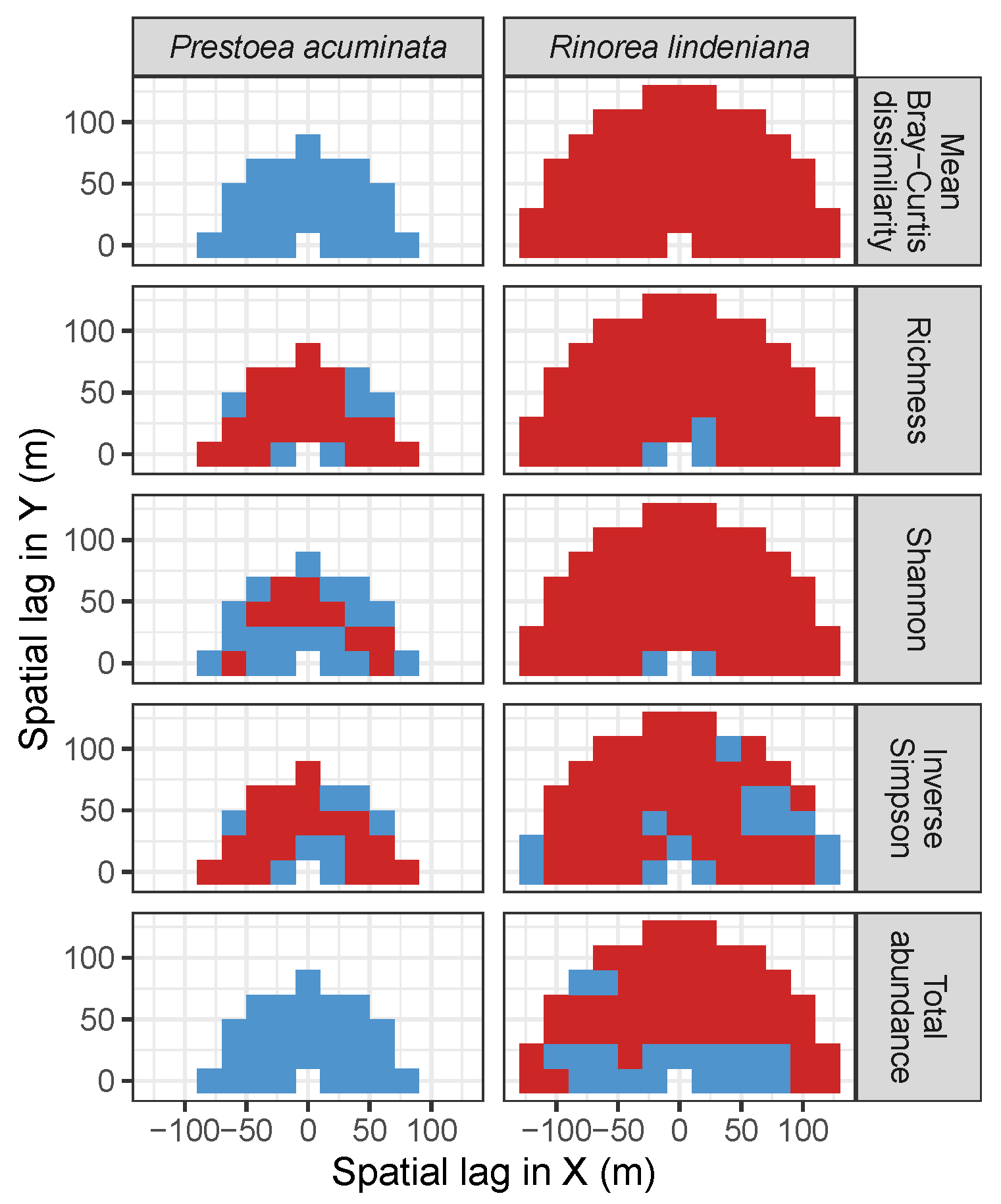
| Site | Species | Diversity Metric | Min | Median | Mean (SD) | Max |
|---|---|---|---|---|---|---|
| WR | Acer circinatum | Bray-Curtis | 0.17 | 0.22 | 0.22 (0.03) | 0.29 |
| Richness | 0.03 | 0.09 | 0.08 (0.03) | 0.14 | ||
| Shannon | 0.1 | 0.14 | 0.15 (0.04) | 0.25 | ||
| Simpson | 0.1 | 0.14 | 0.16 (0.05) | 0.26 | ||
| Abundance | −0.27 | −0.12 | −0.14 (0.05) | −0.06 | ||
| Taxus brevifolia | Bray−Curtis | 0.04 | 0.14 | 0.13 (0.05) | 0.22 | |
| Richness | 0.01 | 0.07 | 0.07 (0.03) | 0.11 | ||
| Shannon | 0.02 | 0.05 | 0.06 (0.03) | 0.1 | ||
| Simpson | 0 | 0.05 | 0.05 (0.03) | 0.12 | ||
| Abundance | 0.03 | 0.12 | 0.11 (0.04) | 0.17 | ||
| TY | Asimina triloba | Bray−Curtis | 0.05 | 0.16 | 0.15 (0.05) | 0.25 |
| Richness | 0.02 | 0.08 | 0.08 (0.04) | 0.16 | ||
| Shannon | −0.12 | 0.01 | 0.02 (0.06) | 0.11 | ||
| Simpson | −0.18 | −0.05 | −0.04 (0.07) | 0.09 | ||
| Abundance | −0.17 | 0.03 | 0.01 (0.11) | 0.18 | ||
| Cornus florida | Bray−Curtis | −0.27 | −0.13 | −0.14 (0.04) | −0.05 | |
| Richness | −0.26 | −0.22 | −0.21 (0.04) | −0.07 | ||
| Shannon | −0.31 | −0.15 | −0.16 (0.07) | −0.02 | ||
| Simpson | −0.2 | −0.07 | −0.08 (0.06) | 0.03 | ||
| Abundance | −0.11 | −0.03 | −0.03 (0.04) | 0.04 | ||
| Frangula caroliniana | Bray−Curtis | −0.04 | 0.08 | 0.07 (0.05) | 0.16 | |
| Richness | −0.07 | 0.05 | 0.07 (0.1) | 0.24 | ||
| Shannon | −0.1 | −0.01 | 0 (0.06) | 0.11 | ||
| Simpson | −0.2 | −0.01 | −0.04 (0.06) | 0.07 | ||
| Abundance | −0.13 | −0.02 | −0.02 (0.07) | 0.16 | ||
| Lindera benzoin | Bray−Curtis | 0.02 | 0.21 | 0.18 (0.09) | 0.33 | |
| Richness | −0.04 | 0.1 | 0.11 (0.08) | 0.25 | ||
| Shannon | 0.02 | 0.08 | 0.08 (0.03) | 0.13 | ||
| Simpson | −0.1 | 0 | 0 (0.05) | 0.07 | ||
| Abundance | −0.1 | 0 | −0.01 (0.05) | 0.08 |
| Site | Species | Diversity Metric | Min | Median | Mean (SD) | Max |
|---|---|---|---|---|---|---|
| LFDP | Prestoea acuminata | Bray-Curtis | −0.04 | 0.05 | 0.04 (0.05) | 0.11 |
| Richness | −0.18 | −0.13 | −0.14 (0.03) | −0.08 | ||
| Shannon | −0.2 | −0.1 | −0.11 (0.04) | −0.05 | ||
| Simpson | −0.22 | −0.13 | −0.12 (0.06) | 0.02 | ||
| Abundance | −0.1 | 0.07 | 0.05 (0.06) | 0.13 | ||
| AM | Rinorea lindeniana | Bray−Curtis | −0.29 | −0.22 | −0.22 (0.04) | −0.08 |
| Richness | −0.02 | 0.19 | 0.19 (0.06) | 0.3 | ||
| Shannon | 0.01 | 0.21 | 0.21 (0.05) | 0.33 | ||
| Simpson | 0 | 0.1 | 0.11 (0.05) | 0.3 | ||
| Abundance | −0.07 | 0.12 | 0.12 (0.06) | 0.21 |
References
- Dayton, P.K. Toward an understanding of community resilience and the potential effects of enrichments to the benthos at McMurdo Sound, Antarctica. In Proceedings of the Colloquium on Conservation Problems in Antarctica, Blacksburg, VA, USA, 10–12 September 1971; Parker, B.C., Ed.; Allen Press: Lawrence, KS, USA, 1972; pp. 81–95. [Google Scholar]
- Ellison, A.M.; Bank, M.S.; Clinton, B.D.; Colburn, E.A.; Elliott, K.; Ford, C.R.; Foster, D.R.; Kloeppel, B.D.; Knoepp, J.D.; Lovett, G.M.; et al. Loss of foundation species: Consequences for the structure and dynamics of forested ecosystems. Front. Ecol. Environ. 2005, 9, 479–486. [Google Scholar] [CrossRef]
- Baiser, B.; Whitaker, N.; Ellison, A.M. Modeling foundation species in food webs. Ecosphere 2013, 4, 146. [Google Scholar] [CrossRef]
- Gaston, K.J.; Fuller, R.A. Biodiversity and extinction: losing the common and the widespread. Prog. Phys. Geog. 2007, 31, 213–225. [Google Scholar] [CrossRef]
- Gaston, K.J.; Fuller, R.A. Commonness, population depletion and conservation biology. Trends Ecol. Evol. 2008, 23, 14–19. [Google Scholar] [CrossRef]
- Ellison, A.M.; Degrassi, A.L. All species are important, but some species are more important than others (Commentary). J. Veg. Sci. 2017, 28, 669–671. [Google Scholar] [CrossRef]
- Paine, R.T. Food web complexity and species diversity. Am. Nat. 1966, 100, 65–75. [Google Scholar] [CrossRef]
- Valls, A.; Coll, M.; Christensen, V. Keystone species: Toward an operational concept for marine biodiversity conservation. Ecol. Monogr. 2015, 85, 29–47. [Google Scholar] [CrossRef]
- Grime, J.P. Dominant and subordinate components of plant communities: implications for succession, stability and diversity. In Colonization, Succession and Stability; Gray, A.J., Crawley, M.J., Edwards, P.J., Eds.; Blackwell Scientific Publications: Oxford, UK, 1987; pp. 413–428. [Google Scholar]
- Leakey, R.; Lewin, R. The Sixth Extinction: Patterns of Life and the Future of Humankind; Anchor Books: New York, NY, USA, 1996. [Google Scholar]
- Short, F.; Carruthers, T.; Dennison, W.; Waycott, M. Global seagrass distribution and diversity: A bioregional model. J. Exp. Mar. Biol. Ecol. 2007, 350, 3–20. [Google Scholar] [CrossRef]
- Fischer, M.; Bossdorf, O.; Gockel, S.; Hänsel, F.; Hemp, A.; Hessenmöller, D.; Korte, G.; Nieschulze, J.; Pfeiffer, S.; Prati, D.; et al. Implementing large-scale and long-term functional biodiversity research: The Biodiversity Exploratories. Basic Appl. Ecol. 2010, 11, 473–485. [Google Scholar] [CrossRef]
- LaManna, J.A.; Mangan, S.A.; Alonso, A.; Bourg, N.A.; Brockelman, W.Y.; Bunyavejchewin, S.; Chang, L.-W.; Chiang, J.-M.; Chuyong, G.B.; Clay, K.; et al. Plant diversity increases with the strength of negative density dependence at the global scale. Science 2017, 356, 1389–1392. [Google Scholar] [CrossRef]
- ReefBase. ReefBase—A Global Information System on Coral Reefs. 2018. Available online: http://www.reefabase.org (accessed on 5 October 2018).
- Ellison, A.M.; Barker Plotkin, A.A.; Khalid, S. Foundation species loss and biodiversity of the herbaceous layer in New England forests. Forests 2016, 7, 9. [Google Scholar] [CrossRef]
- Ellison, A.M. Experiments are revealing a foundation species: A case study of eastern hemlock (Tsuga canadensis). Adv. Ecol. 2014, 2014, 456904. [Google Scholar] [CrossRef]
- Foster, D.R. (Ed.) Hemlock: A Forest Giant on the Edge; Yale University Press: New Haven, CT, USA, 2014. [Google Scholar]
- Orwig, D.A.; Barker Plotkin, A.A.; Davidson, E.A.; Lux, H.; Savage, K.E.; Ellison, A.M. Foundation species loss affects vegetation structure more than ecosystem function in a northeastern USA forest. PeerJ 2013, 1, e41. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, F.; Porcu, E.; Vallejos, R. Study of spatial relationships between two sets off variables: A nonparametric approach. J. Nonparametr. Statist. 2013, 25, 695–714. [Google Scholar] [CrossRef]
- Buckley, H.L.; Case, B.S.; Ellison, A.M. Using codispersion analysis to characterize spatial patterns in species co-occurrences. Ecology 2016, 97, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Buckley, H.L.; Case, B.S.; Zimmermann, J.; Thompson, J.; Myers, J.A.; Ellison, A.M. Using codispersion analysis to quantify and understand spatial patterns in species-environment relationships. New Phytol. 2016, 211, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Case, B.S.; Buckley, H.L.; Barker Plotkin, A.; Ellison, A.M. Using codispersion analysis to quantify temporal changes in the spatial pattern of forest stand structure. Chil. J. Stat. 2016, 7, 3–15. [Google Scholar]
- Case, B.S.; Buckley, H.L.; Barker Plotkin, A.A.; Orwig, D.A.; Ellison, A.M. When a foundation crumbles: Forecasting forest community dynamics associated with the decline of the foundation species. Ecosphere 2017, 8, e01893. [Google Scholar] [CrossRef]
- Anderson-Teixeira, K.J.; Davies, S.J.; Bennett, A.C.; Gonzalez-Akre, E.B.; Muller-Landau, H.C.; Wright, S.J.; Abu Salim, K.; Almeyda Zambrano, A.M.; Alonso, A.; Baltzer, J.L.; et al. CTFS—ForestGEO: A worldwide network monitoring forests in an era of global change. Glob. Change Biol. 2015, 21, 528–549. [Google Scholar] [CrossRef]
- Condit, R. Tropical Forest Census Plots: Methods and Results from Barro Colorado Island, Panama and a Comparison with Other Plots; Springer: Berlin, Germany, 1998. [Google Scholar]
- Lutz, J.A.; Larson, A.J.; Freund, J.A.; Swanson, M.E.; Bible, K.J. The importance of large-diameter trees to forest structural heterogeneity. PLoS ONE 2013, 8, e82784. [Google Scholar] [CrossRef]
- Lutz, J.A.; Furniss, T.J.; Johnson, D.J.; Davies, S.J.; Allen, D.; Alonso, A.; Anderson-Teixeira, K.; Andrade, A.; Baltzer, J.; Becker, K.M.L.; et al. Global importance of large-diameter trees. Global Ecol. Biogeogr. 2018, 27, 849–864. [Google Scholar] [CrossRef]
- Das, A.J.; Larson, A.J.; Lutz, J.A. Individual species-area relationships in temperate coniferous forests. J. Veg. Sci. 2018, 29, 317–324. [Google Scholar] [CrossRef]
- Lutz, J.A.; Larson, A.J.; Furniss, T.J.; Freund, J.A.; Swanson, M.E.; Donato, D.C.; Bible, K.J.; Chen, J.; Franklin, J.F. Spatially non-random tree mortality and ingrowth maintain equilibrium pattern in an old-growth Pseudotsuga-Tsuga forest. Ecology 2014, 95, 2047–2054. [Google Scholar] [CrossRef] [PubMed]
- Griffith, G.E.; Omernik, J.M.; Pierson, S.M.; Kiilsgaard, C.W. The Massachusetts Ecological Regions Project; U.S. Environmental Protection Agency: Corvallis, OR, USA, 1994.
- Westveld, M.; Ashman, R.I.; Baldwin, H.I.; Holdsworth, R.P.; Johnson, R.S.; Lambert, J.H.; Lutz, H.J.; Swain, L.; Standish, M. Natural forest vegetation zones of New England. J. For. 1956, 54, 332–338. [Google Scholar]
- Wang, X.; Wiegand, T.; Anderson-Teixeira, K.J.; Bourg, N.A.; Hao, Z.; Howe, R.; Jin, G.; Orwig, D.A.; Spasojevic, M.J.; Wang, S.; et al. Ecological drivers of spatial community dissimilarity, species replacement and species nestedness across temperate forests. Global Ecol. Biogeogr. 2018, 27, 581–592. [Google Scholar] [CrossRef]
- Raup, H.M.; Carlson, R.E. The history of land use in the Harvard Forest. Harv. For. Bull. 1941, 20, 4–62. [Google Scholar]
- Foster, D.R. Land-use history (1730–1990) and vegetation dynamics in central New England, USA. J. Ecol. 1992, 80, 753–772. [Google Scholar] [CrossRef]
- Foster, D.R.; Zebryk, T.M.; Schoonmaker, P.K.; Lezberg, A.L. Post-settlement history of human land-use and vegetation dynamics of a hemlock woodlot in central New England. J. Ecol. 1992, 80, 773–786. [Google Scholar] [CrossRef]
- Spasojevic, M.J.; Yablon, E.A.; Oberle, B.; Myers, J.A. Ontogenetic trait variation influences tree community assembly across environmental gradients. Ecosphere 2014, 5, 129. [Google Scholar] [CrossRef]
- LaManna, J.A.; Walton, M.L.; Turner, B.L.; Myers, J.A. Negative density dependence is stronger in resource-rich environments and diversifies communities when stronger for common but not rare species. Ecol. Lett. 2016, 19, 657–667. [Google Scholar] [CrossRef]
- Zimmerman, M.; Wagner, W.L. A description of the woody vegetation of oak-hickory forest in the Northern Ozark Highlands. Bull. Torrey Bot. Club 1979, 106, 117–122. [Google Scholar] [CrossRef]
- Thompson, J.; Brokaw, N.; Zimmerman, J.K.; Waide, R.B.; Everham, E.M., III; Lodge, J.; Taylor, C.M.; Garca-Montiel, D.; Fluet, M. Land use history, environment, and tree composition in a tropical forest. Ecol. Appl. 2002, 12, 1344–1363. [Google Scholar] [CrossRef]
- Zimmerman, J.K.; Comita, L.S.; Thompson, J.; Uriarte, M.; Brokaw, N. Patch dynamics and community metastability of a tropical forest: Compound effects of natural disturbance and human land use. Landsc. Ecol. 2010, 25, 1099–1111. [Google Scholar] [CrossRef]
- Hogan, J.A.; Zimmerman, J.K.; Thompson, J.; Nytch, C.J.; Uriarte, M. The interaction of land-use legacies and hurricane disturbance in subtropical wet forest: Twenty-one years of change. Ecosphere 2016, 7, e01405. [Google Scholar] [CrossRef]
- Leigh, E.G., Jr.; Loo de Lao, S.; Condit, R.; Hubbell, S.P.; Foster, R.B.; Pérez, R. Barro Colorado Island Forest Dynamics Plot, Panama. In Tropical Forest Diversity and Dynamism: Findings from a Large-Scale Plot Network; Losos, E.C., Leigh, E.G., Jr., Eds.; University of Chicago Press: Chicago, IL, USA, 2004; pp. 451–463. [Google Scholar]
- Condit, R.; Pérez, R.; Lao, S.; Aguilar, S.; Hubbell, S.P. Demographic trends and climate over 35 years in the Barro Colorado 50 ha plot. For. Ecosyst. 2017, 4, 17. [Google Scholar] [CrossRef]
- Hubbell, S.P.; Foster, R.B.; O’Brien, S.T.; Harms, K.E.; Condit, R.; Wechsler, B.; Wright, S.J.; Loo de Lao, S. Light-gap disturbances, recruitment limitation, and tree diversity in a Neotropical forest. Science 1999, 283, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Zuleta, D.; Russo, S.E.; Barona, A.; Barreto-Silva, J.S.; Carenas, D.; Castaño, N.; Davies, S.J.; Detto, M.; Sua, S.; Turner, B.L.; Duque, A. Importance of topography for tree species habitat distributions in a terra firme forest in the Colombian Amazon. Plant Soil 2018. [Google Scholar] [CrossRef]
- Loehle, C. Species abundance distributions result from body size-energetics relationships. Ecology 2006, 87, 2221–2226. [Google Scholar] [CrossRef]
- Chao, A.; Gotelli, N.J.; Hsieh, T.C.; Snader, E.L.; Ma, K.H.; Colwell, R.K.; Ellison, A.M. Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol. Monogr. 2014, 84, 45–67. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An ordination of upland forest communities of southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R package version 2.5-3. Available online: https://CRAN.R-project.org/package=vegan (accessed on 3 February 2018).
- Vallejos, R.; Buckley, H.; Case, B.; Acosta, J.; Ellison, A.M. Sensitivity of codispersion to noise and error in ecological and environmental data. Forests 2018, 9, 679. [Google Scholar] [CrossRef]
- Osorio, F.; Vallejos, R.; Cuevas, F.; Mancilla, D. SpatialPack Package. R package version 0.3. Available online: https://cran.r-project.org/package=SpatialPack (accessed on 3 February 2018).
- Brantley, S.T.; Ford, C.R.; Vose, J.M. Future species composition will affect forest water use after loss of eastern hemlock from southern Appalachian forests. Ecol. Appl. 2013, 23, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Tomback, D.F.; Resler, L.M.; Keane, R.E.; Pansing, E.R.; Andrade, A.J.; Wagner, A.C. Community structure, biodiversity, and ecosystem services in treeline whitebark pine communities: potential impacts from a non-native pathogen. Forests 2016, 7, 21. [Google Scholar] [CrossRef]
- Whitmore, T.C. Tropical Rain Forests of the Far East; Clarendon: Oxford, UK, 1984. [Google Scholar]
- Iwasa, Y.; Kubo, T.; Sato, K. Maintenance of forest species diversity and latitudinal gradient. Vegetatio 1995, 121, 127–134. [Google Scholar] [CrossRef]
- Lamanna, C.; Blonder, B.; Violle, C.; Kraft, N.J.B.; Sandel, B.; Šímová, I.; Donoghue, J.C., II; Svenning, J.-C.; McGill, B.J.; Boyle, B.; et al. Functional trait space and the latitudinal diversity gradient. Proc. Nat. Acad. Sci. USA 2014, 111, 13745–13750. [Google Scholar] [CrossRef] [PubMed]
- Wieczynski, D.J.; Boyle, B.; Buzzard, V.; Duran, S.M.; Henderson, A.N.; Hulshof, D.M.; Kerkhoff, A.J.; McCarthy, M.C.; Michaletz, S.T.; Swenson, N.G.; et al. Climate shapes and shifts functional biodiversity in forests worldwide. Proc. Nat. Acad. Sci. USA 2019, 116, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Elumeeva, T.G.; Onipchenko, V.G.; Weger, M.J.A. No other species can replace them: evidence for the key role of dominants in an alpine Festuca varia grassland. J. Veg. Sci. 2017, 28, 674–683. [Google Scholar] [CrossRef]
- Hubbell, S.P. Tree dispersion, abundance, and diversity in a tropical dry forest. Science 1979, 203, 1299–1309. [Google Scholar] [CrossRef]
- Hubbell, S.P. The Unified Neutral Theory of Biodiversity and Biogeography; Princeton University Press: Princeton, NJ, USA, 2001. [Google Scholar]
- Leigh, E.G., Jr. Neutral theory: A historical perspective. J. Evol. Biol. 2007, 20, 2075–2091. [Google Scholar] [CrossRef]
- Furniss, T.J.; Larson, A.J.; Lutz, J.A. Reconciling niches and neutrality in a subalpine temperate forest. Ecosphere 2017, 8, e01847. [Google Scholar] [CrossRef]
- Ellison, A.M.; Lavine, M.; Kerson, P.B.; Barker Plotkin, A.A.; Orwig, D.A. Building a foundation: land-use history and dendrochronology reveal temporal dynamics of a Tsuga canadensis (Pinaceae) forest. Rhodora 2014, 116, 377–427. [Google Scholar] [CrossRef]
- Freund, J.A.; Franklin, J.F.; Lutz, J.A. Structure of early old-growth Douglas-fir forests in the Pacific Northwest. Forest Ecol. Manag. 2015, 335, 11–25. [Google Scholar] [CrossRef]
- Lutz, J.A.; Halpern, C.B. Tree mortality during early forest development: A long-term study of rates, causes, and consequences. Ecol. Monogr. 2006, 76, 257–275. [Google Scholar] [CrossRef]
- Halpern, C.B.; Lutz, J.A. Canopy closure exerts weak controls on understory dynamics: A 30-year study of overstory-understory interactions. Ecol. Monogr. 2013, 83, 221–237. [Google Scholar] [CrossRef]
- Basnet, K.; Scatena, F.N.; Likens, G.E.; Lugo, A.E. Ecological consequences of root grafting in tabonuco (Dacryodes excelsa) trees in the Luquillo Experimental Forest, Puerto Rico. Biotropica 1993, 25, 28–35. [Google Scholar] [CrossRef]
- Uriarte, M.; Canham, C.D.; Thompson, J.; Zimmerman, J.K. A neighborhood analysis of tree growth and survival in a hurricane-driven tropical forest. Ecol. Monogr. 2004, 74, 591–614. [Google Scholar] [CrossRef]
- Farris-Lopez, K.; Desnlow, J.S.; Moser, B.; Passmore, H. Influence of a common palm, Oenocarpus mapora on seedling establishment in a tropical moist forest in Panama. J. Trop. Ecol. 2004, 20, 429–438. [Google Scholar] [CrossRef]
- Baker, T.R.; Vela Díaz, D.M.; Chama Moscoso, V.; Navarro, G.; Monteagudo, A.; Pinto, R.; Cangani, K.; Fyllas, N.F.; Lopez Gonzalez, G.; Laurance, W.F.; et al. Consistent, small effects of treefall disturbances on the composition and diversity of four Amazonian forests. J. Ecol. 2016, 104, 497–506. [Google Scholar] [CrossRef]
- Battles, J.J.; Cleavitt, N.L.; Saah, D.S.; Pling, B.T.; Fahey, T.J. Ecological impact of a microburst windstorm in a northern hardwood forest. Can. J. For. Res. 2017, 47, 1695–1701. [Google Scholar] [CrossRef]
- Després, T.; Asselin, H.; Doyon, F.; Drobyshev, I.; Bergeron, Y. Gap dynamics of late successional sugar maple-yellow birch forests at their northern range limit. J. Veg. Sci. 2017, 28, 368–378. [Google Scholar] [CrossRef]
- Dilling, C.; Lambdin, P.; Grant, J.; Buck, L. Insect guild structure associated with eastern hemlock in the southern Appalachians. Env. Entomol. 2007, 36, 1408–1414. [Google Scholar] [CrossRef]
- Rohr, J.; Mahan, C.G.; Kim, K.C. Response of arthropod biodiversity to foundation species declines: the case of the eastern hemlock. Forest Ecol. Manag. 2009, 258, 1503–1510. [Google Scholar] [CrossRef]
- Sackett, T.E.; Record, S.; Bewick, S.; Baiser, B.; Sanders, N.J.; Ellison, A.M. Response of macroarthropod assemblages to the loss of hemlock (Tsuga canadensis), a foundation species. Ecosphere 2011, 2, e74. [Google Scholar] [CrossRef]
- Record, S.; McCabe, T.; Baiser, B.; Ellison, A.M. Identifying foundation species in North American forests using long-term data on ant assemblage structure. Ecosphere 2018, 9, e02139. [Google Scholar] [CrossRef]
- Ruchty, A.; Rosso, A.L.; McCune, B. Changes in epiphyte communities as the shrub, Acer circinatum, develops and ages. The Bryologist 2001, 104, 274–281. [Google Scholar] [CrossRef]
- Schowalter, T.D. Canopy invertebrate community response to disturbance and consequences of herbivory in temperate and tropical forests. Selbyana 1995, 16, 41–48. [Google Scholar]
- Schowalter, T.D. Invertebrate community structure and herbivory in a tropical rain forest canopy in Puerto Rico following Hurricane Hugo. Biotropica 1994, 26, 312–319. [Google Scholar] [CrossRef]
- Weissenhofer, A.; Huber, W.; Wanek, W.; Weber, A. Terrestrial litter trappers in the Golfo Dulce region: Diversity, architecture and ecology of a poorly known group of plant specialists. Stapfia 88, zugleich Kataloge der oberösterreichischen Landesmuseen (Neue Serie) 2008, 80, 143–154. [Google Scholar]
- De Vasconcelos, H.L. Effects of litter collection by understory palms on the associated macroinvertebrate fauna in Central Amazonia. Pedobiologia 1990, 34, 157–160. [Google Scholar]
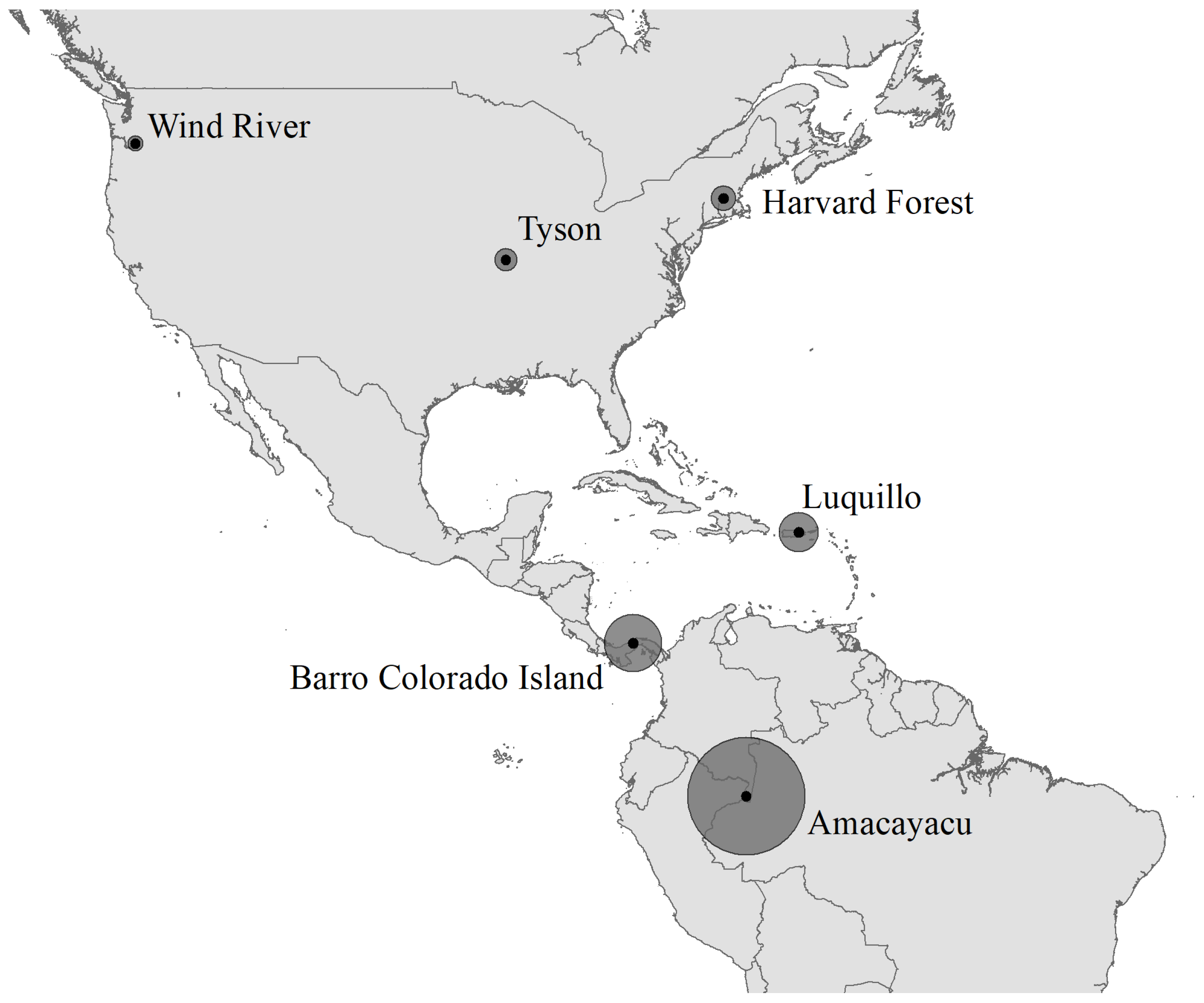


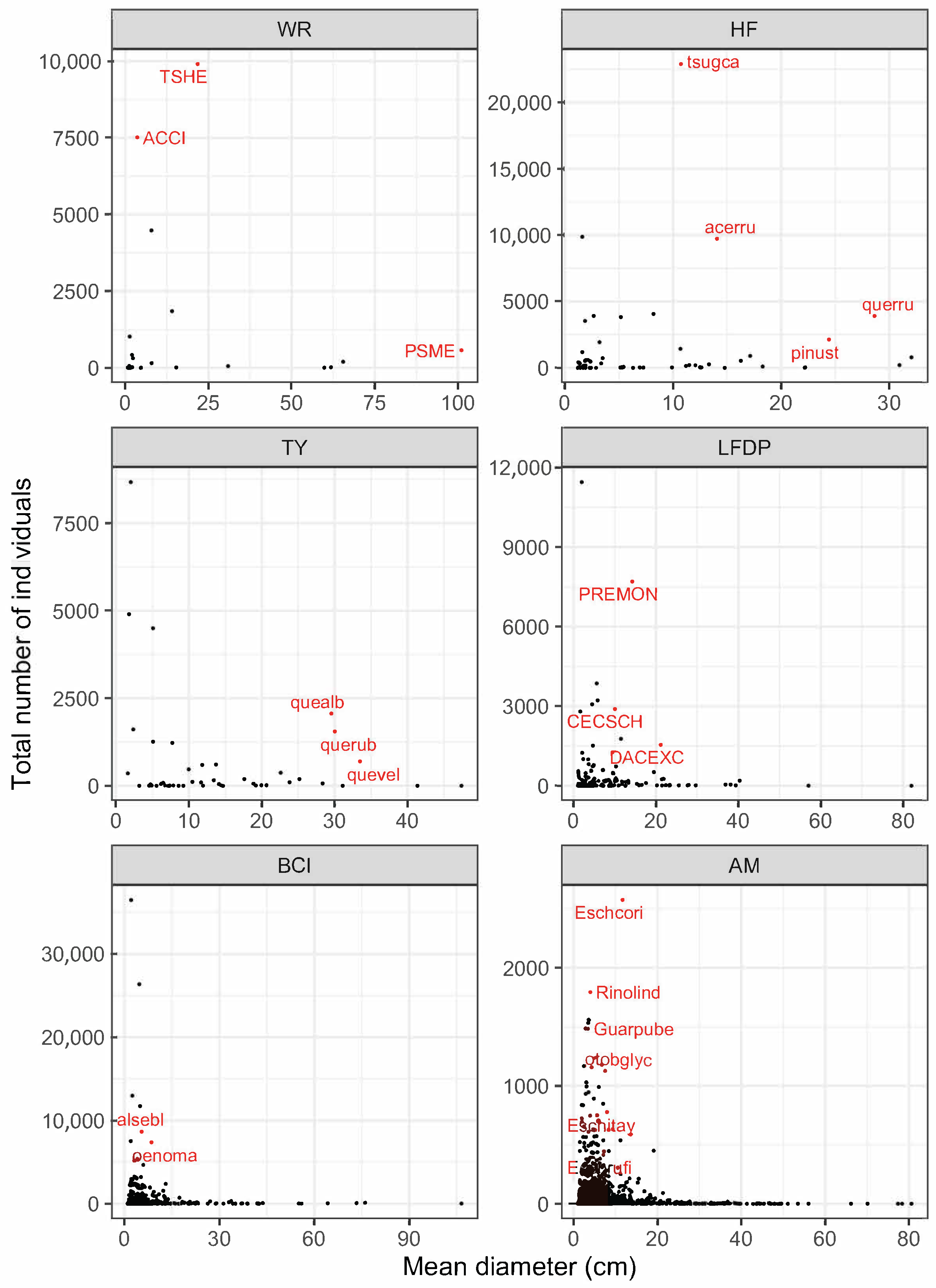
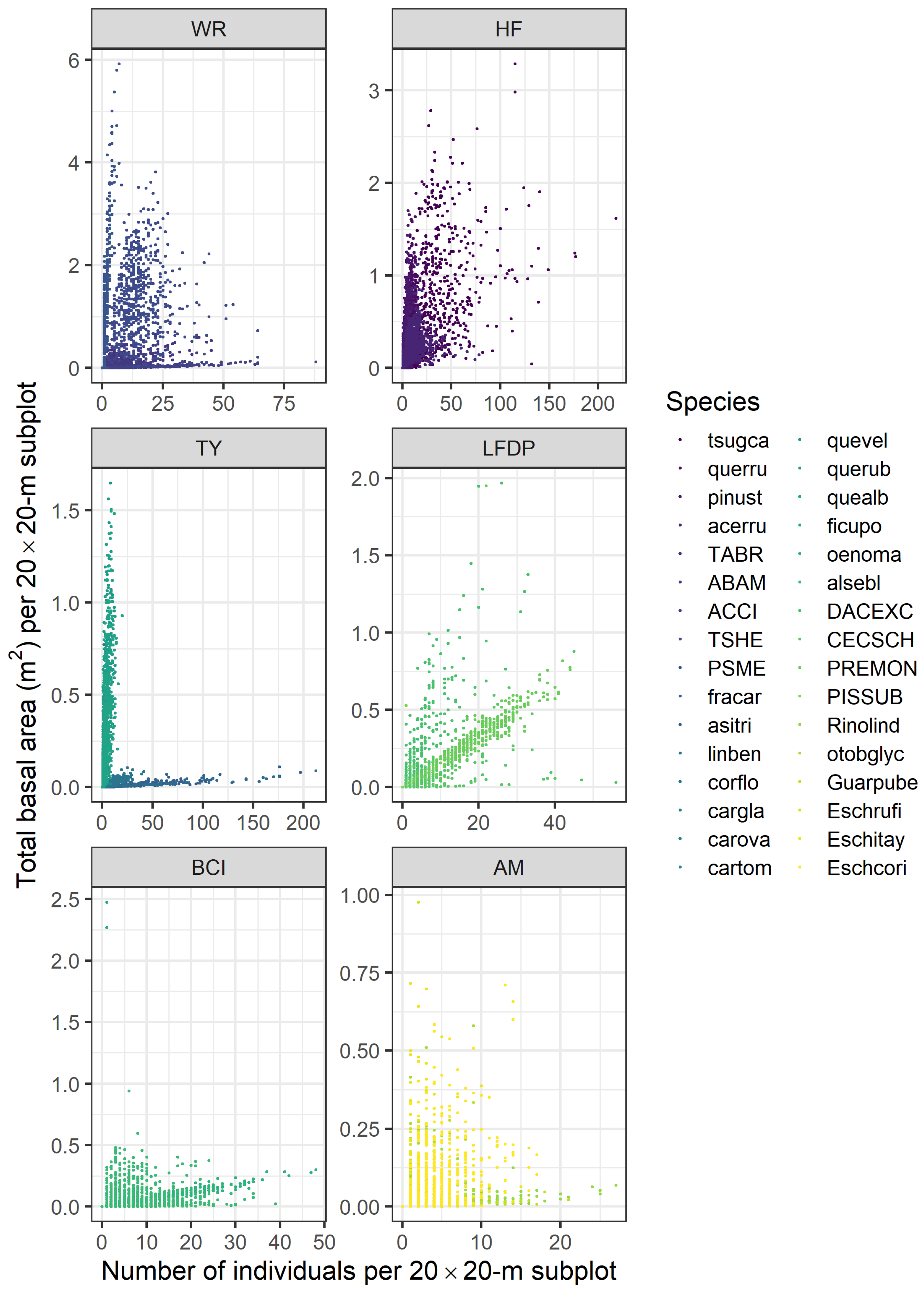
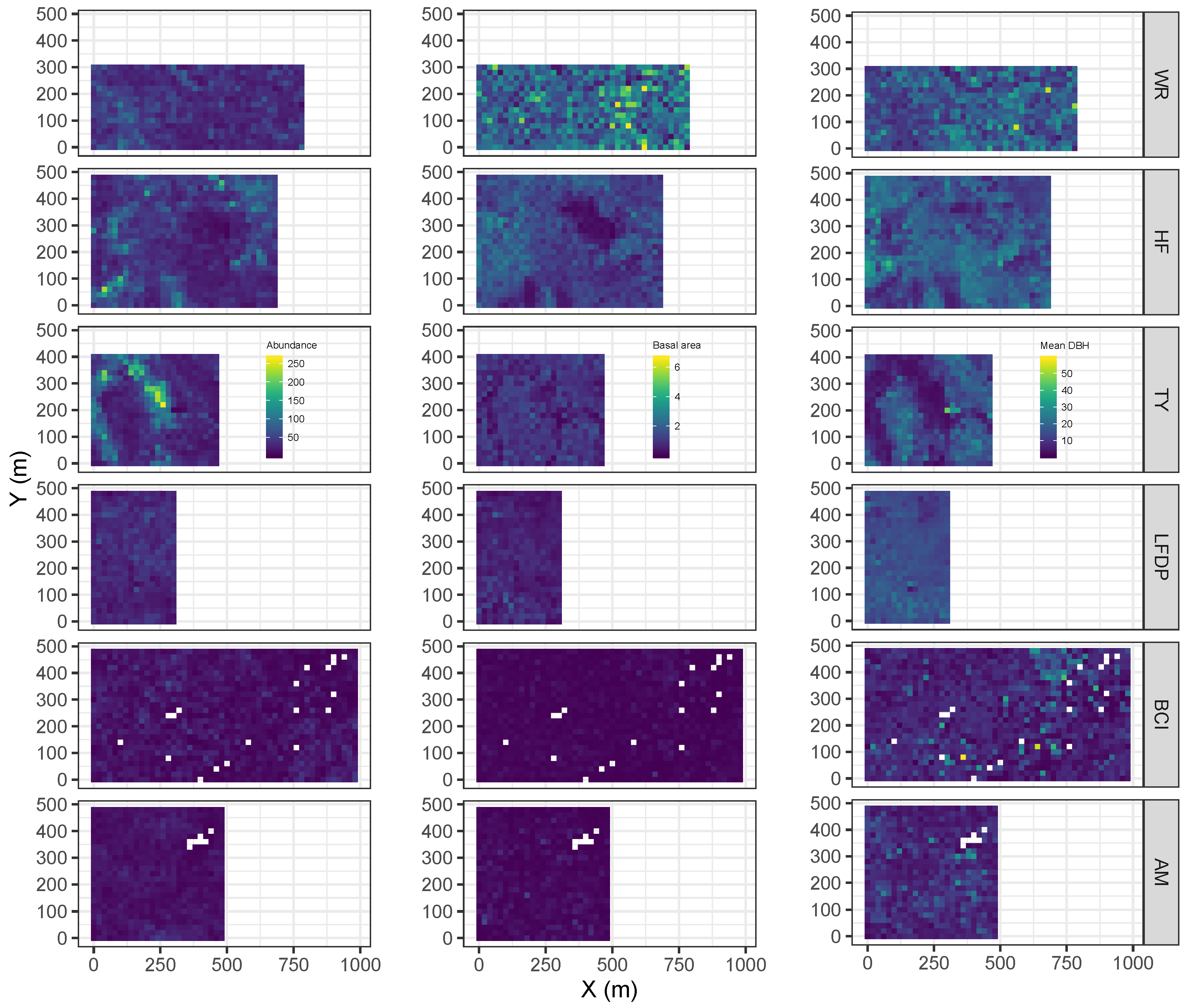
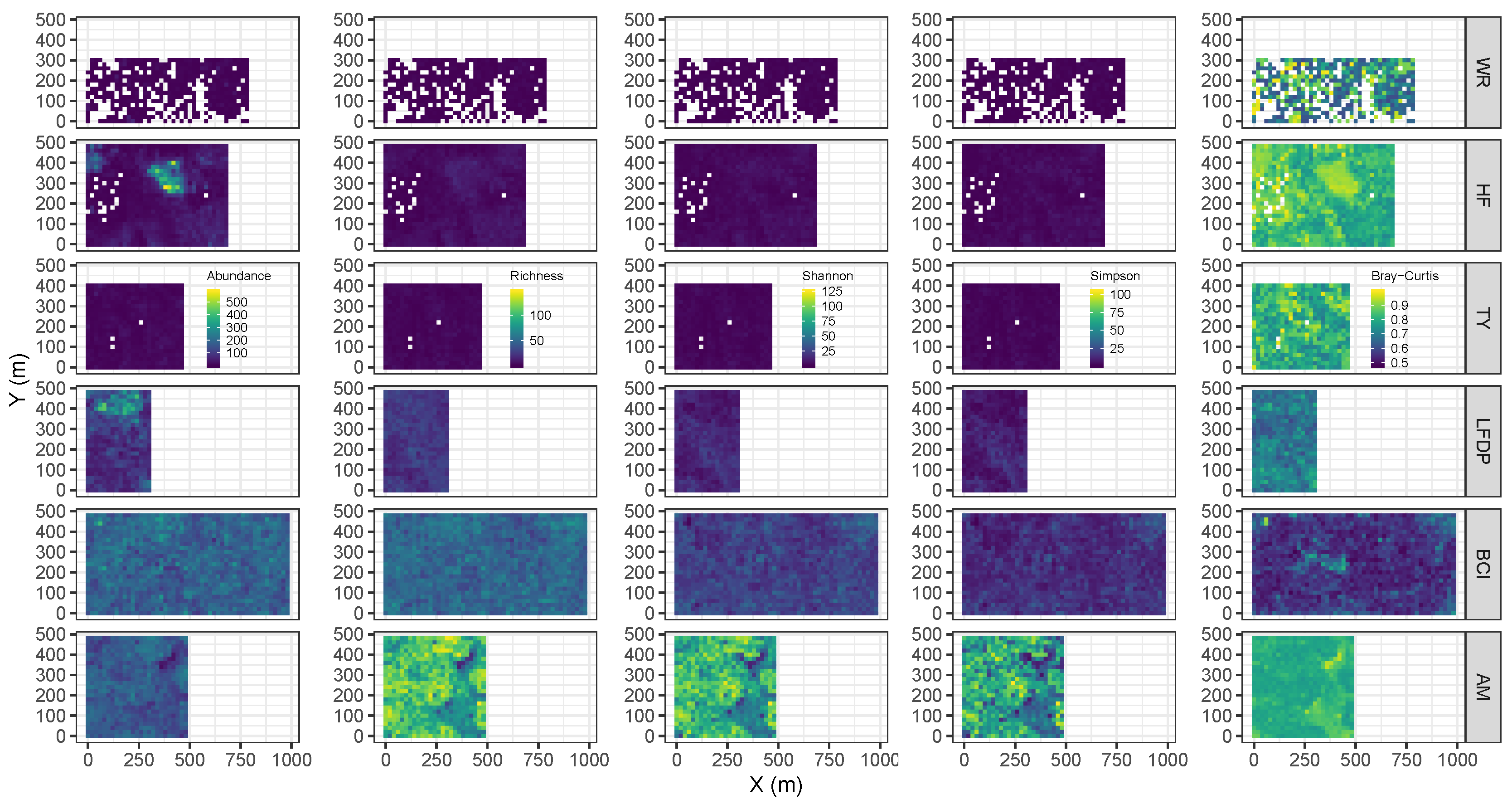


| Site | Species | Abbreviation | N | DBH (SD) | BA |
|---|---|---|---|---|---|
| Wind River | Tsuga heterophylla | TSHE | 9903 | 21.7 (25.35) | 865.90 |
| Pseudotsuga menzeisii | PSME | 564 | 101.1 (26.38) | 483.32 | |
| +Abies amabilis | ABAM | 4481 | 7.9 (9.73) | 55.08 | |
| *Taxus brevifolia | TABR | 1848 | 14.1 (8.15) | 38.30 | |
| *Acer circinatum | ACCI | 7514 | 3.6 (1.99) | 9.93 | |
| Harvard Forest | Tsuga canadensis | tsugca | 22,880 | 10.8 (12.22) | 476.03 |
| Quercus rubra | querru | 3896 | 28.6 (13.38) | 305.79 | |
| Acer rubrum | acerru | 9723 | 14.1 (8.94) | 212.01 | |
| Pinus strobus | pinust | 2126 | 24.4 (18.30) | 155.30 | |
| Tyson Research Center | Quercus alba | quealb | 2061 | 29.6 (16.24) | 184.06 |
| Quercus rubra | querub | 1547 | 30.1 (17.63) | 147.49 | |
| Quercus velutina | quevel | 690 | 33.5 (13.91) | 71.25 | |
| +Carya tomentosa | cartom | 373 | 22.6 (12.76) | 19.72 | |
| +Carya glabra | cargla | 188 | 25.2 (14.96) | 12.66 | |
| +Carya ovata | carova | 190 | 17.6 (11.16) | 6.48 | |
| *Cornus florida | corflo | 4500 | 5.1 (2.43) | 11.23 | |
| *Frangula caroliniana | fracar | 8674 | 2.0 (0.85) | 3.32 | |
| *Lindera benzoin | linben | 4907 | 1.8 (0.66) | 1.47 | |
| *Asimina triloba | asitri | 1610 | 2.4 (1.32) | 0.94 | |
| Luquillo | Dacryodes excelsa | DACEXC | 1544 | 21.2 (15.71) | 84.28 |
| +Cecropia schreberiana | CECSCH | 2902 | 10.0 (6.65) | 32.95 | |
| *Prestoea acuminata | PREMON | 7707 | 14.3 (2.96) | 128.82 | |
| Barro Colorado Island | Alseis blackiana | alsebl | 8680 | 5.4 (8.96) | 74.79 |
| Oenocarpus mapora | oenoma | 7387 | 8.6 (1.84) | 44.57 | |
| Amacayacu | Eschweilera coriacea | Eschcori | 2574 | 11.7 (12.53) | 59.43 |
| Eschweilera itayensis | Eschitay | 776 | 7.9 (13.74) | 15.34 | |
| Otoba glycyarpa | otobglyc | 1124 | 7.5 (10.00) | 13.78 | |
| Eschweilera rufifolia | Eschrufi | 414 | 7.1 (8.72) | 4.10 | |
| Guarea pubescens | Guarpube | 1483 | 3.4 (3.95) | 3.17 | |
| *Rinorea lindeniana | Rinolind | 1793 | 4.0 (2.93) | 3.44 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ellison, A.M.; Buckley, H.L.; Case, B.S.; Cardenas, D.; Duque, Á.J.; Lutz, J.A.; Myers, J.A.; Orwig, D.A.; Zimmerman, J.K. Species Diversity Associated with Foundation Species in Temperate and Tropical Forests. Forests 2019, 10, 128. https://doi.org/10.3390/f10020128
Ellison AM, Buckley HL, Case BS, Cardenas D, Duque ÁJ, Lutz JA, Myers JA, Orwig DA, Zimmerman JK. Species Diversity Associated with Foundation Species in Temperate and Tropical Forests. Forests. 2019; 10(2):128. https://doi.org/10.3390/f10020128
Chicago/Turabian StyleEllison, Aaron M., Hannah L. Buckley, Bradley S. Case, Dairon Cardenas, Álvaro J. Duque, James A. Lutz, Jonathan A. Myers, David A. Orwig, and Jess K. Zimmerman. 2019. "Species Diversity Associated with Foundation Species in Temperate and Tropical Forests" Forests 10, no. 2: 128. https://doi.org/10.3390/f10020128
APA StyleEllison, A. M., Buckley, H. L., Case, B. S., Cardenas, D., Duque, Á. J., Lutz, J. A., Myers, J. A., Orwig, D. A., & Zimmerman, J. K. (2019). Species Diversity Associated with Foundation Species in Temperate and Tropical Forests. Forests, 10(2), 128. https://doi.org/10.3390/f10020128








