Soil Fungal Community in Norway Spruce Forests under Bark Beetle Attack
Abstract
1. Introduction
2. Materials and Methods
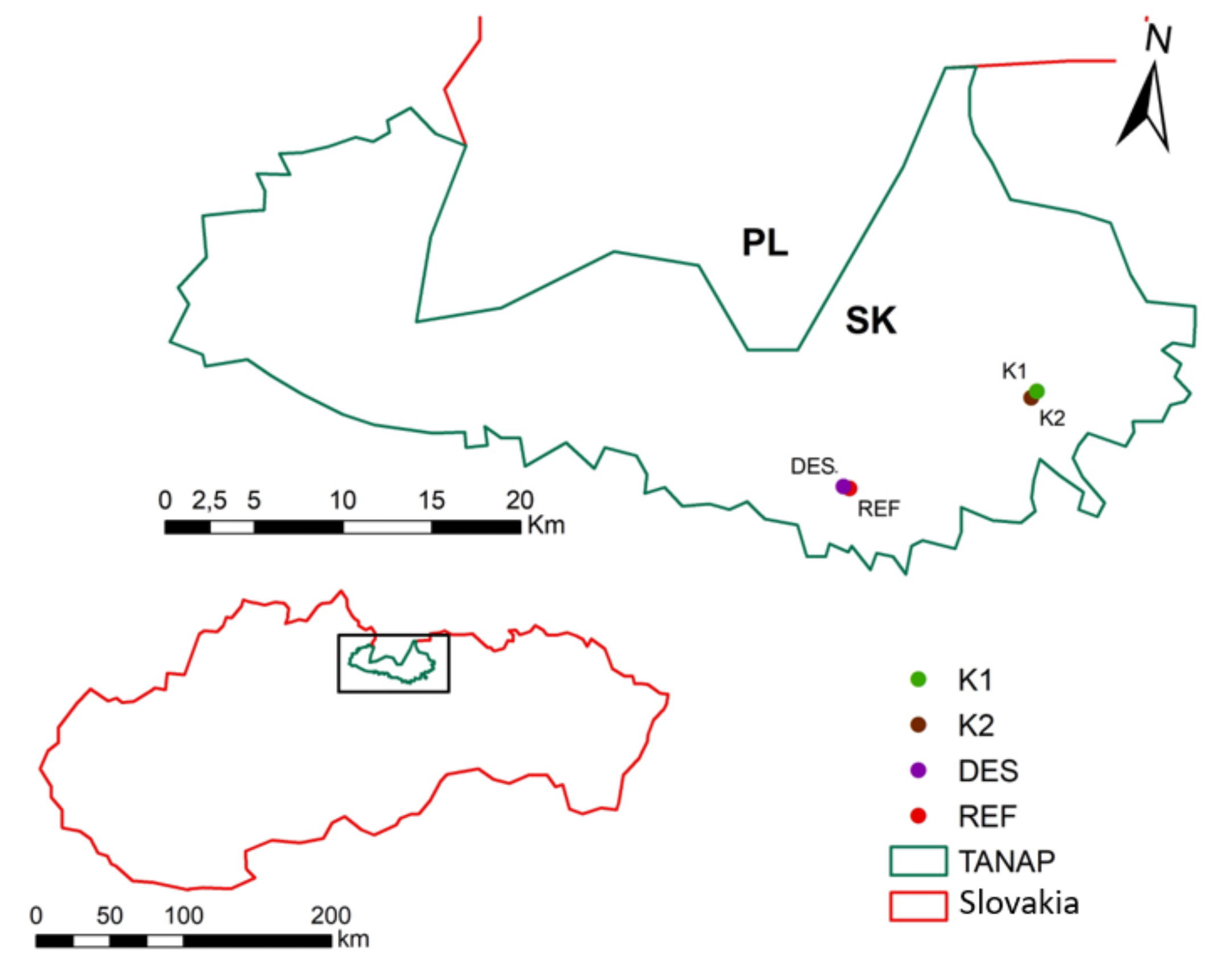
2.1. Study Area and Sample Collection
2.2. Soil Analysis
2.3. DNA Extraction, Amplification, and Sequencing
2.4. Data Processing and Analysis
2.5. Statistical Analysis
3. Results
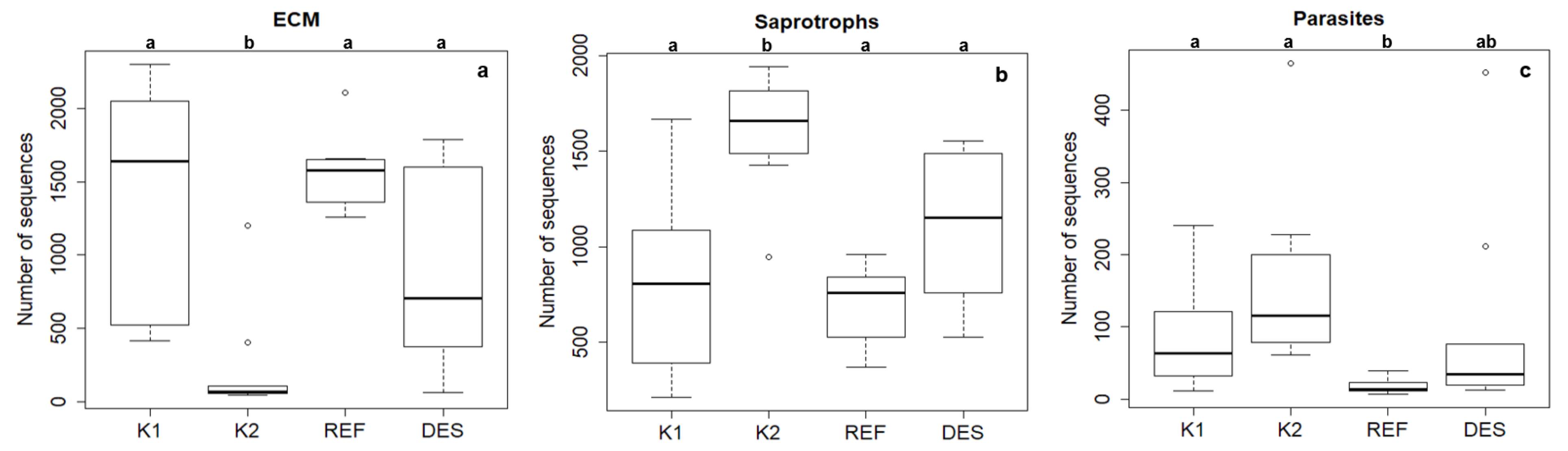
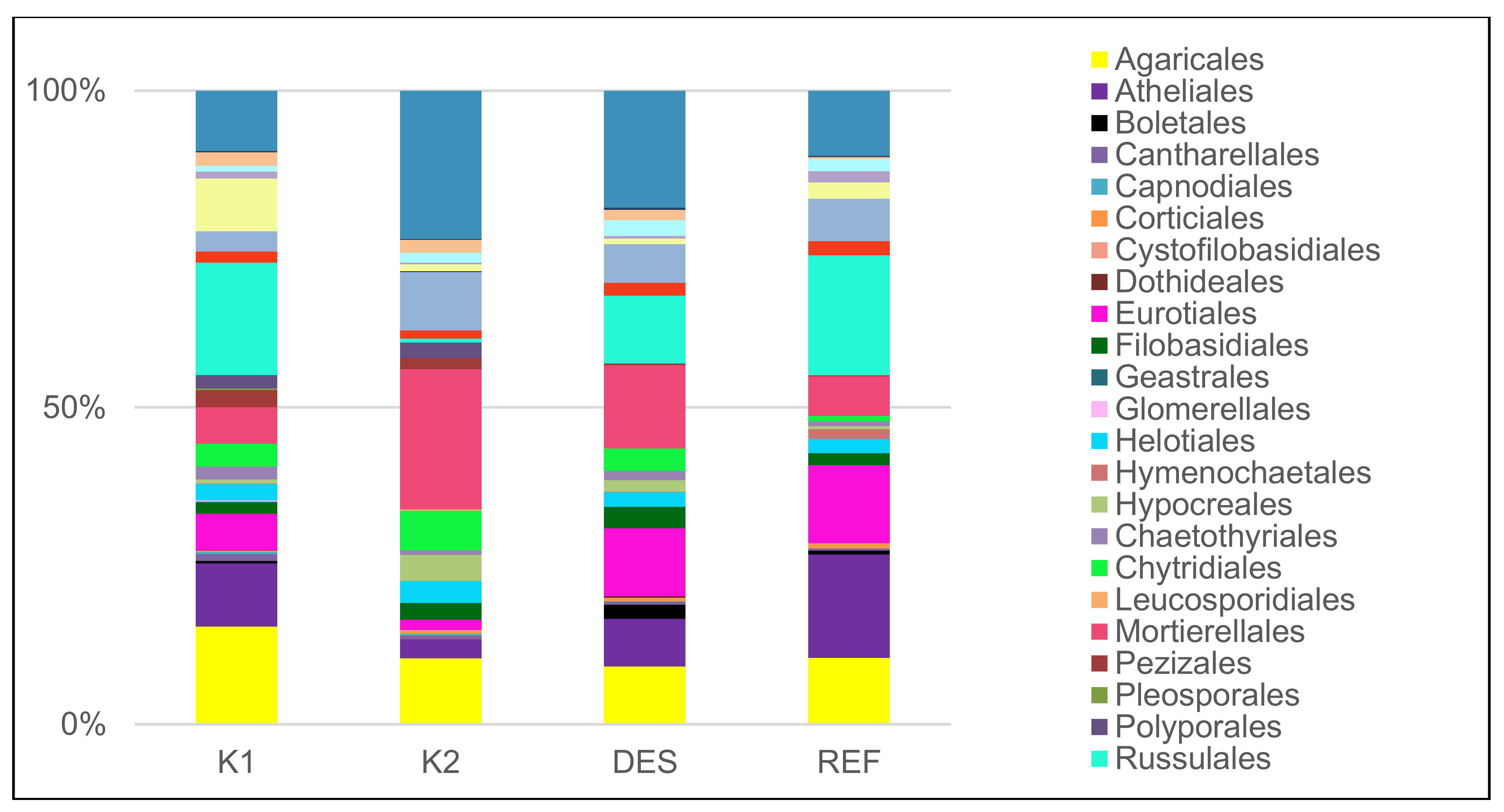
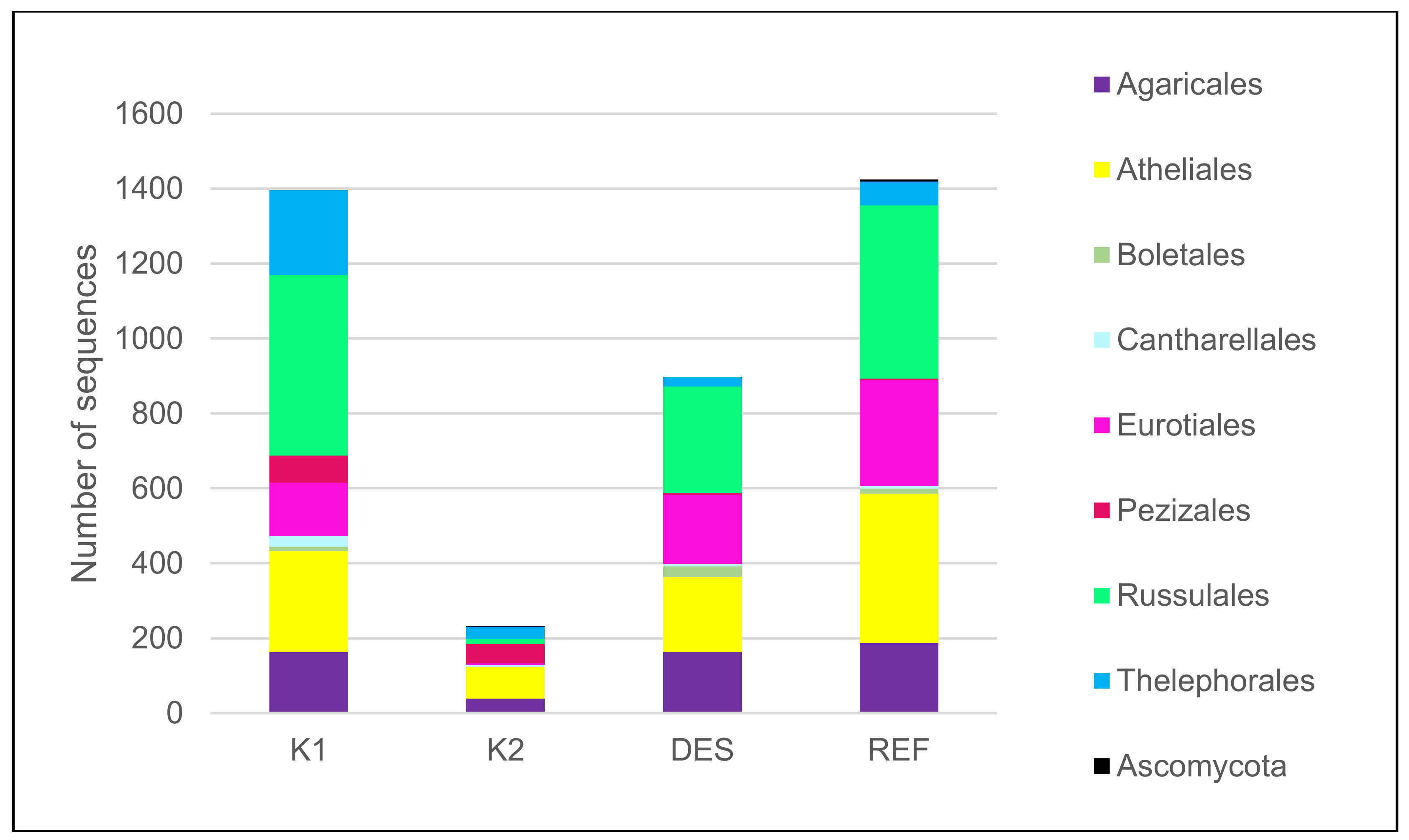
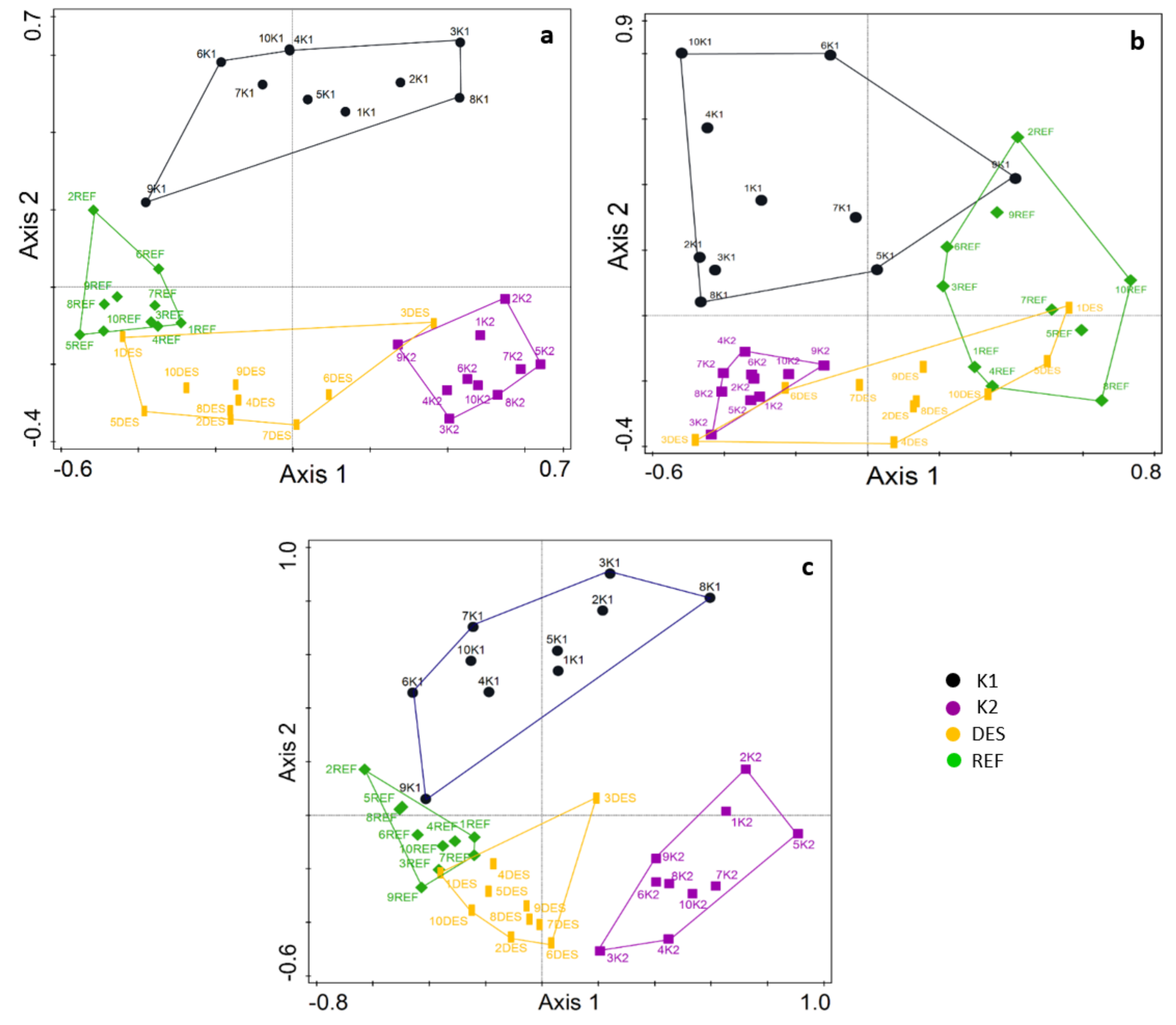
3.1. Fungal Community
3.2. Vegetation
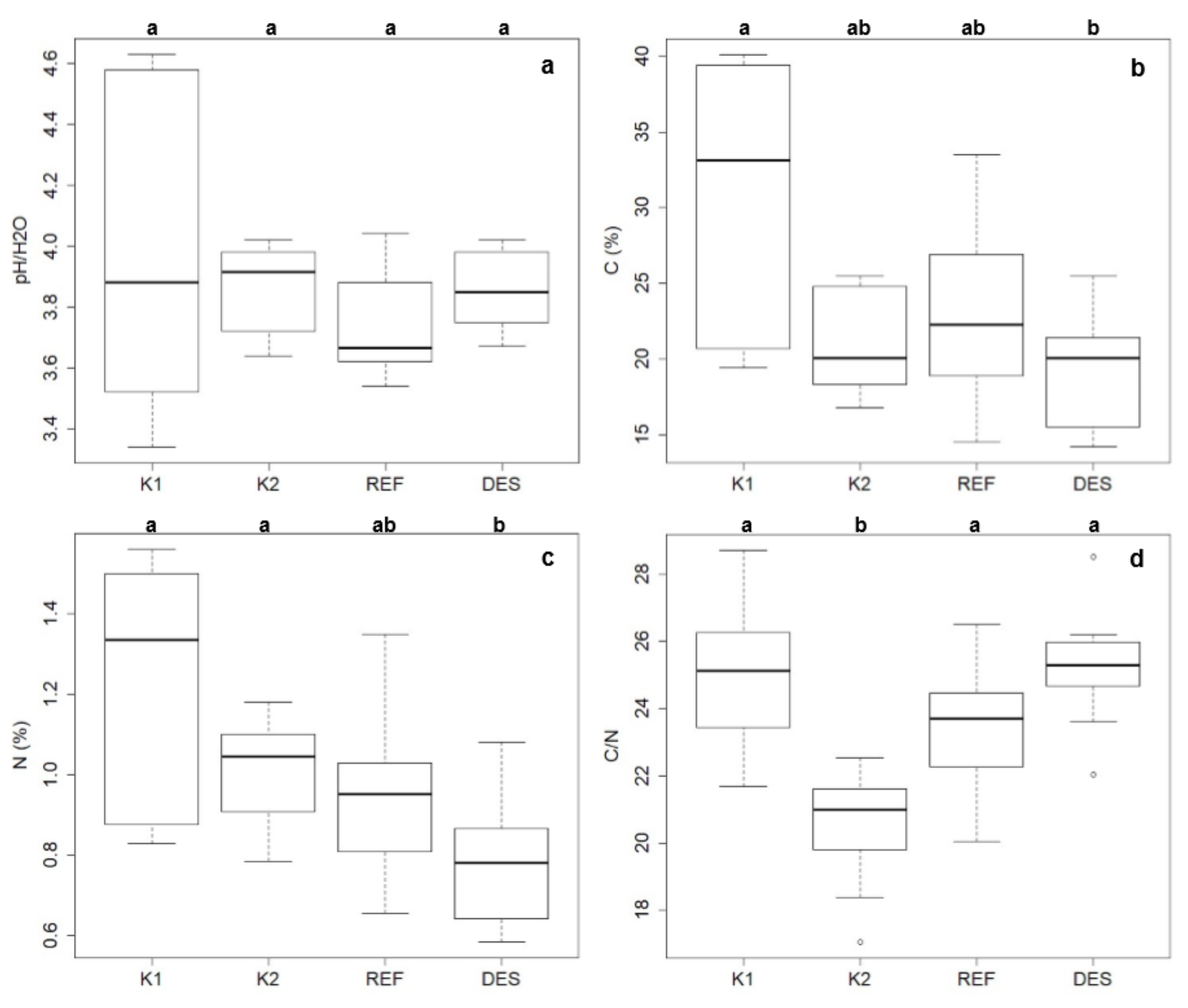
3.3. Soil Properties
3.4. Vegetation and Soil Properties vs. Fungal Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Seidl, R.; Müller, J.; Hothorn, T.; Bässler, C.; Heurich, M.; Kautz, M. Small beetle, large-scale drivers: How regional and landscape factors affect outbreaks of the European spruce bark beetle. J. Appl. Ecol. 2016, 53, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Papaik, M.J.; Canham, C.D. Species resistance and community response to wind disturbance regimes in northern temperate forests. J. Ecol. 2006, 94, 1011–1026. [Google Scholar] [CrossRef]
- Fischer, A.; Marshall, P.; Camp, A. Disturbances in deciduous forest ecosystems of the northern hemisphere: Their effects on both recent and future forest development. Biodivers. Conserv. 2013, 22, 1863–1893. [Google Scholar] [CrossRef]
- Nealis, V.; Peter, B. Risk Assessment of the Threat of Mountain Pine Beetle to Canada’s Boreal and Eastern Pine Forests; Report BC-X-417; Canadian Forest Service, Pacific Forestry Centre: Victoria, BC, Canada, 2008. [Google Scholar]
- Hart, S.J.; Veblen, T.T.; Mietkiewicz, N.; Kulakowsky, D. Negative feedback on bark beetle outbreaks: Widespread and severe spruce beetle infestation restricts subsequent infestation. PLoS ONE 2015, 10, e0127975. [Google Scholar] [CrossRef] [PubMed]
- Raffa, K.F.; Aukema, B.H.; Bentz, B.J.; Carroll, A.L.; Hicke, J.A.; Turner, M.G.; Romme, W.H. Cross-scale drivers of natural disturbances prone to anthropogenic amplification: The dynamics of bark beetle eruptions. BioScience 2008, 58, 501–517. [Google Scholar] [CrossRef]
- Weed, A.S.; Ayres, M.P.; Hicke, J.A. Consequences of climate change for biotic disturbances in North American forests. Ecol. Monogr. 2013, 83, 441–470. [Google Scholar] [CrossRef]
- Hlásny, T.; Turčáni, M. Persisting bark beetle outbreak indicates the unsustainability of secondary Norway spruce forets: Case study from Central Europe. Ann. For. Sci. 2013, 70, 481–491. [Google Scholar] [CrossRef]
- Ayres, M.P.; Lombardero, M.J. Assessing the consequences of global change for forest disturbance from herbivores and pathogens. Sci. Total Environ. 2000, 262, 263–286. [Google Scholar] [CrossRef]
- Rinnan, R.; Bååth, E. Differential utilization of carbon substrates by bacteria and fungi in tundra soil. Appl. Environ. Microbiol. 2009, 75, 3611–3620. [Google Scholar] [CrossRef]
- Iordache, V.; Gherghel, F.; Kothe, E. Assessing the effect of disturbances on ectomycorrhiza diversity. Int. J. Environ. Res. Public Health 2009, 6, 411–432. [Google Scholar] [CrossRef]
- Dahlberg, A. Community ecology of ectomycorrhizal fungi: An advancing interdisciplinary field. New Phytol. 2001, 150, 555–562. [Google Scholar] [CrossRef]
- Haselwandter, K.; Bowen, G.D. Mycorrhizal relation in trees for agroforestry and land rehabilitation. For. Ecol. Manag. 1996, 81, 1–17. [Google Scholar] [CrossRef]
- Gehring, C.; Bennett, A. Mycorrhizal-fungal-plant-insect interactions: The importance of a community approach. Environ. Entomol. 2009, 38, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Saravesi, K.; Markkola, A.; Rautio, P.; Roitto, M.; Tuomi, J. Defoliation causes parallel temporal responses in a host tree and its fungal symbionts. Oecologia 2008, 156, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Štursová, M.; Šnajdr, J.; Cajthaml, T.; Bárta, J.; Šantrůčková, H.; Baldrian, P. When the forest dies: The response of forest soil fungi to a bark beetle-induced tree dieback. ISME J. 2014, 8, 1920. [Google Scholar] [CrossRef] [PubMed]
- Treu, R.; Karst, J.; Randall, M.; Pec, G.J.; Cigan, P.W.; Simard, S.W.; Cooke, J.E.; Erbilgin, N.; Cahill, J.F., Jr. Decline of ectomycorrhizal fungi following a mountain pine beetle epidemic. Ecology 2014, 95, 1096–1103. [Google Scholar] [CrossRef]
- Pec, G.J.; Karst, J.; Taylor, D.L.; Cigan, P.W.; Erbilgin, N.; Cooke, J.E.; Simard, S.W.; Cahill, J.F. Change in soil fungal community structure driven by a decline in ectomycorrhizal fungi following a mountain pine beetle (Dendroctonus ponderosae) outbreak. New Phytol. 2017, 213, 864–873. [Google Scholar] [CrossRef]
- Fleischer, P.; Homolová, Z. Long-term research on ecological condition in the larch-spruce forests in High Tatras after natural disturbance. For. J. 2011, 57, 237–250. [Google Scholar]
- Šebeň, V.; Konopka, B.; Bošela, M.; Pajtík, J. Contrasting development of declining and living larch-spruce stands after a disturbance event: A case study from the High Tatra Mts. For. J. 2015, 61, 157–166. [Google Scholar] [CrossRef]
- Fleischer, P. Windfall research and monitoring in the High Tatra Mts, objectives, principles, methods and current status. Contrib. Geophys. Geod. 2008, 38, 233–248. [Google Scholar]
- Ihrmark, K.; Bödeker, I.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR—Protocols and Applications—A Laboratory Manual. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Větrovský, T.; Baldrian, P. Analysis of soil fungal communities by amplicon pyrosequencing: Current approaches to data analysis and the introduction of the pipeline SEED. Biol. Fertil. Soils 2013, 49, 1027–1037. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. ITSx: Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for use in environmental sequencing. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 2009. [Google Scholar] [CrossRef]
- Kõljalg, U.; Larsson, K.H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Boberg, J.B.; Finlay, R.D.; Stenlid, J.; Ekblad, A.; Lindahl, B.D. Nitrogen and carbon reallocation in fungal mycelia during decomposition of boreal forest litter. PLoS ONE 2014, 9, e92897. [Google Scholar] [CrossRef]
- Carlucci, A.; Raimondo, M.L.; Phillips, A.J.L. Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Personia 2012, 28, 34–48. [Google Scholar] [CrossRef]
- Douhan, G.W.; Rizzo, D.M. Host-parasite relationships among bolete infecting Hypomyces species. Mycol. Res. 2003, 107, 1342–1349. [Google Scholar] [CrossRef]
- Garnier-Delcourt, M.; Marson, G.; Reckinger, C.; Schultheis, B.; Tholl, M. Notes mycologiques luxembourgeoises. VII Bulletin de la Société des Naturalistes Luxembourgeois 2013, 114, 35–54. [Google Scholar]
- Kauserud, H.; Schalchian-Tabrizi, K.; Decock, C. Multilocus sequencing reveald multiple geographically structured lineages of Coniophora arida and C. olivacea (Boletales) in North America. Mycologia 2007, 99, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, H.; Vesterholt, J. Funga Nordica: Agaricoid, Boletoid and Cyphelloid Genera, 1st ed.; Nordsvamp: Copenhagen, Denmark, 2008. [Google Scholar]
- Kohout, P.; Těšilová, T.; Roy, M.; Vohník, M.; Jerásková, J. A diverse fungal community associated with Pseudorchis albida (Orchidaceae) roots. Fungal Ecol. 2013, 6, 50–64. [Google Scholar] [CrossRef]
- Kohout, P.; Charvátová, M.; Štursová, M.; Mašínová, T.; Tomšovský, M.; Baldrian, P. Clearcutting alters decomposition processes and initiates complex restructuring of fungal communities in soil and tree roots. ISME J. 2018, 12, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Langer, G.; Langer, E.; Oberwinkler, F.; Chen, J. Speciation of Botryobasidium subcoronatum (Basidiomycota) collected in Taiwan: Morphology, mating tests, and molecular data. Mycoscience 2000, 41, 201–210. [Google Scholar] [CrossRef]
- Picard, K.T.; Letcher, P.M.; Powell, M.J. Evidence for a facultative mutualist nutritional relationship between the green coccoid alga Bracteacoccus sp. (Chlorophyceae) and the zoosporic fungus Rhizidium phycophilum (Chytridiomycota). Fungal Biol. 2013, 117, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Piel, J.; Atzorn, R.; Gäbler, R.; Kühnemann, F.; Boland, W. Cellulysin from the plant parasitic fungus Trichoderma viride elicits volatile byosynthesis in higher plants via the octadecanoid signalling cascade. FEBS Lett. 1997, 416, 143–148. [Google Scholar] [CrossRef]
- Redhead, S.A. A biogeographic overview of the Canadian mushroom flora. Can. J. Bot. 1989, 67, 3003–3062. [Google Scholar] [CrossRef]
- Tedersoo, L.; Smith, M.E. Lineages of ectomycorrhizal fungi revisited: Foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol. Rev. 2013, 27, 83–99. [Google Scholar] [CrossRef]
- Wachwska, U.; Borowska, J. Antagonistic yeasts competes for iron with winter wheat stem base pathogens. Gesunde Pflanzen 2014, 66, 141–148. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2011; Available online: http://www.R-project.org/ (accessed on 11 February 2018)ISBN 3-900051-07-0.
- Ter Braak, C.J.F.; Šmilauer, P. Canoco Reference Manual and User’s Guide: Software for Ordination, version 5.0; Microcomputer Power: Ithaca, NY, USA, 2012. [Google Scholar]
- Brunner, I.; Bakker, M.R.; Björk, R.G.; Hirano, Y.; Lukac, M.; Aranda, X.; Børja, I.; Eldhuset, T.D.; Helmisaari, H.S.; Jourdan, C.; et al. Fine-root turnover rates of European forests revisited: An analysis of data from sequential coring and ingrowth cores. Plant Soil 2013, 362, 357–372. [Google Scholar] [CrossRef]
- Horton, T.R.; Bruns, T.D. The molecular revolution in ectomycorrhizal ecology: Peeking into the black-box. Mol. Ecol. 2001, 10, 1855–1871. [Google Scholar] [CrossRef] [PubMed]
- Vašutová, M.; Edwars-Jonášová, M.; Veselá, P.; Effenberková, L.; Fleischer, P.; Cudlín, P. Management regime is the most important factor influencing ectomycorrhizal species community in Norway spruce forests after windthrow. Mycorrhiza 2018, 28, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Buscardo, E.; Rodríguez-Echeverría, S.; Freitas, H.; De Angelis, P.; Pereira, J.S.; Muller, L.A. Contrasting soil fungal communities in Mediterranean pine forests subjected to different wildfire frequencies. Fungal Divers. 2015, 70, 85–99. [Google Scholar] [CrossRef]
- Ford, S.A.; Kleinman, J.S.; Hart, J.L. Effects of wind disturbance and salvage harvesting on macrofungal communities in a Pinus Woodland. For. Ecol. Manag. 2018, 407, 31–46. [Google Scholar] [CrossRef]
- Smith, J.E.; McKay, D.; Brenner, G.; McIvers, J.; Spatafora, J.W. Early impacts of forest restoration treatments on the ectomycorrhizal fungal community and fine root biomass in a mixed conifer forest. J. Appl. Ecol. 2005, 42, 526–535. [Google Scholar] [CrossRef]
- Glassman, S.I.; Peay, K.G.; Talbot, J.M.; Smith, D.P.; Chung, J.A.; Taylor, J.W.; Vilgalys, R.; Bruns, T.D. A continental view of pine-associated ectomycorrhizal fungal spore banks: A quiescent functional guild with a strong biogeographic pattern. New Phytol. 2015, 205, 1619–1631. [Google Scholar] [CrossRef]
- Vašutová, M.; Edwards-Jonášová, M.; Baldrian, P.; Čermák, M.; Cudlín, P. Distinct environmental variables drive the community composition of mycorrhizal and saprotrophic fungi at the alpine treeline ecotone. Fungal Ecol. 2017, 27, 116–124. [Google Scholar] [CrossRef]
- Tedersoo, L.; Kõljalg, U.; Hallenberg, N.; Larsson, K.H. Fine scale distribution of ectomycorrhizal fungi and roots across substrate layers including coarse woody debris in a mixed forest. New Phytol. 2003, 159, 153–165. [Google Scholar] [CrossRef]
- Castaño, C.; Alday, J.G.; Lindahl, B.D.; de Aragón, J.M.; de-Miguel, S.; Colinas, C.; Parladé, J.; Pera, J.; Bonet, J.A. Lack of thinning effects over inter-annual changes in soil fungal community and diversity in a Mediterranean pine forest. For. Ecol. Manag. 2018, 424, 420–427. [Google Scholar] [CrossRef]
- Rosenvald, R.; Lõhmus, A. For what, when, and where is green-tree retention better than clear-cutting? A review of the biodiversity aspects. For. Ecol. Manag. 2008, 255, 1–15. [Google Scholar] [CrossRef]
- Sun, H.; Santalahti, M.; Pumpanen, J.; Köster, K.; Berninger, F.; Raffaello, T.; Jumpponen, A.; Asiegbu, F.O.; Heinonsalo, J. Fungal community shifts in structure and function across a boreal forest fire chronosequence. Appl. Environ. Microbiol. 2015, 81, 7869–7880. [Google Scholar] [CrossRef] [PubMed]
- Treseder, K.K.; Lennon, J.T. Fungal traits that drive ecosystem dynamics on land. Microbiol. Mol. Biol. Rev. 2015, 79, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Picard, K.T.; Letcher, P.M.; Powell, M.J. Rhizidium phycophilum, a new species in Chytridiales. Mycologia 2009, 101, 696–706. [Google Scholar] [CrossRef] [PubMed]
| K1 | K2 | DES | REF | |
|---|---|---|---|---|
| average species richness | 123 ± 4.3 | 135 ± 3.8 | 115 ± 2.4 | 119 ± 2.7 |
| mycorrhizal sequences proportion* | 62 ± 8 | 15 ± 5 | 45 ± 9 | 70 ± 3 |
| saprotrophic sequence proportion* | 34 ± 6 | 77 ± 4 | 50 ± 6 | 29 ± 2 |
| parasitic sequence proportion* | 4 ± 1 | 8 ± 2 | 4 ± 2 | 1 ± 0.1 |
| total number of mycorrhizal species | 52 | 16 | 27 | 46 |
| total number of saprotrophic species | 52 | 52 | 47 | 44 |
| total number of parasitic species | 5 | 10 | 7 | 3 |
| dominant ECM species* (5 dominant species) | Russula decolorans (Fr.) Fr. 1838 (14.3), Pseudotomentella mucidula (P. Karst.) Svrček (8.1), Elaphomyces sp. (7.4), R. puellaris Fr. (6.2), Thelephora terrestris Ehrh. (5.4) | Piloderma olivaceum (Parmasto) Hjortstam (23.6), Inocybe soluta Velen. (12.6), Wilcoxina sp. (11.9), T. terrestris (8.2), Tylospora asterophora (Bonord.) Donk (5.8) | Elaphomyces sp. (20), R. vesca Fr. 1836 (17.3), R. mustelina Fr. 1838 (9.8), T. asterophora (9.7), Inocybe umbrina Bres. (7.9) | Elaphomyces sp. (16.3), R. puellaris (13.8), T. asterophora (7.3) and R. vesca (6.9), Amanita fulva Pers. 1818 (3.7) |
| dominant saprotrophic species* (5 dominant species) | Rhodotorula sp. (10.5), Mycena pura (Pers.) P. Kumm. 1871 (9), Rhizidium phycophilum K.T. Picard (8.8), Physisporinus sanguinolentus (Alb. & Schwein.) Donk 1966 (6.2), Cryptococcus terricola T.A. Pedersen (5.5) | Rhodotorula sp. (15.1), Mortierella macrocystis W. Gams (8.8), M. humilis Linnem. (7.5), R. phycophilum (6.5), Mycena galopus (Pers.) P. Kumm. 1871 (5.0) | Rhodotorula sp. (14.5), M. humilis (9.1), C. terricola (8.2), M. macrocystis (6.9), R. phycophilum (6.6) | Rhodotorula sp. (24.8), Cryptococcus sp. (7.1), M. humilis (6.9), C. terricola (6.8), M. macrocystis (4.5) |
| K1 | K2 | DES | REF | |
|---|---|---|---|---|
| tree cover * | 43 ± 10.9 | 0 | 5 ± 3 | 43 ± 9.2 |
| cover of dominant herbs (more than 10%)* | Vaccinium myrtillus (36 ± 11), Oxalis acetosella (25 ± 7.4), Dryopteris sp. (17 ± 8.7) | Callamagrostis villosa (31 ± 9.6), Vaccinium myrtillus (24 ± 6.2), Rubus idaeus (23 ± 5.3) | Vaccinium myrtillus (32 ± 5.9), Callamagrostis villosa (18 ± 5.8), Avenella flexuosa (17 ± 4.9) | Vaccinium myrtillus (39 ± 11), Avenella flexuosa (14 ± 6.2), Calamagrostis villosa (13 ± 5.4) |
| spruce seedling cover* | 1.4 ± 0.6 | 1.2 ± 0.5 | 3 ± 1 | 10 ± 8 |
| dead wood cover* | lying logs (4.4 ± 2.9) | lying logs (15 ± 4.4) | piles of branches (22 ± 5.9) | piles of branches (14.5 ± 7.9), lying logs (3.2 ± 3) |
| Explanatory Variables | Explained Variation Adjusted (%) | Pseudo-F | p |
|---|---|---|---|
| Dataset all | |||
| a) forest type | 18.7 | 4.0 | 0.001 |
| b) tree cover, Vaccinium myrtillus, Oxalis acetosella, Dryopteris sp., N%, C%, N° | 22.2 | 2.6 | 0.001 |
| ECM dataset | |||
| a) forest type | 13.4 | 3.0 | 0.001 |
| c) tree cover, Dryopteris sp., N° | 15.3 | 3.3 | 0.001 |
| Saprotroph dataset | |||
| a) forest type | 23.3 | 5.0 | 0.001 |
| b) tree cover, Sorbus aucuparia, Calamagrostis villosa, N%, C%, pH/H2O | 23.5 | 3.0 | 0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veselá, P.; Vašutová, M.; Edwards-Jonášová, M.; Cudlín, P. Soil Fungal Community in Norway Spruce Forests under Bark Beetle Attack. Forests 2019, 10, 109. https://doi.org/10.3390/f10020109
Veselá P, Vašutová M, Edwards-Jonášová M, Cudlín P. Soil Fungal Community in Norway Spruce Forests under Bark Beetle Attack. Forests. 2019; 10(2):109. https://doi.org/10.3390/f10020109
Chicago/Turabian StyleVeselá, Petra, Martina Vašutová, Magda Edwards-Jonášová, and Pavel Cudlín. 2019. "Soil Fungal Community in Norway Spruce Forests under Bark Beetle Attack" Forests 10, no. 2: 109. https://doi.org/10.3390/f10020109
APA StyleVeselá, P., Vašutová, M., Edwards-Jonášová, M., & Cudlín, P. (2019). Soil Fungal Community in Norway Spruce Forests under Bark Beetle Attack. Forests, 10(2), 109. https://doi.org/10.3390/f10020109






