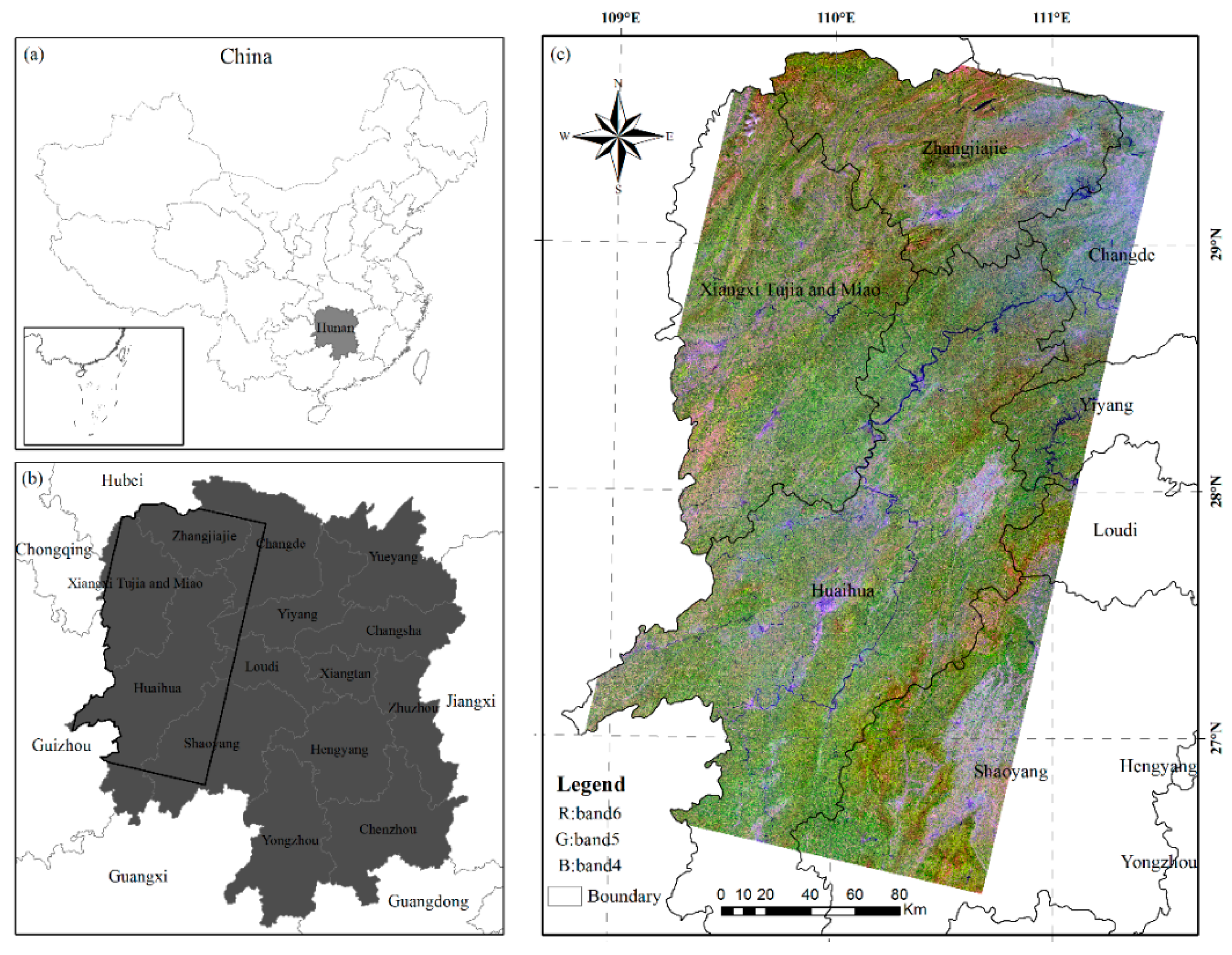
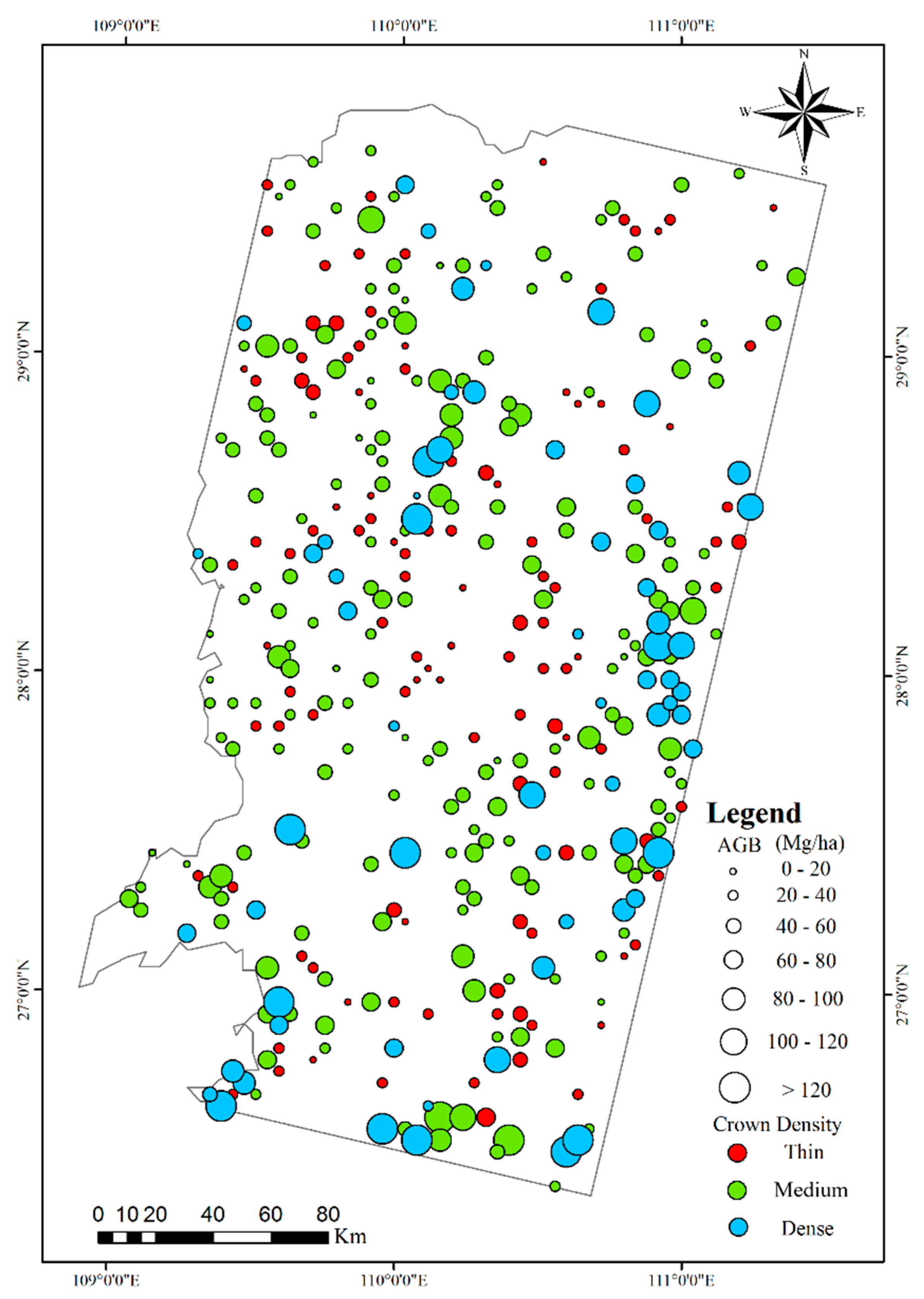
1. Introduction
Forest ecosystems provide important ecosystem services and are an important component of the earth’s energy cycle. Forest biomass is a fundamental parameter for describing the structure and function of forest ecosystems [
1,
2]. Many ecosystem processes are impacted by forest biomass and, in turn, forest biomass is impacted by these processes [
3]. Forests provide important terrestrial carbon storage. Studies on forest biomass are essential for determining the carbon storage, carbon balance, and carbon cycling at the regional and global levels.
Due to difficulties in measuring forest belowground biomass, the majority of previous studies have mostly focused on forest aboveground biomass (AGB). The estimation of AGB is an essential task for assessing carbon stocks and carbon balance [
4]. In past studies, three main approaches have been used to estimate forest AGB, namely: process-based ecosystem models, field measurements, and a combination of forest inventory plots and remotely sensed data [
5,
6]. The remote-sensing-based method has been commonly used in the last decades for several reasons: (1) Remote sensing data covers large areas, allowing for the assessment of the spatial variation of vegetation and making it possible to determine the spatial distribution and pattern of biomass in large areas and complex forest landscapes; (2) multiple sensors and multiple spatial resolutions can be used for forest biomass research at different scales; and (3) multi-temporal remote sensing images provide long-term, dynamic, and continuous AGB observations [
7,
8].
The rapid development of remote sensing technology has provided a wide variety of remotely sensed imagery data for AGB estimation. The data can be divided into three categories: (1) optical remote sensing data such as Landsat, Systeme Probatoire d’Observation de la Terre (SPOT), moderate-resolution imaging spectroradiometer (MODIS), QuickBird, ASTER, Advanced Very High-Resolution Radiometer (AVHRR), and China-brazil earth resource satellite (CBERS); (2) active remote sensing data including Radar and Lidar; and (3) the integration of multisource remote sensing data [
5,
9,
10,
11,
12,
13]. In particular, Landsat has been commonly used for forest biomass estimation in combination with sample plots because the images can be freely downloaded, have medium spatial (30 m × 30 m) and temporal (16 days) resolutions, and have wide coverage [
14,
15]. In many countries, the spatial resolution of Landsat is similar to the size of sample plots in national forest inventories, thus reducing the spatial errors in matching the pixels and the sample plots [
8].
Generally, forest stands with different biomass have different forest structures and different biophysical parameters. These features are reflected in remote sensing images as different colors, structures, and textures. Using feature extraction methods, the image parameters that are closely related to forest biomass can be extracted from the remote sensing images, and forest biomass can be estimated. Vegetation information in remote sensing images is mainly reflected by the spectral characteristics. The spectral differences in leaves and vegetation canopies and their changes over time differ in different spectral bands [
9,
16,
17]. Vegetation parameters derived from optical remote sensing include vegetation indices, leaf area index, absorbed photosynthetically active radiation (APAR), and various image transformations [
18,
19,
20]. Landsat images can be used to derive spectral information that can be correlated with forest inventory AGB data [
21]. The remote sensing information is strongly related to several forest parameters and the use of spectral variables in modeling forest biomass has a long history. The Landsat variables that have been commonly used include spectral bands, vegetation indices (e.g., normalized differential vegetation index (NDVI), Enhanced Vegetation Index (EVI)), image transformations (e.g., principal component analysis (PCA) and tasseled cap transformation (TCT)), and texture images [
5,
15,
22,
23,
24,
25,
26].
Parametric algorithms and nonparametric algorithms have been applied for AGB estimation [
27]. In parametric algorithms, it is assumed that the direct or indirect relationships between the remotely sensed parameters and the forest AGB can be expressed using regression models. The application of parametric algorithms over large areas requires the assumption of spatially homogeneous relationships between the ground-based information and remote-sensing data. Parametric algorithms are easy to apply but are weak in terms of describing the complex relationship between AGB and remote sensing data. In addition, the accuracy of the algorithms largely relies on the statistical robustness. In contrast to parametric algorithms, nonparametric algorithms do not have explicit equations [
28] and do not assume a normal distribution of the independent and dependent variables. Nonparametric algorithms are more flexible to describe the nonlinear relationship between AGB and image data, but the physical mechanisms of the models are not clear and there are risks of over-fitting.
The linear model was frequently used in forest biomass estimation based on remote sensing. In previous studies, when the linear models were built for estimating AGB, the remote sensing factors were directly considered as fixed effect variables. The linear models did not consider the effects of forest characteristics, effects which may influence the independent variables and the model fitting, which in turn affect the fitting accuracy of the models. In this study, based on the analysis of the differences between the independent variables and AGB of different vegetation types in different crown density classes, the basic AGB linear models using remote sensing were built. The crown density classes which were considered as the influencing factor (random effect or dummy variable) were introduced into the model, and the linear dummy variable model and linear mixed-effects model were fitted to estimate AGB. The accuracies of the linear model, linear dummy variable model, and linear mixed-effects model were compared.
3. Results
The Pearson correlation coefficients between all spectral variables and the AGB were calculated and 30 variables had significant correlation with the AGB of four vegetation types. The correlation coefficients are listed in
Table 3. The result showed that the correlation coefficients were not higher than 0.260 for all the 30 spectral variables, and 11 texture features had significant correlation with the AGB.
Three types of models for each dependent variable (i.e., AGB of total vegetation, AGB of pine forest, AGB of fir forest, and AGB of mixed forest) were developed using the spectral variables which were selected by stepwise regression as the independent variable (
Table 4 and
Table 5). Twelve models were obtained. Parameter estimates of models 1–3 for different vegetation types are presented in
Table 4 and
Table 5. The independent variables of the total vegetation AGB were dominated by the image texture information, and the independent variables of the pine, fir, and mixed forests were dominated by the image texture information and spectral features. The model standard coefficients of the linear models showed that the texture information contributed more to the AGB estimation than the spectral features, which indicated that the texture information was important for AGB estimation in this study.
The fitting results of models 1–3 are summarized in
Table 6 and
Table 7. For the different vegetation types, the
R2 and
R2adj of model 2 and model 3 were larger than those of model 1, and the RMSE values were smaller than those of model 1. These results indicate that the performances of model 2 and model 3 were better than that of model 1. The
R2adj of model 2 and model 3 for pine forest had the smallest increase compared with model 1; the value of
R2adj increased by 0.16, and the RMSE values were smaller for model 2 and model 3 than for model 1. For the fir forest, model 2 and model 3 had the largest
R2adj values, and compared with model 1, the values increased more than 0.39. For the mixed forest and total vegetation, the
R2adj and RMSE values of model 2 and model 3 were better than those of model 1. These results show that model 2 and model 3, which were considered the crown density classes, had higher accuracies of AGB estimation than model 1.
To further test whether model 2 and model 3 significantly improved the accuracy of model 1, the F-test was used for determining the differences between model 1 and model 2 and between model 1 and model 3 (
Table 8). The F-test results show that, except for model 3 of the mixed forest, there were significant differences between model 2 and model 1 and between model 3 and model 1. This indicated that the performances of model 2 and model 3 were significantly better than that of model 1. The fitting results of the model 2 and model 3 had no significant differences.
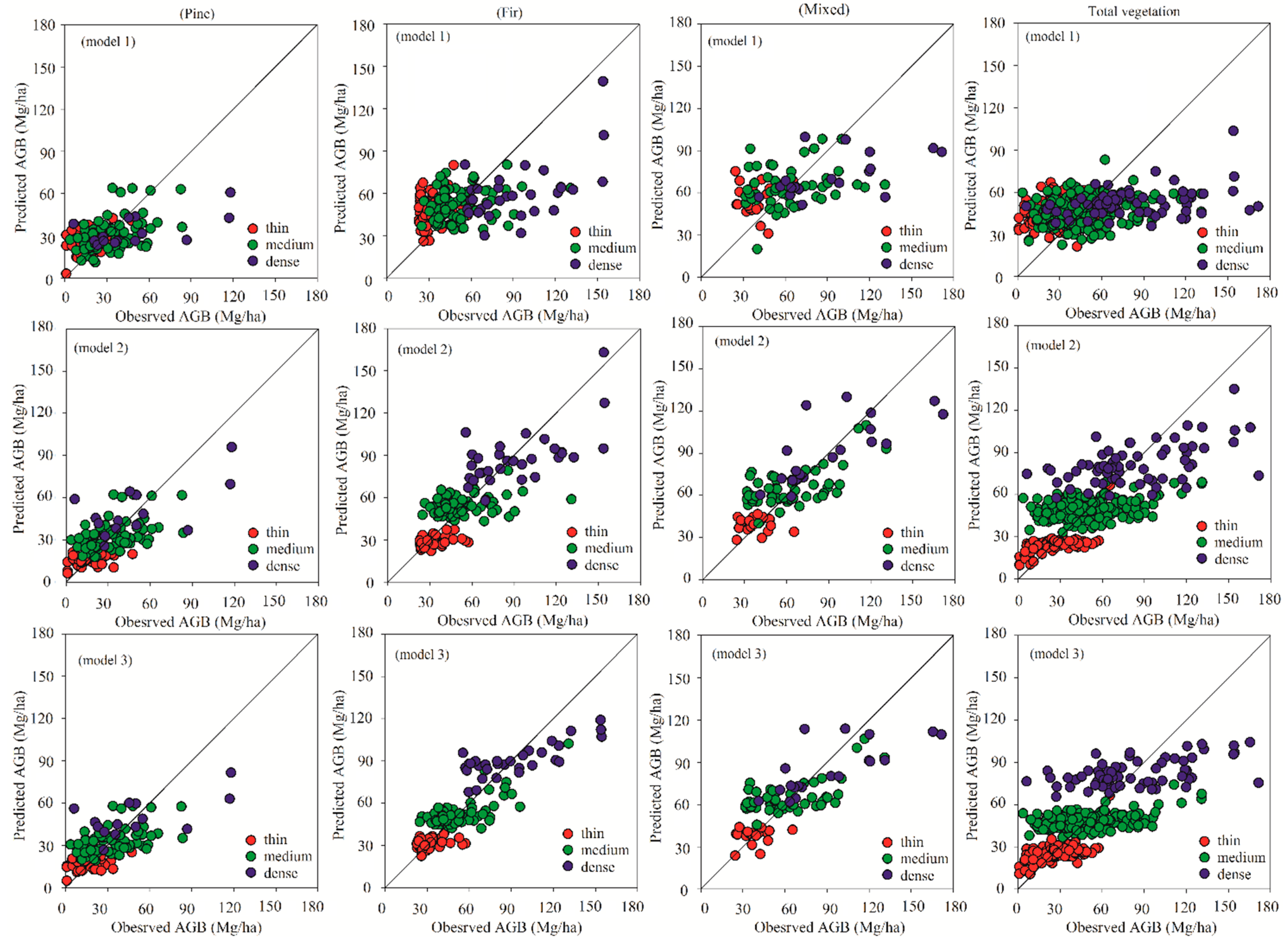
The performance of the predictions could be explained with the scatterplots showing the relationships between the predicted AGB values and observed AGB values (
Figure 3). It indicates that the overestimation and underestimation problems were obvious for the linear model (model 1) for each vegetation type. This situation, especially, became worse for all the vegetation types in thin and dense plots. For model 2 and model 3, the overestimations and underestimations in thin and dense crown density plots were alleviated for four vegetation types, and the estimates were more accurate than model 1 (
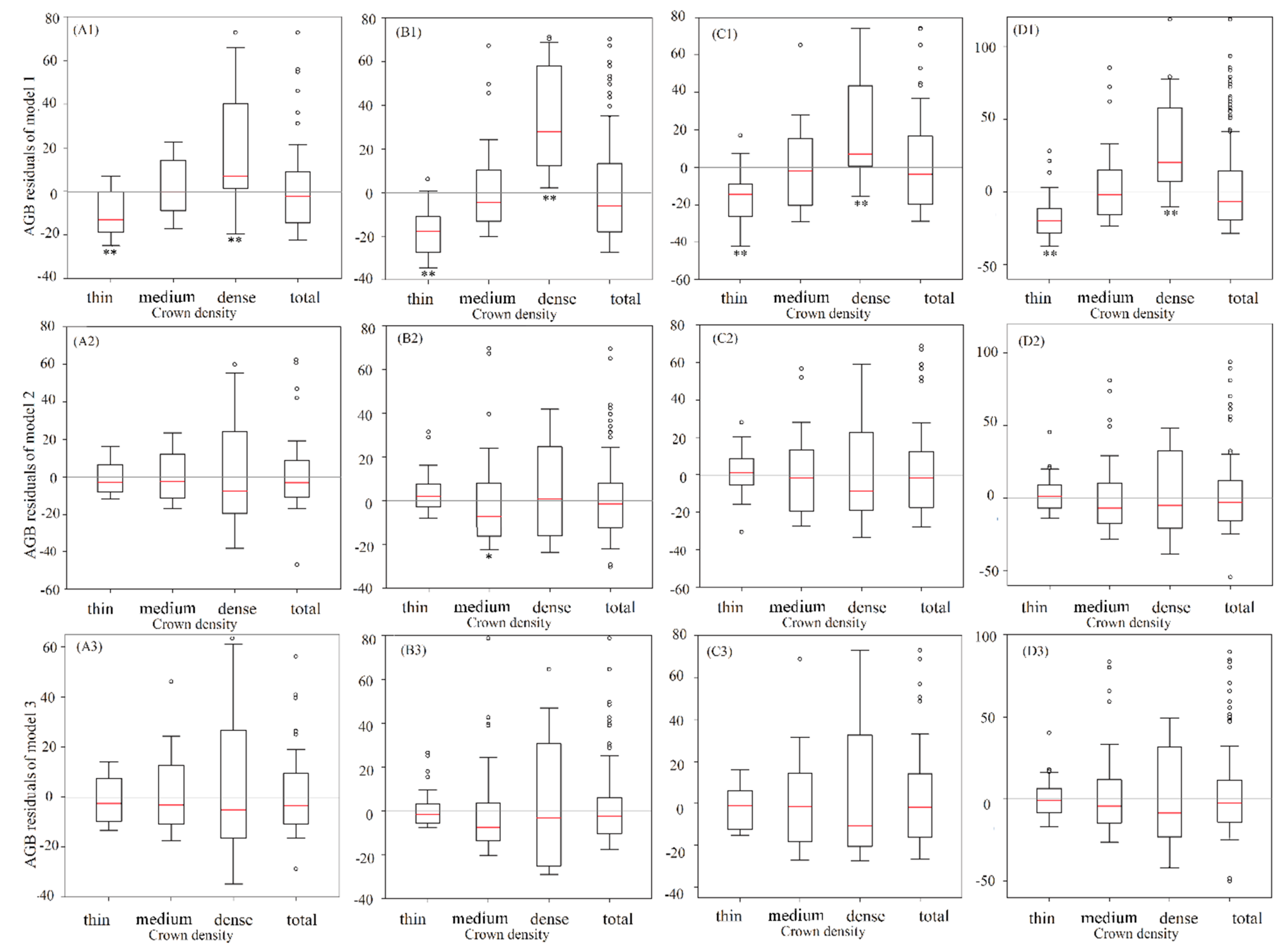
Figure 3). A single-sample
t-test was used to compare the model residuals of models 1, 2, and 3 (
Figure 4). In model 1, there were no significant differences between the residuals and 0 for the total plots and medium crown density plots for each vegetation type (
Figure 4). In the thin crown density plots, the residual values of model 1 were significantly smaller than 0, and in the dense crown density plots, the residual values of model 1 were significantly larger than 0 (
Figure 4). These results indicate that there were significant inaccuracies in the AGB estimations of the thin and dense plots of model 1 (the former was overestimated and the latter was underestimated) (
Figure 4). The residuals of model 2 were significantly different from 0 only in the thin and medium crown density plots for the fir forest, whereas the other three vegetation types exhibited no significant differences in each crown density class. The residuals of model 3 were not significantly different from 0 for all vegetation types for the different crown density classes (
Figure 4). The residual results indicate that model 2 and model 3 had higher accuracies of AGB estimation than model 1 for the different crown density classes.
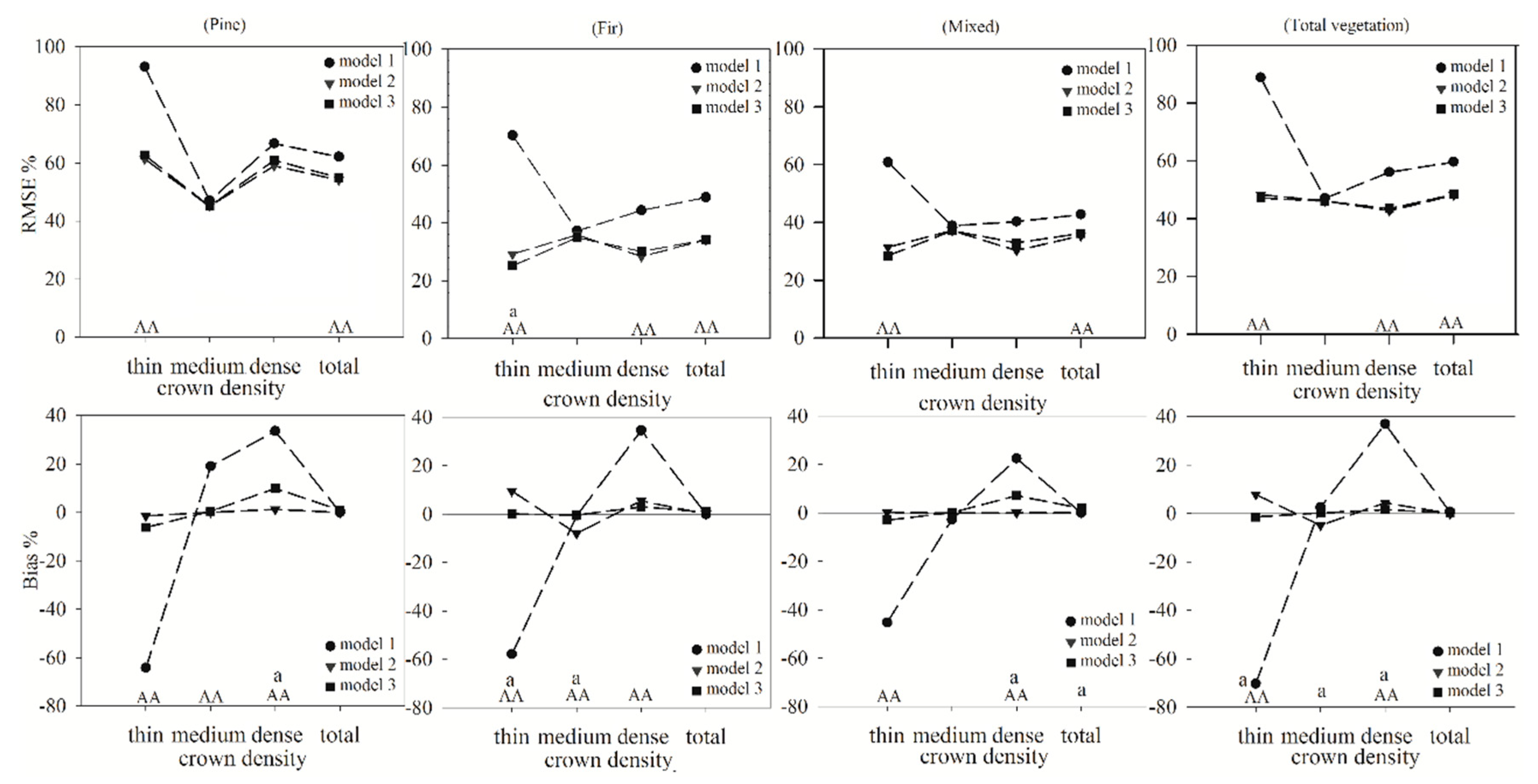
In this study, the RMSE% and Bias% of the three models of the different crown density classes were calculated for further comparison of the models (
Figure 5). Generally, the RMSE% of model 2 and model 3 were lower than those of model 1 in the total plots for all vegetation types, and the differences in the RMSE% between model 1 and model 2 and between model 1 and model 3 were all significant. For the thin crown density plots, the differences in the RMSE% exceeded 27%, and both values were significantly different from the RMSE% of model 1. For the medium crown density plots, the RMSE% of model 2 and model 3 were smaller than those of model 1, but the differences between them were not significant. For the dense crown density plots, the differences in the RMSE% exceeded 5%, and the differences between model 2 and model 1 and between model 3 and model 1 were significant for the fir forest and total vegetation. In the thin and dense plots, the values of the Bias% for model 2 and model 3 were nearer to 0 than those of model 1, and the differences between model 2 and model 1 and between model 3 and model 1 were significant, indicating that model 2 and model 3 were more accurate than model 1 in these two crown density classes. In the medium crown density plots, the trends of the Bias% between model 1 and model 2 and between model 1 and model 3 were not clear, and significant decreases only existed in model 2 and model 3 of the pine forest. The total Bias% values were not significantly different between the three models for the different vegetation types, indicating that the overall estimated values obtained from models 1, 2, and 3 were not significantly different. The differences between model 2 and model 3 for the different vegetation types were compared. The overall RMSE% and Bias% values of model 2 and model 3 were not significantly different, and model 2 was slightly better than model 3, but the performances of model 2 and model 3 were different among the thin, medium, and dense crown density classes.
4. Discussion
The choice of the independent variables is important for remote-sensing-based AGB estimation models, and potential variables from the images, such as single bands, vegetation indices, transformed images, textural information were applied because of the correlation with forest biomass. The correlation analysis results of over 300 spectral variables and the AGB of different vegetation types indicated that only 30 spectral variables simultaneously had significant correlation with AGB. This indicated that a large amount of remote sensing information does not fully reflect the forest characteristics. During the modelling process, stepwise regression was used to select the independent variables that were closely related to AGB. Although this variable selection method depended on the degree of linear correlation, the variables with low correlation coefficients may have been selected and thus affected the accuracy of the model.
Linear stepwise regression models have been widely used for AGB estimation using remote sensing [
7,
23]. In this study, the
R2 of the linear model (model 1) for the four vegetation types ranged from 0.1 to 0.3, indicating that the model had low accuracy. In addition, model 1 exhibited overestimation in the low crown density class and underestimation in the high crown density class of all vegetation types. The overestimations and underestimations of AGB were also investigated by Zhao et al., who determined that they were caused by the “global model (stepwise regression)” [
40]. In addition, overestimations and underestimations have been observed when AGB was estimated using nonparametric models such as random forest, decision tree, and K-nearest neighbor methods [
41,
42,
43]. In this study, the significant overestimations and underestimations of the linear model occurred in the thin (crown density < 0.4) and dense (crown density ≥ 0.7) plots, respectively. There were no significant overestimations or underestimations for model 2 and model 3 in the thin and dense plots. In addition, there were no significant differences between the linear dummy variable model (model 2) and linear mixed-effects model (model 3) except for the mixed forests (
Table 8). However, in comparison with the model 1, model 2 and model 3 performed significantly better, and the results of the F-test and residuals verified the significant differences. The AGB estimation results of the three models were evaluated in the crown density classes and the results showed that the overestimation in the thin plots and underestimation in the dense plots of model 1 were not observed in model 2 and model 3.
The average AGB estimates of the sample plots for the total vegetation in the “Greater Xiangxi” varied from 47.4 Mg/ha to 47.7 Mg/ha, which were very close to the referenced value (47.7 Mg/ha) of the plots measured, and the average AGB estimates of pine forest, fir forest, and mixed forest were also very close to those of the referenced values. In Hunan province, the average AGB value of pine forest in 2011 was 31.61 Mg/ha, and the average AGB value of fir forest in the forest average AGB values obtained from the sample plots of the 4th and 8th national forest inventories in 1990 and 2009 were 31.76 Mg/ha [
44] and 27.56 Mg/ha, respectively. This implied that the AGB values of forests in the “Greater Xiangxi” were larger than those of the whole Hunan mainly because the study area was a key forestry area and had various protected forests.
A comparison of the
R2adj and RMSE of the three models indicated that the performances of model 2 and model 3 were better than that of model 1. The dummy variable model considered the group differences as special fixed parameters. The purpose of using the dummy variable model in this study was to introduce the parameter of crown density class into the intercept of the model so that the degree of freedom of the error was increased and the variance of the error was decreased, thereby improving the precision of the model [
45]. The linear mixed-effects model considered the group differences as two parts: One part was the difference caused by different groups, and the other was the difference caused by random effects. Since the error and the random effect of the variance-covariance structures was considered, the model had high precision. Some studies compared dummy variable models with mixed-effects models for the estimation of large-scale forest growth models and the determination of biomass allometric growth equations. The linear mixed-effects model was a compromise between the dummy variable model and the linear model; in most cases, the dummy variable model was slightly better than the mixed-effects model, but this often depended on the sample size [
45,
46]. In this study, the sample plots were divided into the three categories of thin, medium, and dense crown density. The overall RMSE% and Bias% of model 2 were better than model 3, which supported the aforementioned results. In the past, the application of dummy variable models and mixed-effects models focused on the determination of allometric growth equations, whereas in this study, we considered whether the partition of the crown density classes improved the estimation accuracy of AGB using remote sensing data.
In statistics and biometrics, it is often debated whether the dummy variable model or mixed-effects model should be selected [
46]. The choice often depends on the number of groups (random effects/dummy variables, crown density classes in this study) and the number of samples in each group. For a small group size (less than 10), the dummy variable model is commonly preferred; otherwise, the mixed-effects model is more appropriate [
37,
47]. Unlike in most other studies, we not only compared the overall differences between the linear dummy variable model and linear mixed-effects model but also the differences in model performance among different groups. Although the overall RMSE% and Bias% were better in model 2 than in model 3, this trend was not always the same for the different crown density classes. In the fir forest and the total vegetation, groups that had a large number of samples, the RMSE% and Bias% were smaller for model 3 than model 2 for all crown density classes. In pine and mixed forests, which had a small number of samples in each group, the RMSE% and Bias% were smaller for model 2 than model 3 for all crown density classes. Therefore, regardless of which of the models was chosen, we believe that if the overall differences between the two models are not significant, the fitting effects of the groups should be compared and the model with good performance in each group should be selected.
The climate of this region is a typical subtropical monsoon humid climate, and the typical forests are evergreen broad leaf forests and evergreen coniferous forests. In this study, the mixed forests were almost evergreen coniferous forests, and the seasonal variation of the vegetation types were not obvious. Many studies analyzed the variation of different vegetation types (NDVI) in the subtropical regions of China. They demonstrated that the NDVI of evergreen forests (evergreen broad leaf forest and evergreen coniferous forest) had no obviously seasonal variation [
48]. Besides, the seasonal variations of leaf area index (LAI) and clumping index (CI) were very small because the canopy structure of evergreen forests were stable through the year [
49,
50], and texture information which referred the forest structure were relatively stable in the imagery. The spectral characteristics of remote sensing images are influenced by the soil, topography, vegetation type, forest structure, and other factors. It is important to choose appropriate spectral variables as independent variables in AGB estimation using remote-sensing-based methods [
5,
51]. Many studies have shown that when only spectral indices were used in AGB estimation, saturation occurred and caused inaccuracies of AGB estimation. Texture information calculated from a small neighborhood of pixels [
26] may have a stronger correlation with AGB than spectral indices, and in some regions, AGB may only be closely correlated with texture information rather than spectral information. Texture information has been demonstrated to be an important factor in remote-sensing-based AGB estimation [
52,
53].
The independent variables of the linear model in this study illustrated that texture information had considerable influences on the accuracy of the AGB estimation in our study area. The linear models had low accuracy for the thin and dense crown density classes, and the linear dummy variable models and linear mixed-effects models had higher accuracy because the crown density classes were considered. The results indicate that the crown density class may be an important factor affecting the accuracy of AGB estimation. The sensitivity to the stand information decreased with increasing crown density in the dense stands; the spectral information may be affected by other non-forest characteristics in thin stands with low AGB, causing the low accuracy of the AGB estimation model. Many studies have demonstrated that a complex stand structure and high crown density caused saturation in remote sensing images and low crown density and sparse trees increased the occurrence of soil/vegetation mixed pixels [
6,
54,
55]. The saturation and mixed pixels problems have attracted increased attention for remote-sensing-based AGB estimation. In this study, we demonstrated that the crown density classes influenced the accuracy of AGB estimation; however, the underlying mechanisms and relationships should be studied in more detail in the future.
In this study, the models for AGB estimation were explored combining sample plot data and remote sensing, and the results illustrated that the crown density was a factor that influences model accuracy. The crown density data incorporated in the linear dummy variable model and linear mixed-effects model were the most accurate. The aim of this study was to demonstrate that the crown density is an important factor that influences the accuracy of the models. A large amount of research has explored the potential of using satellite imagery for exploring remote-sensing-based methods of crown density, and there are more precise results [
56]. This should be examined in future research for mapping large-scale AGB using our models when the crown density data were not available.