Genome-Wide Identification and Characterization of Heat-Shock Transcription Factors in Rubber Tree
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. Stress Treatments
2.3. Real-Time Quantitative PCR Analysis
2.4. Bioinformatics Analysis
2.5. Subcellular-Localization Analysis
2.6. Predicted Protein–Protein Interaction-Network Construction
2.7. Statistical Analysis
3. Results
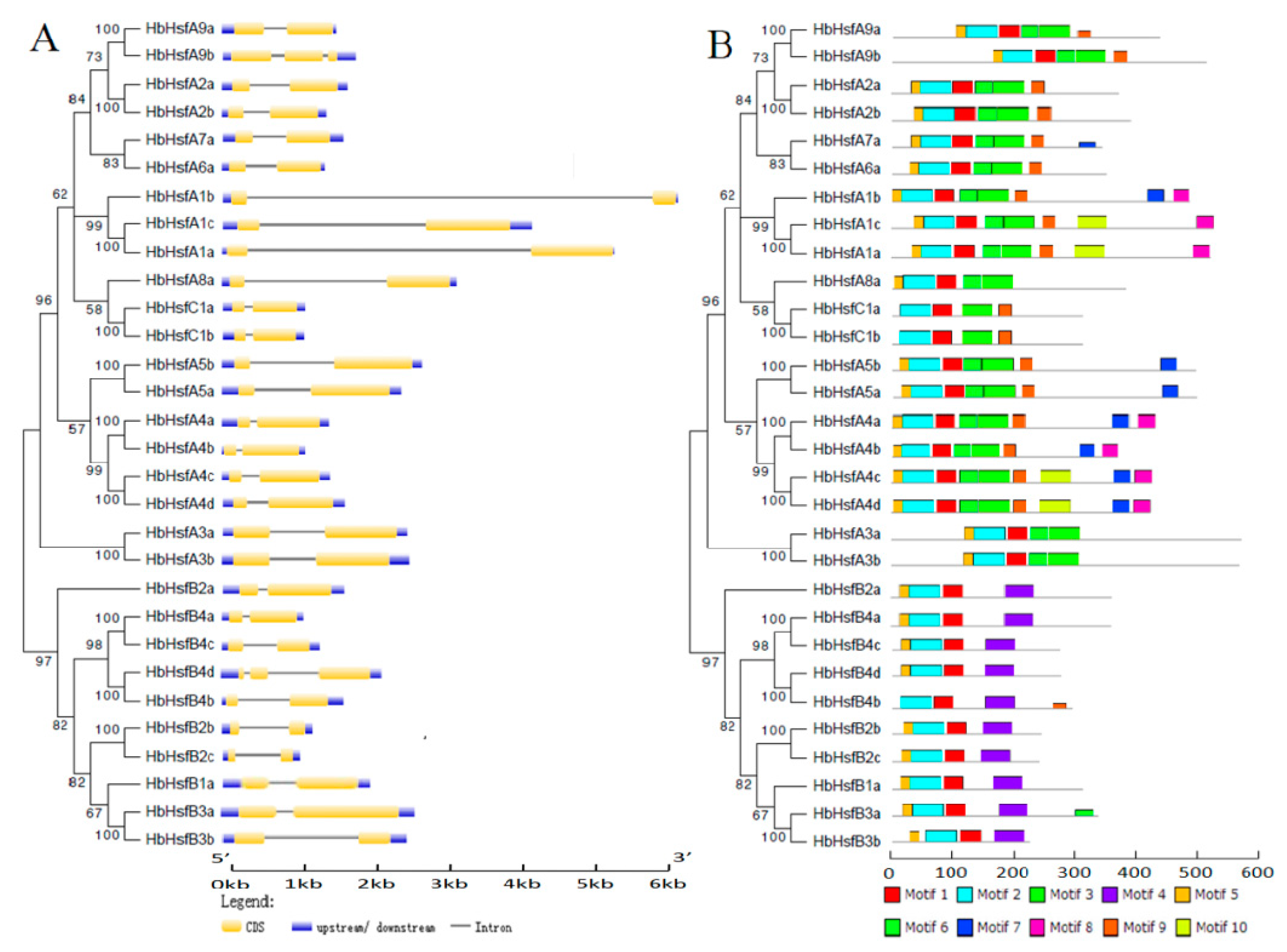
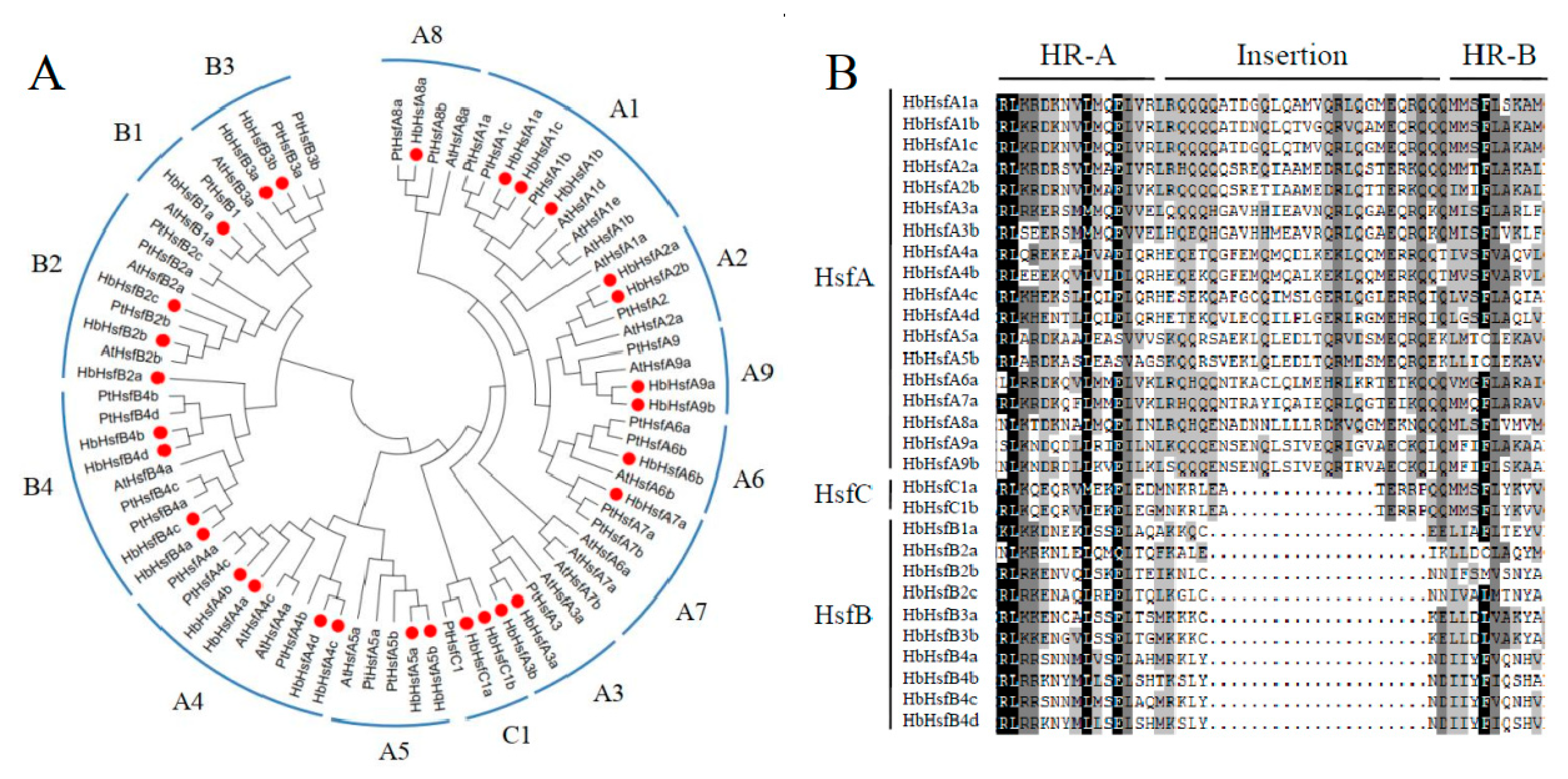
3.1. Isolation and Identification of Hsf Genes in Rubber Tree
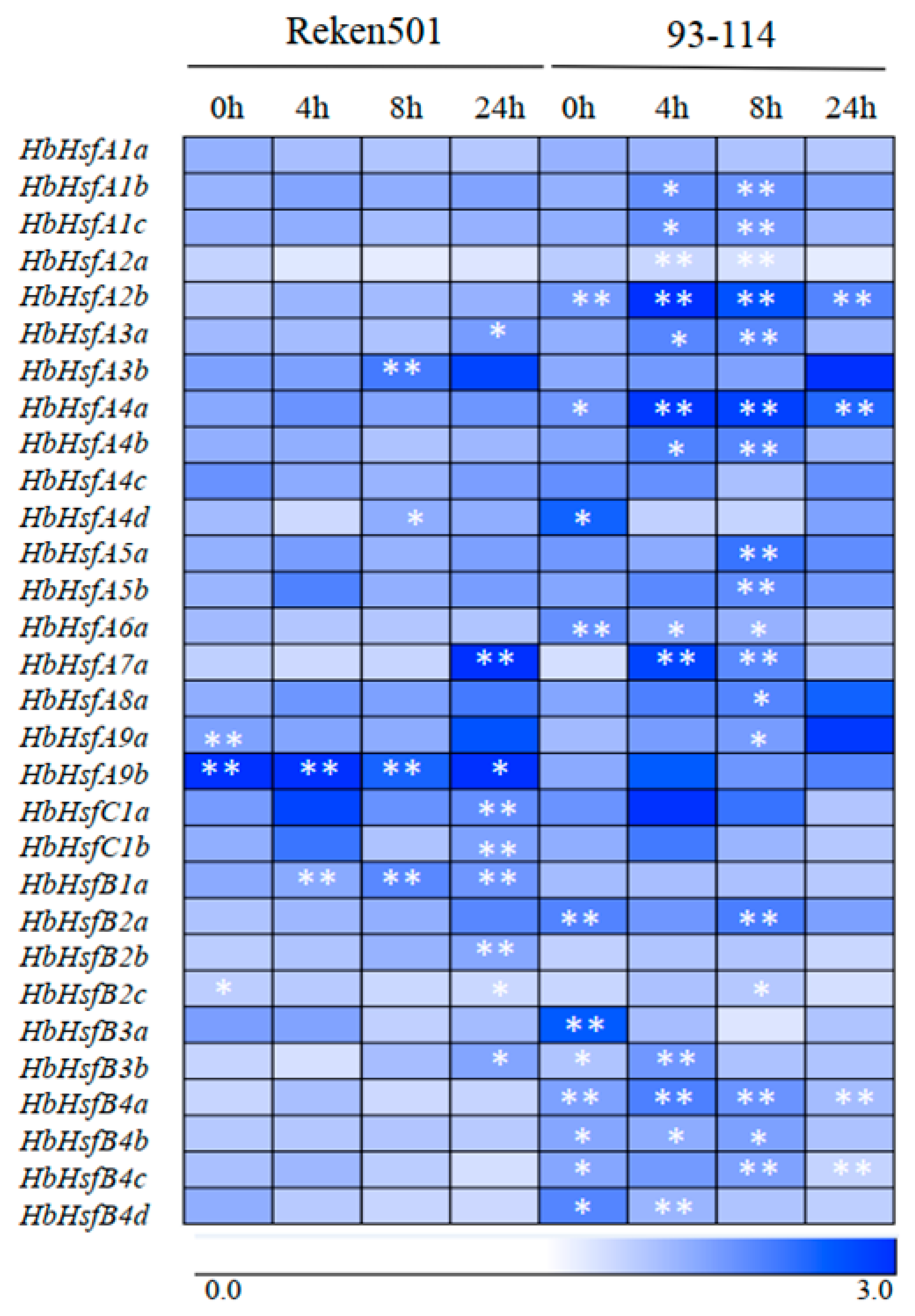
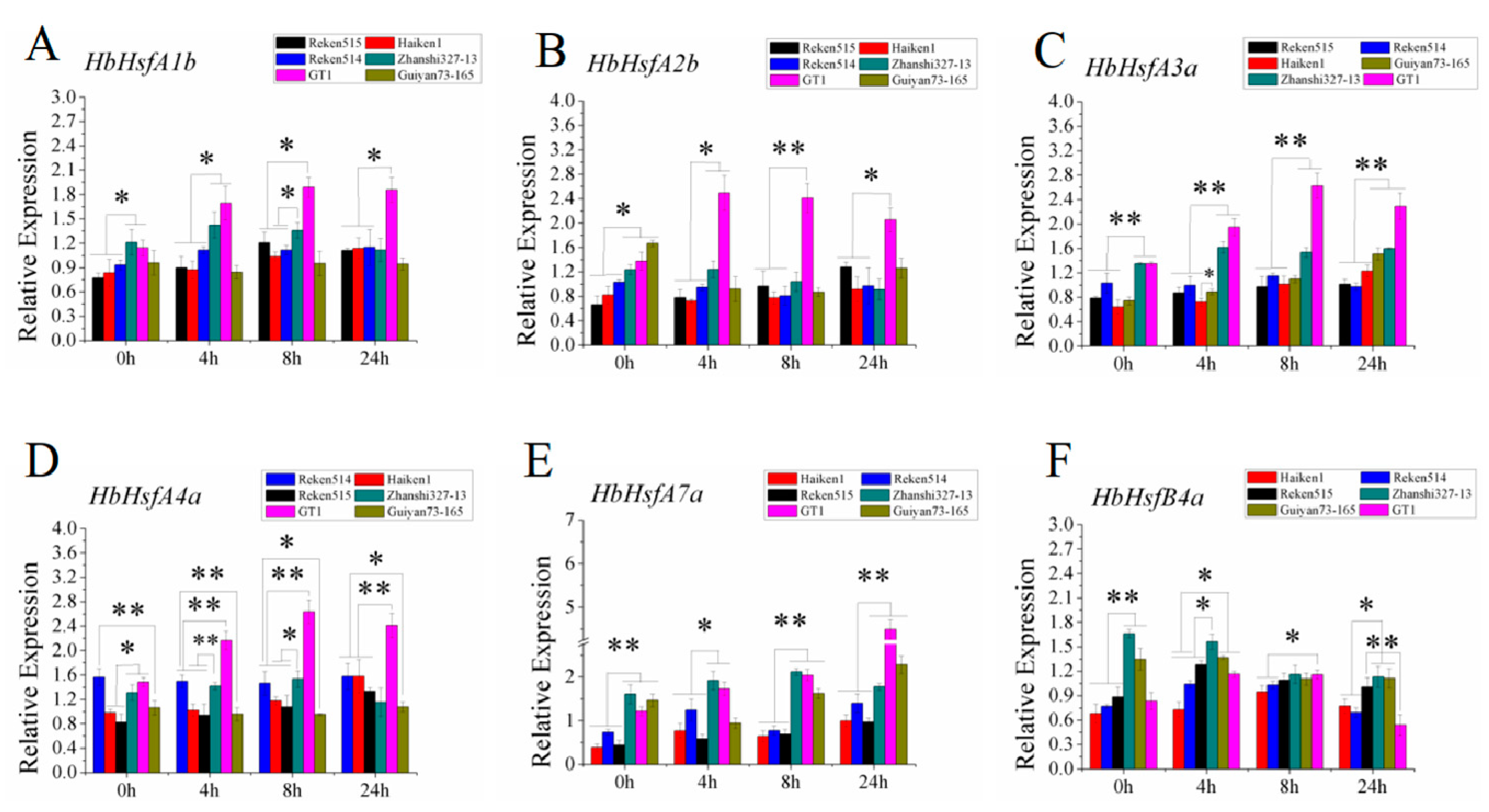
3.2. Expression Patterns of Hbhsfs in Cold-Tolerant and Cold-Sensitive Rubber-Tree Clones in Response to Cold Stress
3.3. Subcellular Localization and Predicted Protein–Protein Interactions of HbHsfA2b, HbHsfA3a, and HbHsfA7a
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. BBA Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Von Koskull-Döring, P.; Scharf, K.D.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Wiederrecht, G.; Seto, D.; Parker, C.S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 1988, 54, 841–853. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperon 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Chauhan, H.; Khurana, N.; Agarwal, P.; Khurana, P. Heat shock factors in rice (Oryza sativa L.): Genome-wide expression analysis during reproductive development and abiotic stress. Mol. Genet. Genom. 2011, 286, 171–187. [Google Scholar] [CrossRef]
- Mittal, D.; Chakrabarti, S.; Sarkar, A.; Singh, A.; Grover, A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 2009, 47, 785–795. [Google Scholar] [CrossRef]
- Giorno, F.; Guerriero, G.; Baric, S.; Mariani, C. Heat shock transcriptional factors in Malus domestica: Identification, classification and expression analysis. BMC Genom. 2012, 13, 639. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.; Kim, K.M.; Lee, J.H. Genome-wide analysis and molecular characterization of heat shock transcription factor family in Glycine max. J. Genet. Genom. 2013, 40, 127–135. [Google Scholar] [CrossRef]
- Xue, G.P.; Sadat, S.; Drenth, J.; McIntyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef]
- Huang, X.Y.; Tao, P.; Li, B.Y.; Wang, W.H.; Yue, Z.C.; Lei, J.L.; Zhong, X.M. Genome-wide identification, classification, and analysis of heat shock transcription factor family in Chinese cabbage (Brassica rapa pekinensis). Genet. Mol. Res. 2015, 14, 2189–2204. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Han, Y.T.; Wei, W.; Li, Y.J.; Zhang, K.; Gao, Y.R.; Zhao, F.L.; Feng, J.Y. Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragariavesca. Front. Plant Sci. 2015, 6, 736. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Hu, W.; Xia, F.; Zeng, H.; Li, X.; Yan, Y.; He, C.; Shi, H. Heat shock transcription factors in banana: Genome-wide characterization and expression profile analysis during development and stress response. Sci. Rep. 2016, 6, 36864. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Huang, C.; Zhang, L.; Liu, H.; Yu, J.; Hu, Z.; Hua, W. Systematic analysis of Hsf family genes in the Brassica napus genome reveals novel responses to heat, drought and high CO2 stresses. Front. Plant Sci. 2017, 8, 1174. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Yang, J.; Guo, C.; Bao, M.; Zhang, J. Genome-wide identification and classification of the Hsf and sHsp gene families in Prunus mume, and transcriptional analysis under heat stress. Peer J. 2019, 7, e7312. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Wang, J.; Li, Y.; Wu, S.; Yang, S.; Chao, J.; Chen, Y.; Zhang, S.; Shi, M.; Tian, W. Comparative transcriptome analysis reveals phytohormone signalings, heat shock module and ROS scavenger mediate the cold-tolerance of rubber tree. Sci. Rep. 2018, 8, 4931. [Google Scholar] [CrossRef]
- Deng, D.; Wang, J.X.; Wang, J.; Tian, W.M. Two HbHsfA1 and HbHsfB1 genes from the tropical woody plant rubber tree confer cold stress tolerance in Saccharomyces cerevisiae. Braz. J. Bot. 2018, 41, 711–724. [Google Scholar] [CrossRef]
- Lau, N.S.; Makita, Y.; Kawashima, M.; Taylor, T.D.; Kondo, S.; Othman, A.S.; Shu-Chien, A.C.; Matsui, M. The rubber tree genome shows expansion of gene family associated with rubber biosynthesis. Sci. Rep. 2016, 6, 28594. [Google Scholar] [CrossRef]
- Pootakham, W.; Sonthirod, C.; Naktang, C.; Ruang-Areerate, P.; Yoocha, T.; Sangsrakru, D.; Theerawattanasuk, K.; Rattanawong, R.; Lekawipat, N.; Tangphatsornruang, S. De novo hybrid assembly of the rubber tree genome reveals evidence of paleotetraploidy in Hevea species. Sci. Rep. 2017, 7, 41457. [Google Scholar] [CrossRef]
- Rahman, A.Y.; Usharraj, A.O.; Misra, B.B.; Thottathil, G.P.; Jayasekaran, K.; Feng, Y.; Hou, S.; Ong, S.Y.; Ng, F.L.; Lee, L.S.; et al. Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genom. 2013, 14, 75. [Google Scholar] [CrossRef]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qin, Y.; Xiao, X.; Tang, C. Screening of valid reference genes for real-time RT-PCR data normalization in Hevea brasiliensis and expression validation of a sucrose transporter gene HbSUT3. Plant Sci. 2011, 181, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Huang, X.; Wu, M.; Wang, Y.; Chang, Y.; Liu, K.; Zhang, J.; Zhang, Y.; Zhang, F.; Yi, L.; et al. A rapid, highly efficient and economical method of Agrobacterium-mediated in planta transient transformation in living onion epidermis. PLoS ONE 2014, 9, e83556. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Liu, G.; Chang, Y.; He, C.; Shi, H. Heat shock transcription factor 3 regulates plant immune response through modulation of salicylic acid accumulation and signaling in cassava. Mol. Plant Pathol. 2018, 19, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, N.; Deng, T.; Zhang, L.; Zuo, K. Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genom. 2014, 15, 961. [Google Scholar] [CrossRef]
- Wang, P.; Song, H.; Li, C.; Li, P.; Li, A.; Guan, H.; Hou, L.; Wang, X. Genome-wide dissection of the heat shock transcription factor family genes in Arachis. Front. Plant Sci. 2017, 8, 106. [Google Scholar] [CrossRef]
- Zou, Z.; Yang, L.; Gong, J.; Mo, Y.; Wang, J.; Cao, J.; An, F.; Xie, G. Genome-wide identification of Jatropha curcas aquaporin genes and the comparative analysis provides insights into the gene family expansion and evolution in Hevea brasiliensis. Front. Plant Sci. 2016, 7, 395. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Li, J.; Zhang, L.; Wang, Y.; Zheng, H.; Lu, M.; Chen, J. Hsf and Hsp gene families in Populus: Genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015, 16, 181. [Google Scholar] [CrossRef]
- Yu, X.Y.; Yao, Y.; Hong, Y.H.; Hou, P.Y.; Li, C.X.; Xia, Z.Q.; Geng, M.T.; Chen, Y.H. Differential expression of the Hsf family in cassava under biotic and abiotic stresses. Genome 2019, 62, 563–569. [Google Scholar] [CrossRef]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; Von Koskull-Döring, P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Mittler, R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. 2006, 98, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, W.; Yang, Z.; Liu, J.; Huang, B. Transcriptional regulation of heat shock proteins and ascorbate peroxidase by CtHsfA2b from African bermudagrass conferring heat tolerance in Arabidopsis. Sci. Rep. 2016, 6, 28021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Y.; Xing, D.; Gao, C. Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. J. Exp. Bot. 2009, 60, 2073–2091. [Google Scholar] [CrossRef]
- Wang, J.X.; Li, Y.; Tian, W.M. Physiological responses of two rubber tree clones with differential cold-tolerant potential to cold stress. J. Rubber Res. 2016, 20, 117–129. [Google Scholar] [CrossRef]
- Usman, M.G.; Rafii, M.Y.; Martini, M.Y.; Yusuff, O.A.; Ismail, M.R.; Miah, G. Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnol. Genet. Eng. Rev. 2017, 33, 26–39. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotech. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Meiri, D.; Breiman, A. Arabidopsis ROF1(FKBP62) modulates thermotolerance by interaction with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009, 59, 387–399. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yu, W.; Chen, Y.; Yang, S.; Wu, S.; Chao, J.; Wang, X.; Tian, W.-M. Genome-Wide Identification and Characterization of Heat-Shock Transcription Factors in Rubber Tree. Forests 2019, 10, 1157. https://doi.org/10.3390/f10121157
Li Y, Yu W, Chen Y, Yang S, Wu S, Chao J, Wang X, Tian W-M. Genome-Wide Identification and Characterization of Heat-Shock Transcription Factors in Rubber Tree. Forests. 2019; 10(12):1157. https://doi.org/10.3390/f10121157
Chicago/Turabian StyleLi, Yan, Wencai Yu, Yueyi Chen, Shuguang Yang, Shaohua Wu, Jinquan Chao, Xinlong Wang, and Wei-Min Tian. 2019. "Genome-Wide Identification and Characterization of Heat-Shock Transcription Factors in Rubber Tree" Forests 10, no. 12: 1157. https://doi.org/10.3390/f10121157
APA StyleLi, Y., Yu, W., Chen, Y., Yang, S., Wu, S., Chao, J., Wang, X., & Tian, W.-M. (2019). Genome-Wide Identification and Characterization of Heat-Shock Transcription Factors in Rubber Tree. Forests, 10(12), 1157. https://doi.org/10.3390/f10121157





