Diversity and Functional Potential of Soil Bacterial Communities in Different Types of Farmland Shelterbelts in Mid-Western Heilongjiang, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Plot Characteristics and Sample Collection
2.2. DNA Extraction and PCR Amplification
2.3. Library Construction and Illumina Miseq Sequencing
2.4. Data Processing
3. Results and Analysis
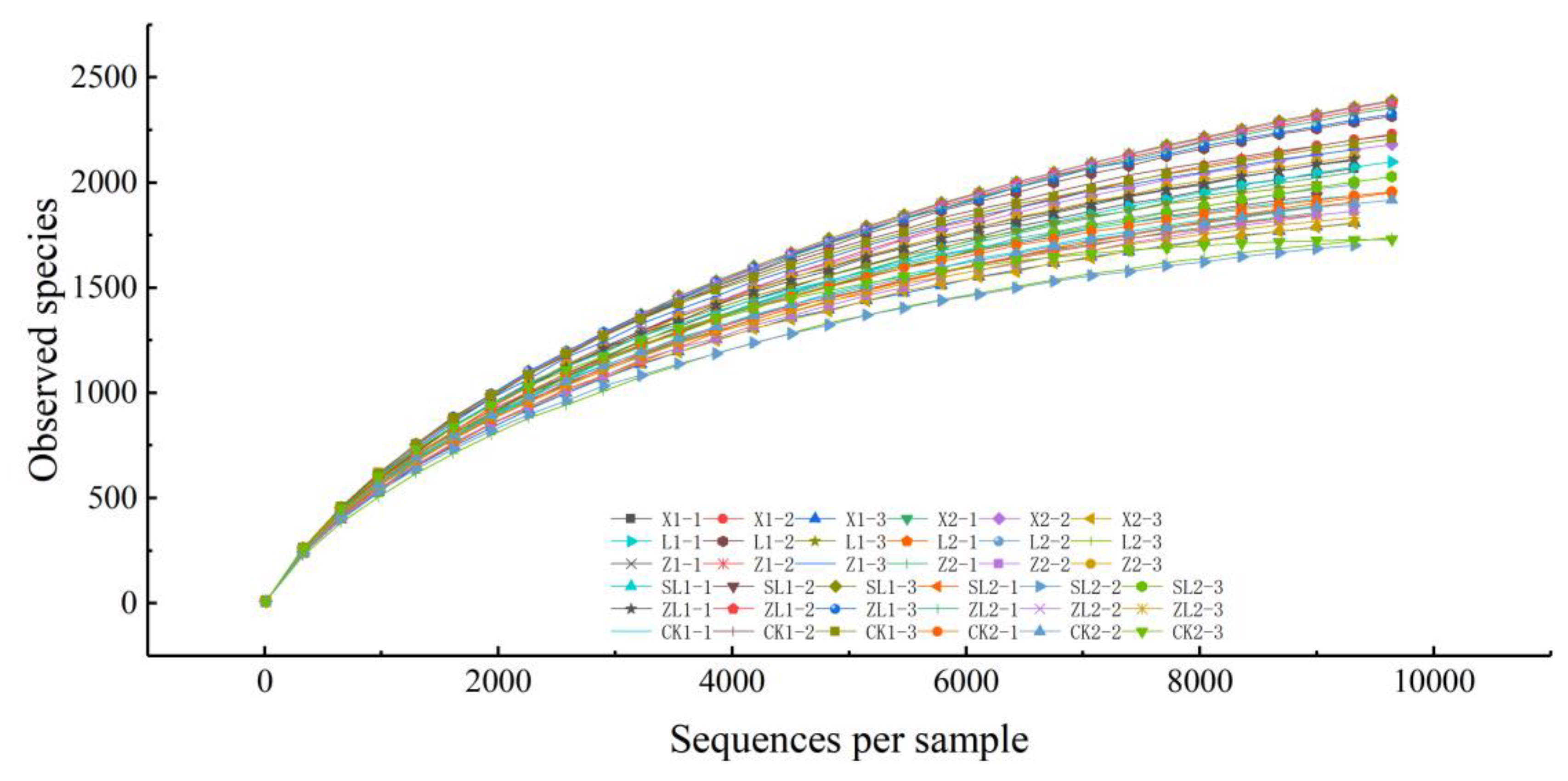
3.1. Sequencing Effectiveness and OTU Statistics for Study Samples
3.2. Bacterial OTU Alpha Diversity
3.3. Bacterial Community Composition
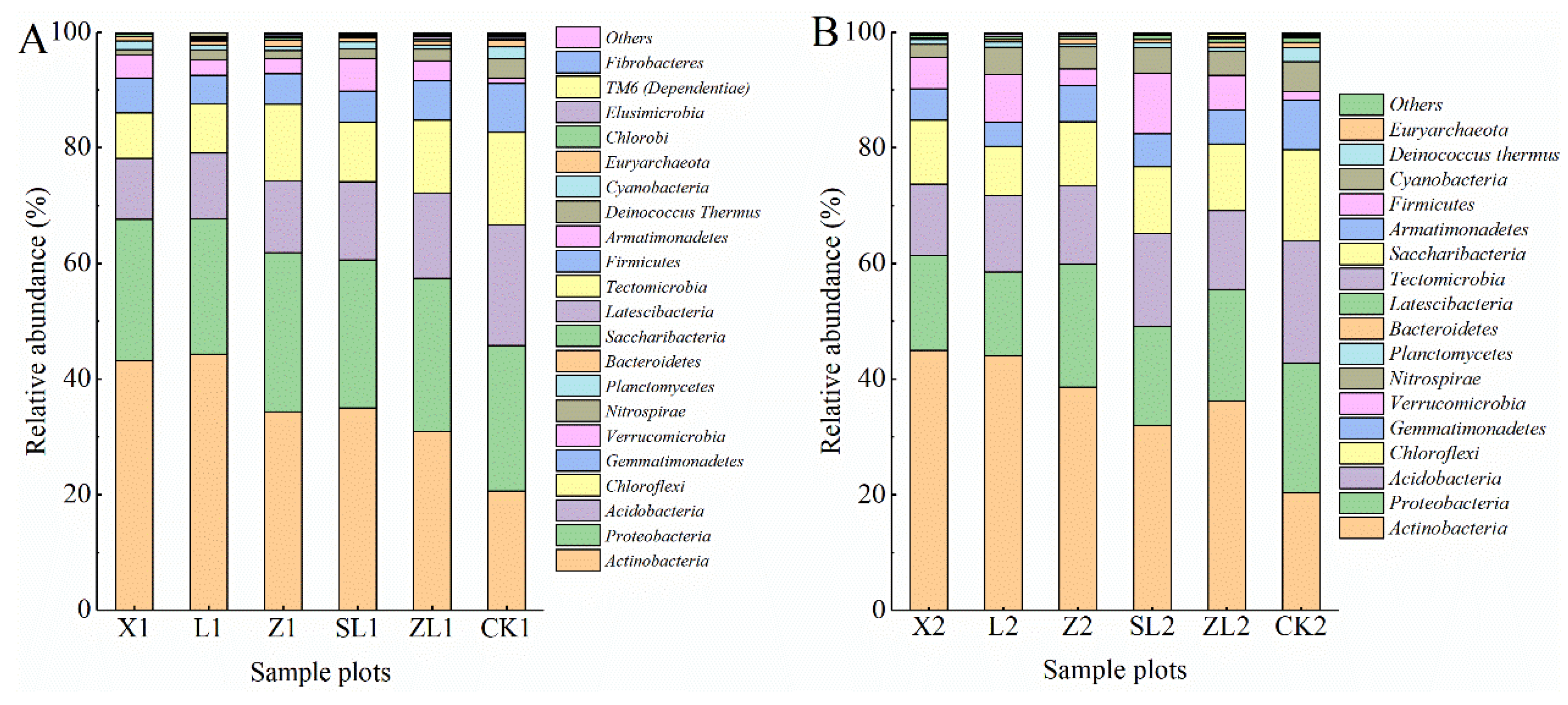
3.3.1. Composition of Soil Bacterial Communities at Each Taxonomic Level
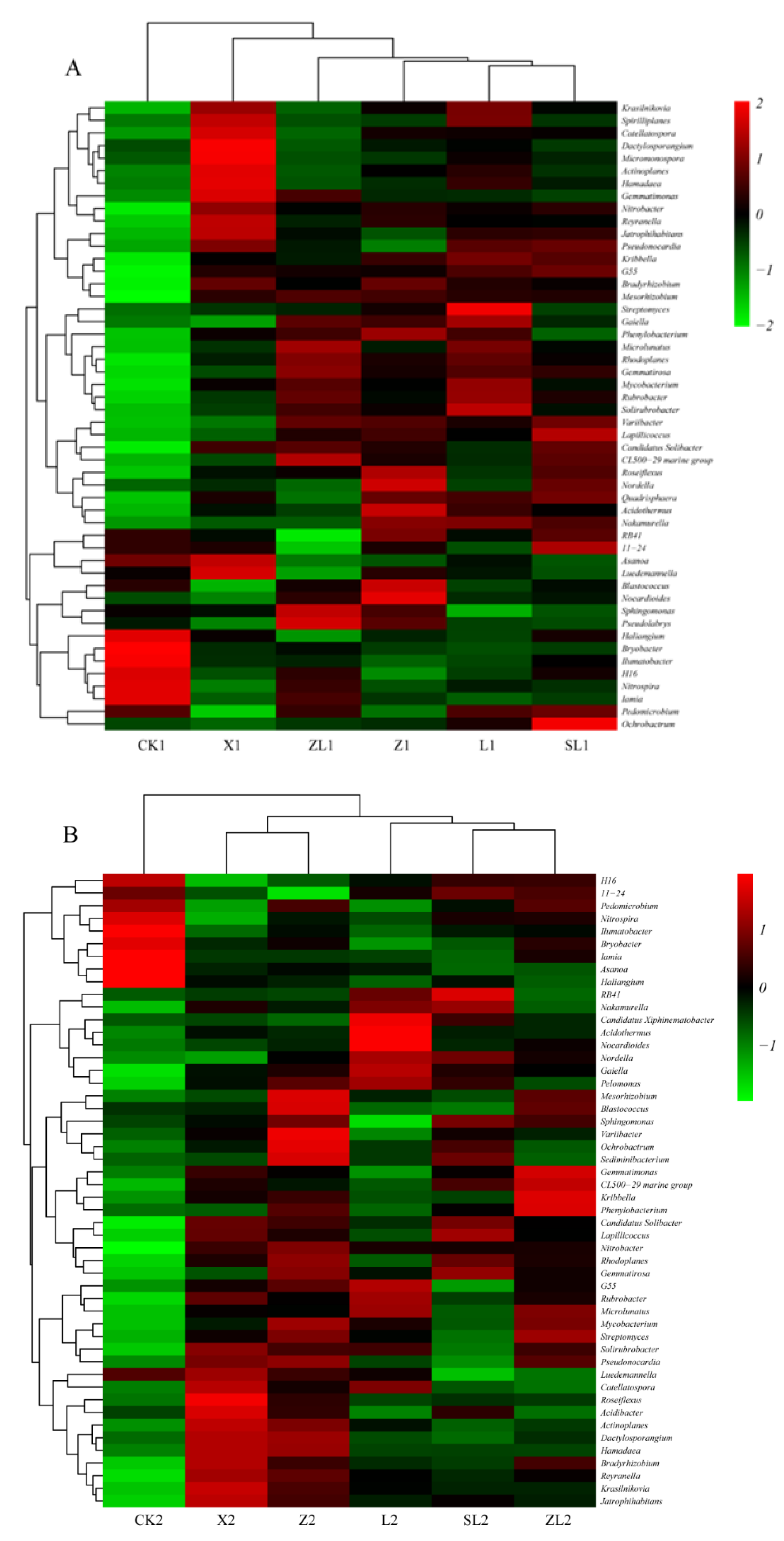
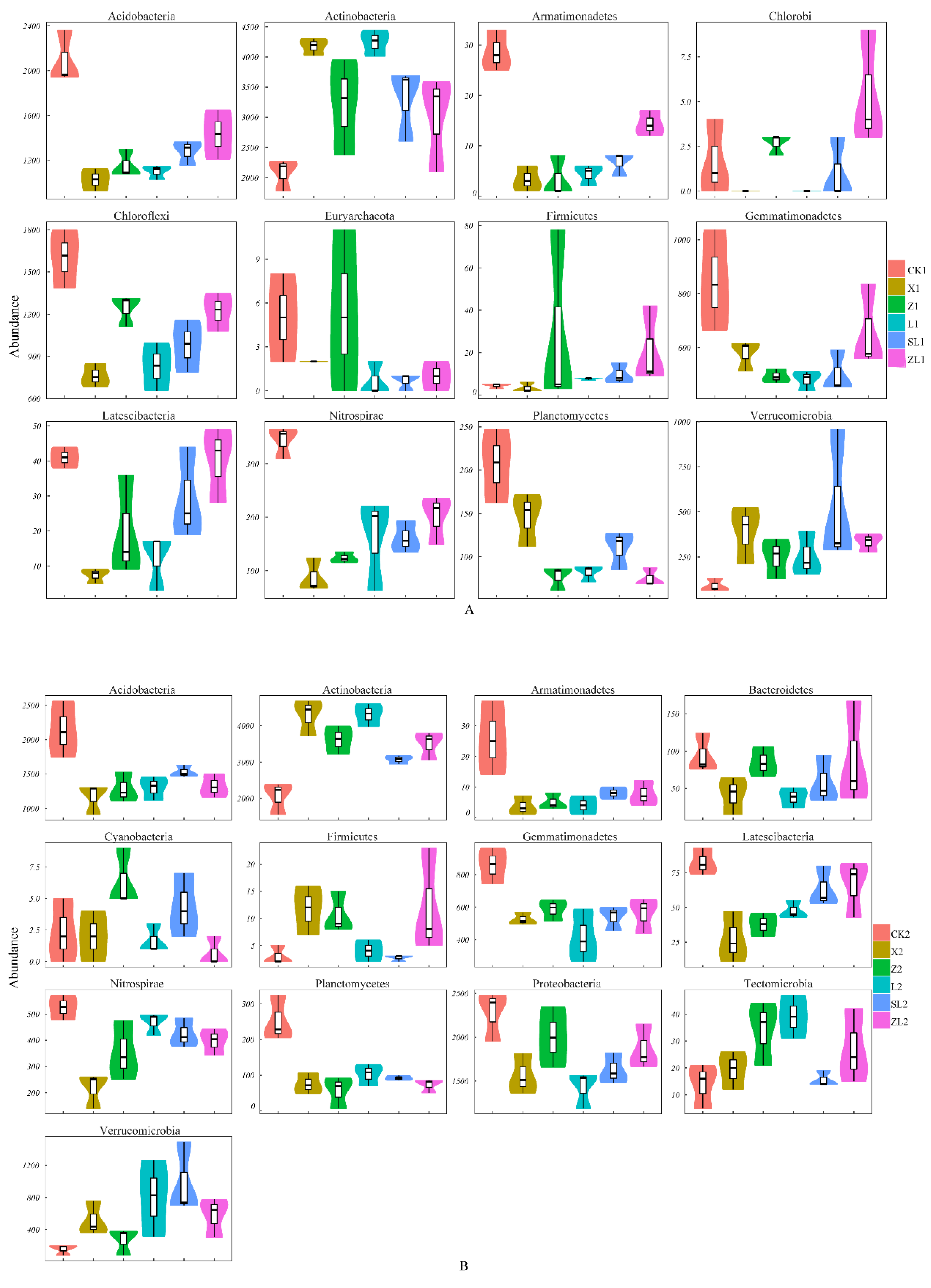
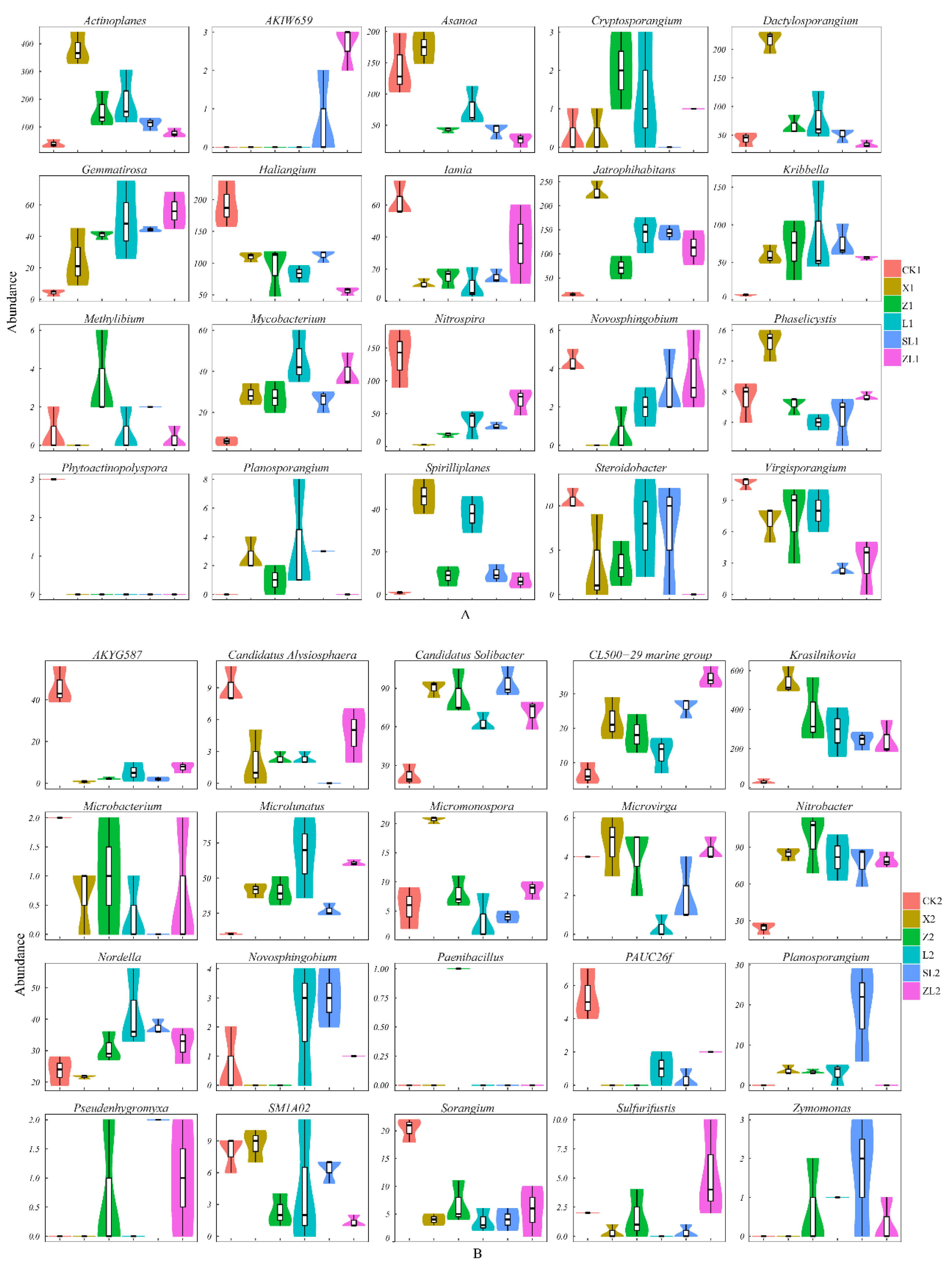
3.3.2. Differences in Taxonomic Composition Among Study Sites
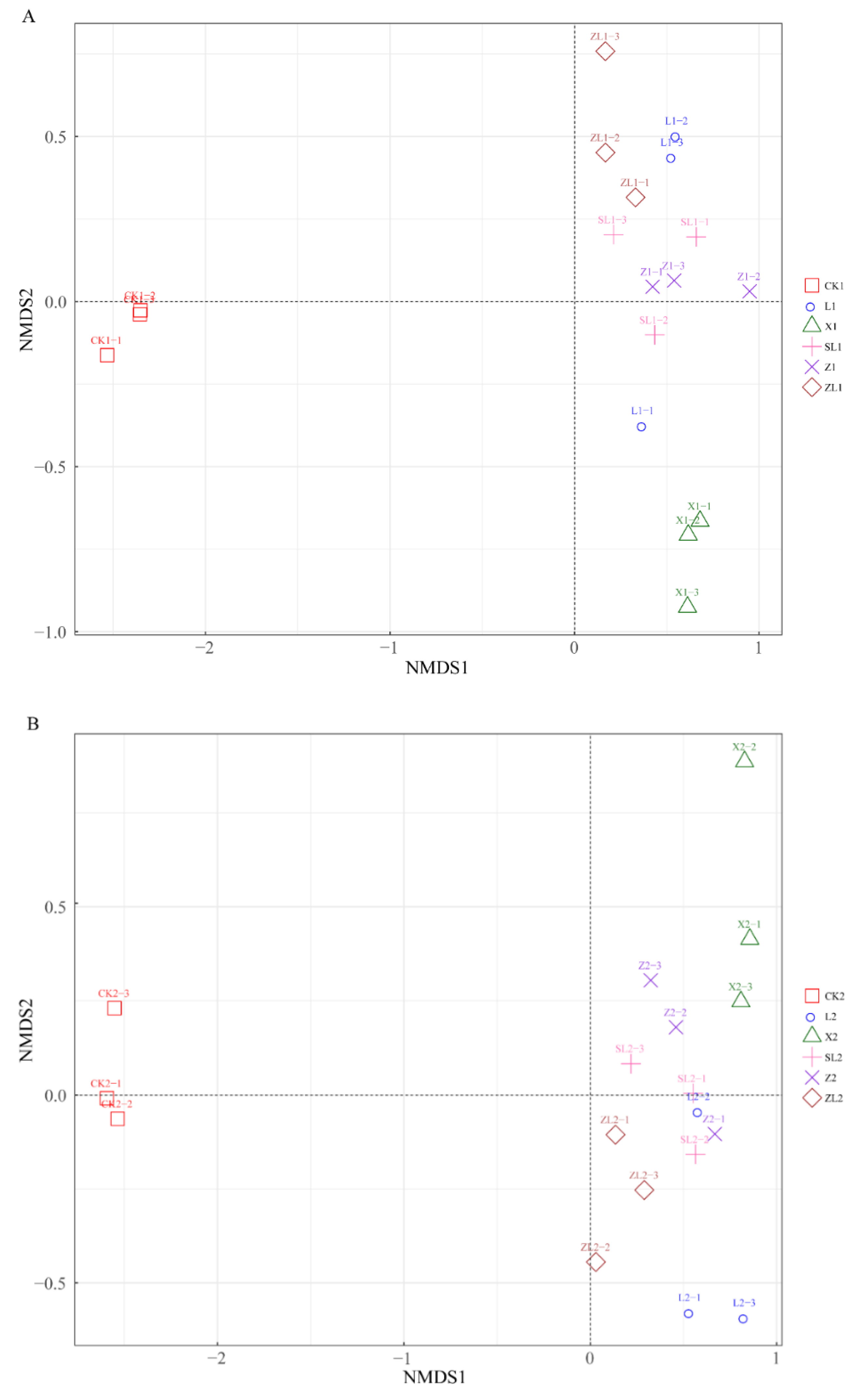
3.4. Bacterial OTU Beta Diversity
3.5. Network Analysis of Associations Among Genera
3.6. Microbial Community Function
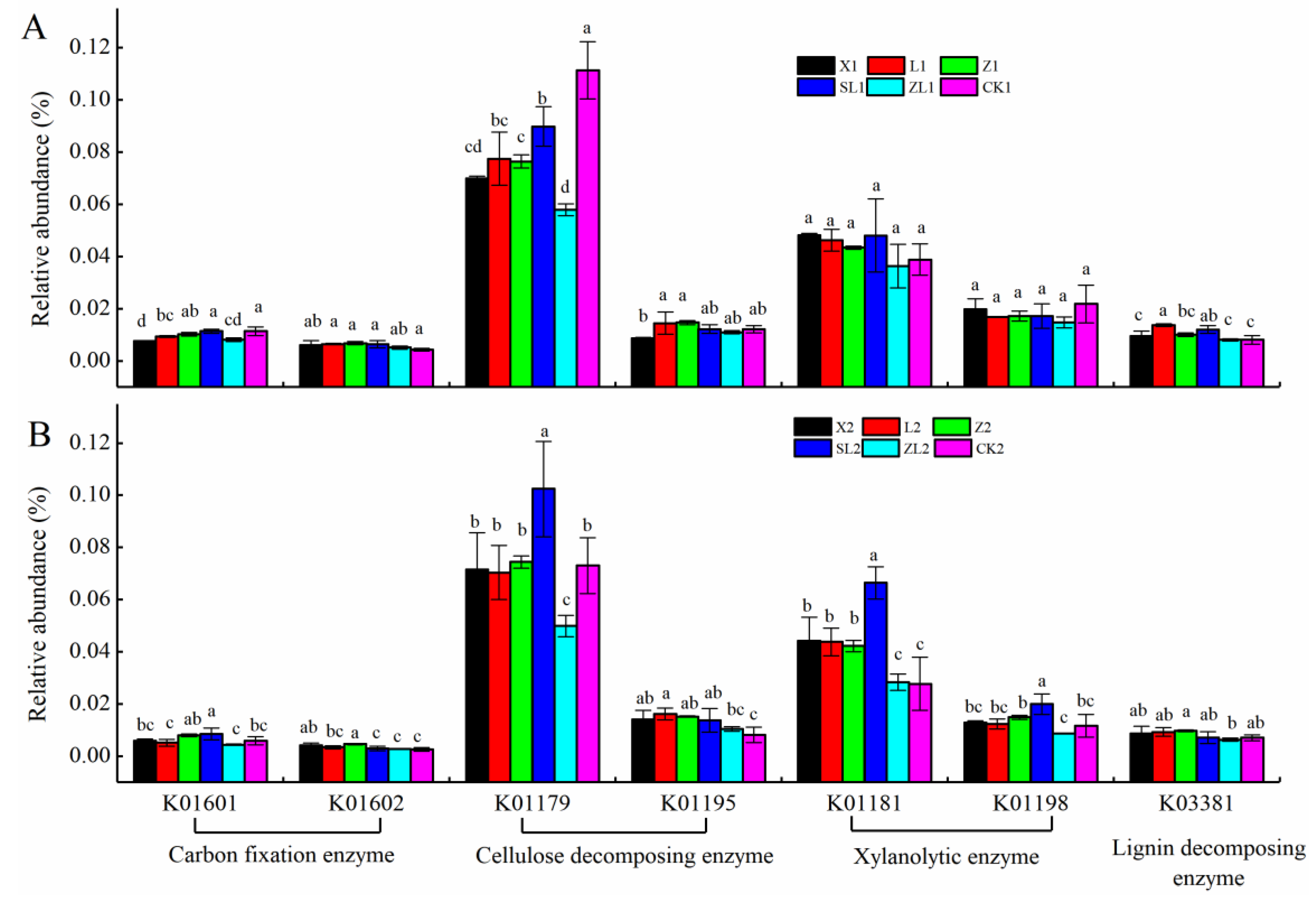
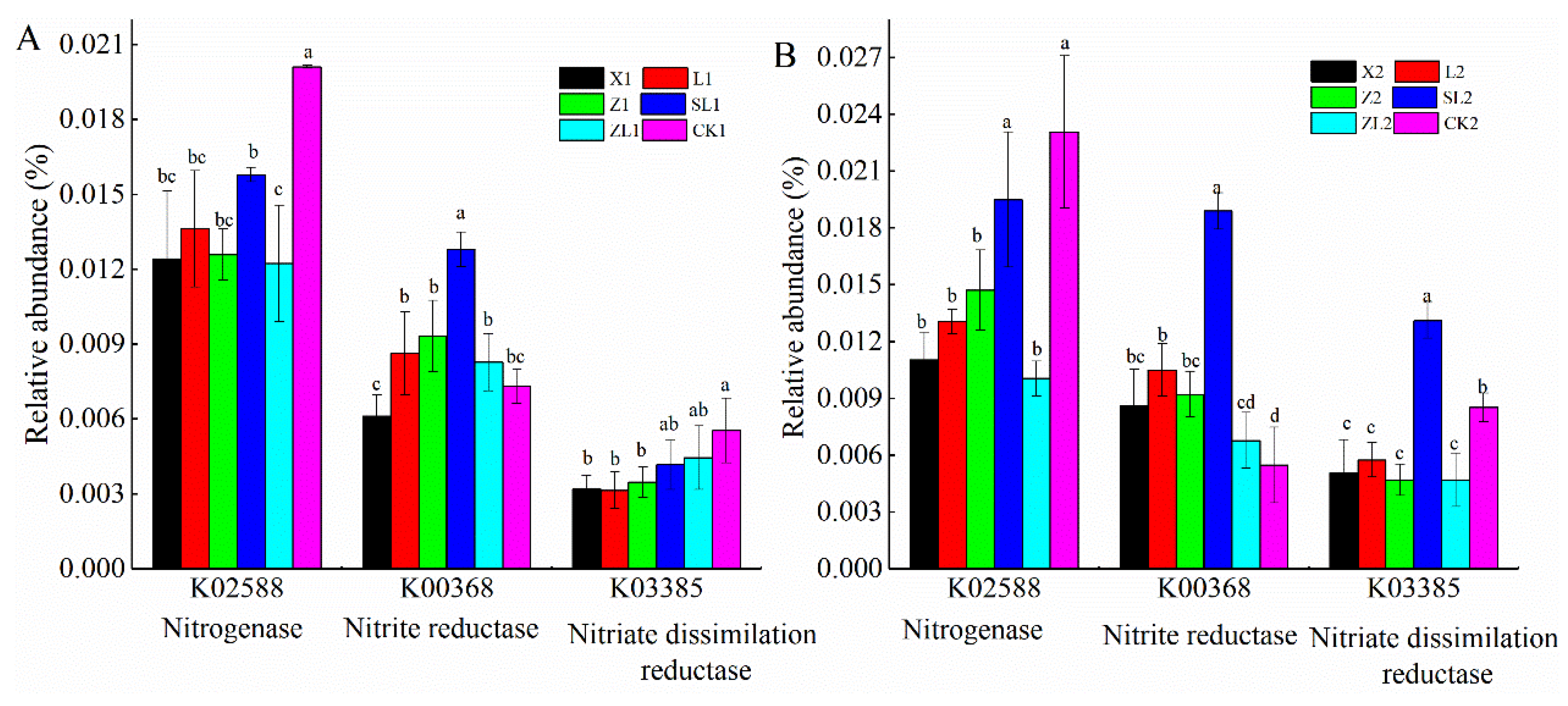
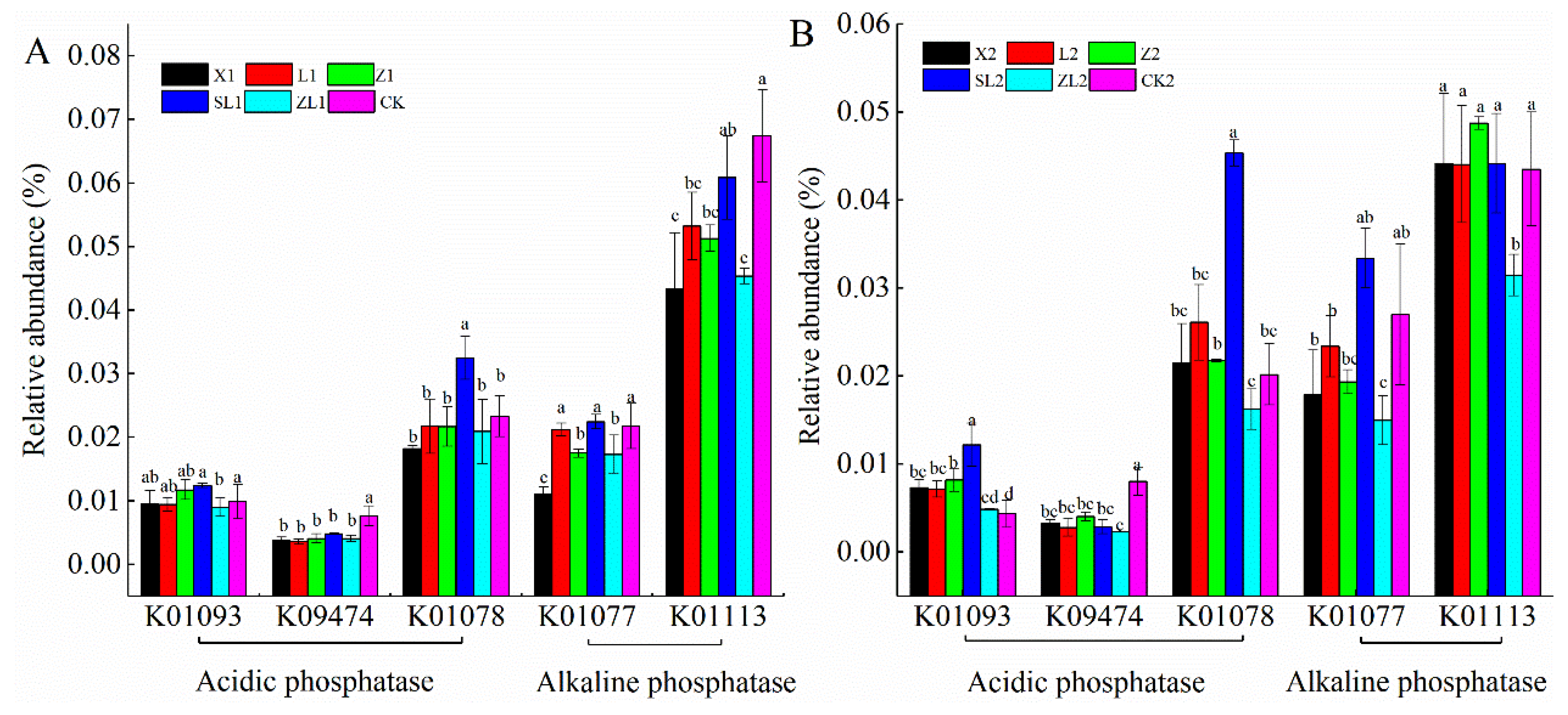
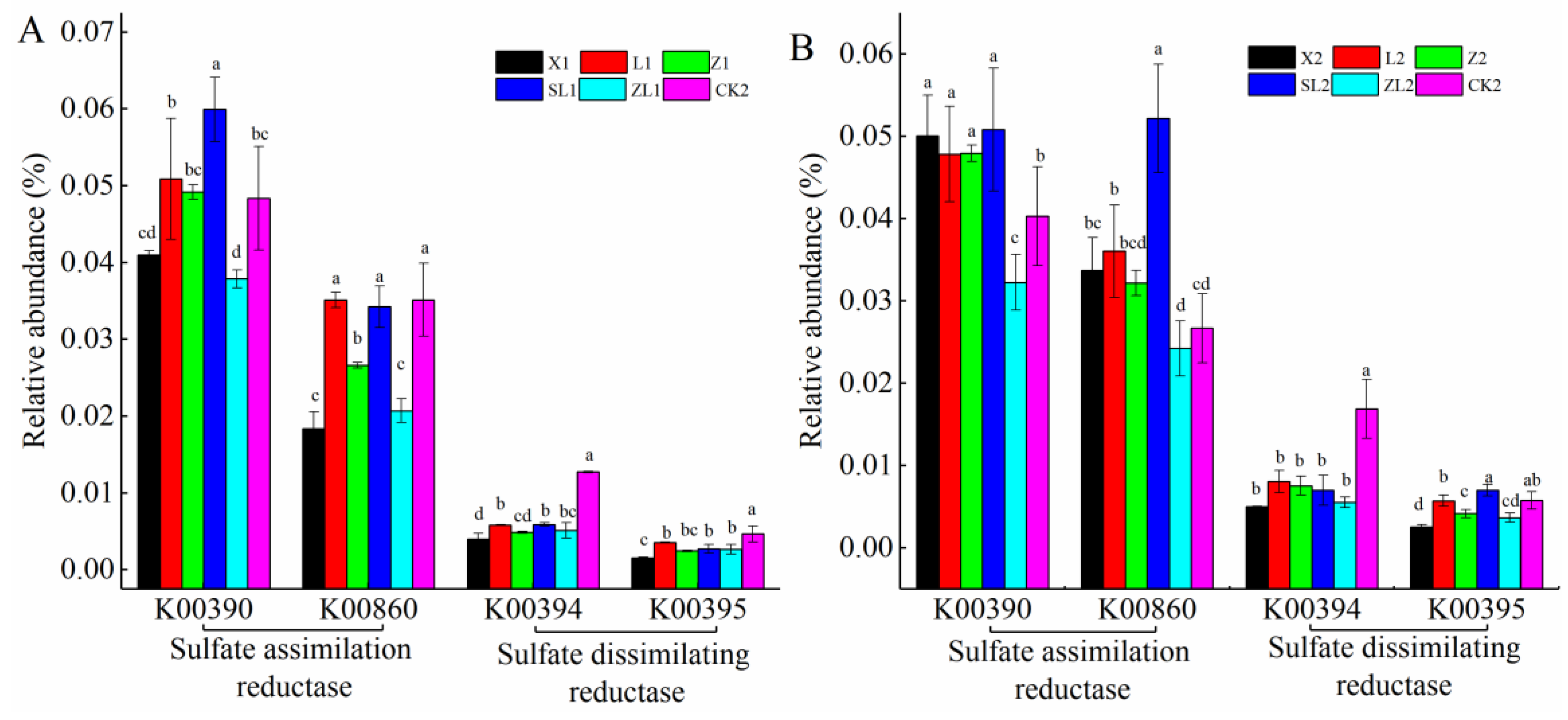
3.6.1. Microbial Metabolism
3.6.2. Functional Potential of Bacterial Homologous Gene Clusters
4. Discussion
4.1. Diversity of Soil Bacterial Communities in Shelterbelts of Different Forest Types
4.2. Function Potential of Soil Bacterial Community in Shelterbelts of Different Forest Types
4.3. Suggestions for Future Research
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sun, L.B.; Chang, X.M.; Yu, X.X.; Jia, G.D.; Chen, L.H.; Liu, Z.Q.; Zhu, X.H. Precipitation and soil water thresholds associated with drought-induced mortality of farmland shelter forests in a semi-arid area. Agric. Ecosyst. Environ. 2019, 284, 106595. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellin, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Schadt, C.W. Towards a holistic understanding of the beneficial interactions across the Populus microbiome. New Phytol. 2015, 205, 1424–1430. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.A.; Lennon, J.T. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. USA 2012, 109, 14058–14062. [Google Scholar] [CrossRef] [PubMed]
- Zolla, G.; Badri, D.V.; Bakker, M.G.; Manter, D.K.; Viyanco, J.M. Soil microbiomes vary in their ability to confer drought tolerance to Arabidopsis. Appl. Soil Ecol. 2013, 68, 1–9. [Google Scholar] [CrossRef]
- Yergeau, E.; Bell, T.H.; Champagne, J.; Maynard, C.; Tardif, S.; Tremblay, J.; Greer, C.W. Transplanting soil microbiomes leads to lasting effects on willow growth, but not on the rhizosphere microbiome. Front. Microbiol. 2015, 6, 1436. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; Coleman Derr, D.; Tringe, S.G.; Dangl, J.L.; Mitchell Olds, T. Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild Arabidopsis relative. Ecol. Lett. 2015, 18, 218–220. [Google Scholar] [CrossRef]
- Bennett, A.E.; Grussu, D.; Kam, J.; Caul, S.; Halpin, C. Plant lignin content altered by soil microbial community. New Phytol. 2015, 206, 166–174. [Google Scholar] [CrossRef]
- Qin, Y.; Pan, X.Y.; Jin, W.; Chen, L.Q.; Yuan, Z.L. Comparison of four extraction methods of soil microbiome in poplar plantation. Sci. Silvae Sin. 2018, 54, 169–176. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Araujo, R.A.; Costa, C.S.; Lima, A.J.T.; Oliveira, M.E.; Cutrim, J.A.A.; Santos, F.N.S.; Araujo, J.S.; Santos, V.M.; Araujo, A.S.F. Soil microbial biomass in an agroforestry system of Northeast Brazil. Trop. Grassl. Forrajes Trop. 2015, 3, 41–48. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Schlaeppi, K.; Bulgarelli, D. The plant microbiome at work. Mol. Plant Microbe Interact. 2015, 28, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, S.G.; Magbanua, Z.V.; Williams, M.A.; Jangid, K.; Whitman, W.B.; Peterson, D.G.; Kingery, W.L. Erratum to: Bacterial diversity patterns differ in soils developing in sub-tropical and cool-temperate ecosystems. Microb. Ecol. 2017, 73, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Cookson, W.R.; Murphy, D.V.; Roper, M.M. Characterizing the relationships between soil organic matter components and microbial function and composition along a tillage disturbance gradient. Soil Biol. Biochem. 2008, 40, 763–777. [Google Scholar] [CrossRef]
- Pace, N.R. A molecular view of microbial diversity and the biosphere. Science 1997, 276, 734–740. [Google Scholar] [CrossRef]
- Jansson, J.K.; Prosser, J.I. Microbiology: The life beneath our feet. Nature 2013, 494, 40–41. [Google Scholar] [CrossRef]
- Mi, L.; Wang, G.H.; Jin, J.; Sui, Y.Y.; Liu, J.D.; Liu, X.B. Comparison of microbial community structures in four Black soils along. Can. J. Soil Sci. 2012, 92, 543–549. [Google Scholar] [CrossRef]
- Barriuso, J.; Marin, S.; Mellado, R.P. Effect of the herbicide glyphosate on glyphosate-tolerant maize rhizobacterial communities: A comparison with pre-emergency applied herbicide consisting of a combination of acetochlor and terbuthylazine. Environ. Microbiol. 2010, 12, 1021–1030. [Google Scholar] [CrossRef]
- Qin, J.J.; Li, R.Q.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–67. [Google Scholar] [CrossRef]
- Singh, A.K.; Dubey, S.K. Current trends in Bt crops and their fate on associated microbial community dynamics: A review. Protoplasma 2016, 253, 663–681. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Mark Welch, D.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef] [PubMed]
- Breulmann, M.; Schulz, E.; Weisshuhn, K.; Buscot, F. Impact of the plant community composition on labile soil organic carbon, soil microbial activity and community structure in semi-natural grassland ecosystems of different productivity. Plant Soil 2012, 352, 253–265. [Google Scholar] [CrossRef]
- Fang, X.M.; Yu, D.P.; Zhou, W.M.; Zhou, L.; Dai, L.M. The effects of forest type on soil microbial activity in Changbai Mountain, Northeast China. Ann. Forest Sci. 2016, 73, 473–482. [Google Scholar] [CrossRef]
- Bezemer, T.M.; Wang, X.G.; Li, H.; Hao, Z.Q. Drivers of bacterial beta diversity in two temperate forests. Ecol. Res. 2016, 31, 57–64. [Google Scholar] [CrossRef]
- Liu, J.J.; Sui, Y.Y.; Yu, Z.H.; Yao, Q.; Shi, Y.; Chu, H.Y.; Jin, J.; Liu, X.B.; Wang, G.H. Diversity and distribution patterns of Acidobacterial communities in the black soil zone of northeast China. Soil Biol. Biochem. 2016, 95, 212–222. [Google Scholar] [CrossRef]
- Sun, J.X.; Zhao, Y.S.; Xin, Y. Soil nutrient changes of different stage of poplar farmland shelterbelts in black soil region. J. Northeast For. Univ. 2018, 46, 13. [Google Scholar] [CrossRef]
- Deng, J.J.; Zhang, Y.; Yin, Y.; Zhu, X.; Zhu, W.X.; Zhou, Y.B. Comparison of soil bacterial community and functional characteristics following afforestation in the semi-arid areas. PeerJ 2019, 7, e7141. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Blaxter, M.; Mann, J.; Chapman, T.; Thomas, F.; Whitton, C.; Floyd, R.; Abebe, E. Defining operational taxonomic units using DNA barcode data. Philos. Trans. R. Soc. B 2005, 360, 1935–1943. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Buffington, S.A.; Di Prisco, G.V.; Auchtung, T.A.; Ajami, N.J.; Petrosino, J.F.; Costa-Mattioli, M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 2016, 165, 1762–1775. [Google Scholar] [CrossRef]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Chu, H.Y.; Fierer, N.; Lauber, C.L.; Caporaso, J.G.; Knight, R.; Grogan, P.; Lauber, C.L.; Grogan, P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 2010, 12, 2998–3006. [Google Scholar] [CrossRef]
- Finlay, B.J. Global Dispersal of Free-Living Microbial Eukaryote Species. Science 2002, 296, 1061–1063. [Google Scholar] [CrossRef]
- Griffiths, R.I.; Thomson, B.C.; James, P.; Bell, T.; Bailey, M.; Whiteley, A.S. The bacterial biogeography of British soils. Environ. Microbiol. 2011, 13, 1642–1654. [Google Scholar] [CrossRef]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B.H. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; He, Z.; Wilson, M.J.; Campbell, C.D. Microbial Biomass and Community Structure in a Sequence of Soils with Increasing Fertility and Changing Land Use. Microb. Ecol. 2000, 40, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Sui, Y.Y.; Yu, Z.H.; Shi, Y.; Chu, H.Y.; Jin, J.; Liu, X.B.; Wang, G.H. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Yang, X.T.; Ning, G.H.; Dong, H.Y.; Li, Y. Soil microbial characters under different vegetation communities in Taihang Mountain area. Chin. J. Appl. Ecol. 2006, 17, 1761–1764. [Google Scholar]
- Niu, X.Y.; Liu, Z.Q.; Zhao, J.J.; Wang, Y.Q.; Chen, Y.Q.; Du, H.; Zhang, C.F. Impact of forest succession on soil microbial diversity after fire in Greater Khingan Mountains. Microbiology 2017, 44, 1825–1833. [Google Scholar] [CrossRef]
- Carnovale, D.; Bissett, A.; Thrall, P.H.; Baker, G. Plant genus (Acacia and Eucalyptus) alters soil microbial community structure and relative abundance within revegetated shelterbelts. Appl. Soil Ecol. 2019, 133, 1–11. [Google Scholar] [CrossRef]
- Li, P.; Shi, R.J.; Zhao, F.; Yu, J.H.; Cui, X.Y.; Hu, J.G.; Zhang, Y. Soil bacterial community structure and predicted functions in the larch forest during succession at the Greater Khingan Mountains of Northeast China. Chin. J. Appl. Ecol. 2019, 30, 95–107. [Google Scholar] [CrossRef]
- Preem, J.K.; Truu, J.; Truu, M.; Mander, U.; Oopkaup, K.; Lohmus, K.; Helmisaari, H.S.; Uri, V.; Zobel, M. Bacterial community structure and its relationship to soil physico-chemical characteristics in alder stands with different management histories. Ecol. Eng. 2012, 49, 10–17. [Google Scholar] [CrossRef]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Zhao, B.X.; Pan, F.R.; Han, X.R. Bacterial community development based on Illumina amplicon sequencing of 16S rDNA in the cherry rhizosphere. Chin. J. Soil Sci. 2018, 49, 596–601. [Google Scholar] [CrossRef]
- Yang, L.B.; Sui, X.; Cui, F.X.; Zhu, D.G.; Song, H.L.; Ni, H.W. Soil bacterial diversity between different forest wet successional stages in Tangwanghe National Park. Res. Environ. Sci. 2019, 32, 458–464. [Google Scholar] [CrossRef]
- Kang, H.Z.; Gao, H.H.; Yu, W.J.; Yi, Y.; Wang, Y.; Ning, M.L. Changes in soil microbial community structure and function after afforestation depend on species and age: Case study in a subtropical alluvial island. Sci. Total Environ. 2018, 625, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Zang, H.D.; Mortimer, P.; Shi, L.L.; Li, Y.J.; Xu, J.C.; Ostermann, A. Tree species and recovery time drives soil restoration after mining: A chronosequence study. Land Degrad. Dev. 2018, 29, 1738–1747. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, A.A.; Kuramae, E.E.; de Hollander, M.; Pijl, A.S.; van Veen, J.A.; Tsai, S.M. Acidobacterial community responses to agricultural management of soybean in Amazon forest soils. FEMS Microbiol. Ecol. 2013, 83, 607–621. [Google Scholar] [CrossRef] [PubMed]
- DeBruyn, J.M.; Nixon, L.T.; Fawaz, M.N.; Johnson, A.M.; Radosevich, M. Global Biogeography and Quantitative Seasonal Dynamics of Gemmatimonadetes in Soil. Appl. Environ. Microb. 2011, 77, 6295–6300. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.A.; Chase, J.M.; Jiménez, I.; Jørgensen, P.M.; Araujo-Murakami, A.; Paniagua-Zambrana, N.; Seidel, R.; Cornell, H. Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecol. Lett. 2013, 16, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.J.; Zhao, F.Z.; Kang, D.; Yang, G.H.; Han, X.H.; Tong, X.G.; Feng, Y.Z.; Ren, G.X. Linkages of C:N:P stoichiometry and bacterial community in soil following afforestation of former farmland. Forest Ecol. Manag. 2016, 376, 59–66. [Google Scholar] [CrossRef]
- Gunina, A.; Smith, A.R.; Godbold, D.L.; Jones, D.L.; Kuzyakov, Y. Response of soil microbial community to afforestation with pure and mixed species. Plant Soil 2017, 412, 357–368. [Google Scholar] [CrossRef]
- Bardgett, R.; Wardle, D.A. Aboveground-belowground linkages: Biotic interactions, ecosystem processes, and global change. Eos Trans. Am. Geophys. Union 2011, 92, 222. [Google Scholar] [CrossRef]
- Kaiser, K.; Wemheuer, B.; Korolkow, V.; Wemheuer, F.; Nacke, H.; Schoning, I.; Schrumpf, M.; Daniel, R. Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci. Rep. UK 2016, 6, 33696. [Google Scholar] [CrossRef] [PubMed]
- Bezemer, T.M.; Fountain, M.T.; Barea, J.M.; Christensen, S.; Dekker, S.C.; Duyts, H.; van Hal, R.; Harvey, J.A.; Hedlund, K.; Maraun, M. Divergent composition but similar function of soil food webs of individual plants: Plant species and community effects. Ecology 2010, 91, 3027–3036. [Google Scholar] [CrossRef]
- Li, S.J.; Zhu, T.H.; Liu, Z.X. Effects of two models of forest rehabilitation on dominant groups of soil microbes. Bull. Soil Water Conserv. 2014, 34, 186–191. [Google Scholar] [CrossRef]
- Palomo, A.; Pedersen, A.G.; Fowler, S.J.; Dechesne, A.; Sicheritz-Pontén, T.; Smets, B.F. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 2017, 12, 1779–1793. [Google Scholar] [CrossRef]
- Ushiki, N.; Jinno, M.; Fujitani, H.; Suenaga, T.; Terada, A.; Tsuneda, S. Nitrite oxidation kinetics of two Nitrospira strains: The quest for competition and ecological niche differentiation. J. Biosci. Bioeng. 2017, 123, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, L.E.; Barbosa, O. Metagenomic analysis exploring taxonomic and functional diversity of soil microbial communities in Chilean vineyards and surrounding native forests. PeerJ 2017, 5, e3098. [Google Scholar] [CrossRef]
- Finlay, B.J.; Clarke, K.J. Ubiquitous dispersal of microbial species. Nature 1999, 400, 828. [Google Scholar] [CrossRef]
- Xu, F.; Cao, F.Q.; Kong, Q.; Zhou, L.L.; Yuan, Q.; Zhu, Y.J.; Wang, Q.; Du, Y.D.; Wang, Z.D. Electricity production and evolution of microbial community in the constructed wetland-microbial fuel cell. Chem. Eng. J. 2018, 339, 479–486. [Google Scholar] [CrossRef]
- Gelaw, A.M.; Singh, B.R.; Lal, R. Soil organic carbon and total nitrogen stocks under different land uses in a semi-arid watershed in Tigray, Northern Ethiopia. Agric. Ecosyst. Environ. 2014, 188, 256–263. [Google Scholar] [CrossRef]
- Yan, X.Y.; Cai, Z.C.; Wang, S.W.; Smith, P. Direct measurement of soil organic carbon content change in the croplands of China. Glob. Chang. Biol. 2011, 17, 1487–1496. [Google Scholar] [CrossRef]
- Cao, X.B.; Lin, D.; Cai, L.; Jiang, Y.M.; Zhu, D. Effects of different vegetation communities on soil carbon fraction, RubisCO activity and cbbl genes in Nanjishan wetland of Poyang Lake. Acta Pedol. Sin. 2017, 54, 1269–1279. [Google Scholar] [CrossRef]
- Claus, H. Laccases: Structure, reactions, distribution. Micron 2004, 35, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Ebersberger, D.; Niklaus, P.A.; Kandeler, E. Long term CO2 enrichment stimulates N-mineralisation and enzyme activities in calcareous grassland. Soil Biol. Biochem. 2003, 35, 965–972. [Google Scholar] [CrossRef]
- Liang, B.C.; Wang, X.L.; Ma, B.L. Maize root-induced change in soil organic carbon pools. Soil Sci. Soc. Am. J. 2002, 66, 845–847. [Google Scholar] [CrossRef]
- Lu, J.L.; Shen, G.; Wang, Q.; Ren, M.L.; Pei, Z.X.; Wei, C.H.; Wang, W.J. Effect of urban tree species on soil physicochemical properties in Harbi, Northeastern China, and afforestation implications. Bull. Bot. Res. 2016, 36, 549–555. [Google Scholar] [CrossRef]
- Wang, W.B.; Wang, Y.P.; Wang, H.T.; Ma, X.S.; Yi, W.H. Effects of different continuous cropping and rotation of poplar plantation on soil nitrogen bacteria community and nitrogen metabolism. Sci. Silvae Sin. 2016, 52, 45–54. [Google Scholar] [CrossRef]
- Yoon, S.; Cruz-Garcia, C.; Sanford, R.; Ritalahti, K.M.; Loffler, F.E. Denitrification versus respiratory ammonification: Environmental controls of two competing dissimilatory NO3−/NO2− reduction pathways in Shewanella loihica strain PV-4. ISME J. 2015, 9, 1093–1104. [Google Scholar] [CrossRef]
- Bent, E.; Nemeth, D.; Wagner-Riddle, C.; Dunfield, K. Residue management leading to higher field-scale N2O flux is associated with different soil bacterial nitrifier and denitrifier gene community structures. Appl. Soil Ecol. 2016, 108, 288–299. [Google Scholar] [CrossRef]
- Kyaing, S.M.; Gu, L.H.; Cheng, H.M. The role of nitrate reductase and nitrite reductase in plant. Curr. Biotechnol. 2011, 1, 159–164. [Google Scholar] [CrossRef]
- Li, X.B.; Xia, L.L.; Yan, X.Y. Application of membrane inlet mass spectrometry to directly quantify denitrification in flooded rice paddy soil. Biol. Fertil. Soils 2014, 50, 891–900. [Google Scholar] [CrossRef]
- Ma, X.S.; Wang, W.B.; Wang, Y.P.; Wang, H.T.; Yi, W.H. Characteristics of phosphate-solubilizing microbial community in the soil of poplar plantations under successive-planting and rotation. Chin. J. Appl. Ecol. 2016, 27, 1877–1885. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus uptake by plants: From soil to cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Krey, T.; Vassilev, N.; Baum, C.; Eichler-Löbermann, B. Effects of long-term phosphorus application and plant-growth promoting rhizobacteria on maize phosphorus nutrition under field conditions. Eur. J. Soil Biol. 2013, 55, 124–130. [Google Scholar] [CrossRef]
- Tang, W.P.; Li, J.Y.; Qi, L.H.; Hu, X.Y.; Cui, H.X. Effects of poplar successive plantation on soil chemical characteristics in Jianghan Plain. J. Cent. South Univ. For. Technol. 2009, 29, 72–76. [Google Scholar]
- Wu, Y.; Wang, W.J.; Wang, Q.; Zhong, Z.L.; Pei, Z.X.; Wang, H.M.; Yao, Y.L. Impact of poplar shelterbelt plantations on surface soil properties in northeastern China. Can. J. Forest Res. 2018, 48, 559–567. [Google Scholar] [CrossRef]
- Chang, L.F. Isolation, Purification and Properties of APS Reductase and Sulfate Reductase from Sulfate-Reducing Bacterium; Inner Mongolia Normal University: Huhehaote, China, 2008. [Google Scholar]
- Bick, J.A.; Dennis, J.J.; Zylstra, G.J.; Nowack, J.; Leustek, T. Identification of a new class of 5′-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria. J. Bacteriol. 2000, 182, 135–142. [Google Scholar] [CrossRef]
- Biderre Petit, C.; Boucher, D.; Kuever, J.; Alberic, P.; Jézéquel, D.; Chebance, B.; Borrel, G.; Fonty, G.; Peyret, P. Identification of sulfur-cycle prokaryotes in a low-sulfate lake (Lake Pavin) using aprA and 16S rRNA gene markers. Microb. Ecol. 2011, 61, 313–327. [Google Scholar] [CrossRef]
- Nair, A.; Ngouajio, M. Soil microbial biomass, functional microbial diversity, and nematode community structure as affected by cover crops and compost in an organic vegetable production system. Appl. Soil Ecol. 2012, 58, 45–55. [Google Scholar] [CrossRef]
- Kubartova, A.; Moukoumi, J.; Beguiristain, T.; Ranger, J.; Berthelin, J. Microbial diversity during cellulose decomposition in different forest stands: I. Microbial communities and environmental conditions. Microb. Ecol. 2007, 54, 393–405. [Google Scholar] [CrossRef]
- Bergkemper, F.; Welzl, G.; Lang, F.; Kruger, J.; Schloter, M.; Schulz, S. The importance of C, N and P as driver for bacterial community structure in German beech dominated forest soils. J. Plant Nutr. Soil. Sci. 2016, 179, 472–480. [Google Scholar] [CrossRef]


| Plot Types | Height /m | DBH /cm | Spacing /m | Longitude and Latitude | Altitude /m |
|---|---|---|---|---|---|
| X | 17.86 ± 1.77 | 16.64 ± 2.03 | 2 × 1.5 | 125°51′12.4″E—125°51′38.0″E, 47°38′22.8″N—47°38′26.2″N | 241~241 |
| L | 9.44 ± 1.55 | 11.00 ± 2.51 | 2 × 1.5 | 125°48′59.7″E—125°49′03.5″E, 47°38′38.2″N—47°38′55.3″N | 239~267 |
| Z | 8.18 ± 1.29 | 12.59 ± 1.67 | 2 × 1.5 | 125°51′37.9″E—125°51′41.4″E, 47°38′11.9″N—47°38′24.8″N | 248~251 |
| SL | 9.76 ± 1.47 | 10.99 ± 2.69 | 2 × 3 | 125°49′03.3″E—125°49′06.4″E, 47°38′23.6″N—47°38′36.8″N | 242~252 |
| ZL | 11.08 ± 1.40 | 13.96 ± 2.98 | 2 × 1.5 | 125°47′15.5″E—125°47′17.4″E, 47°39′09.6″N—47°39′30.5″N | 249~256 |
| CK | —— | —— | —— | 125°48′28.1″E—125°48′29.9″E, 47°38′35.6″N—47°38′36.4″N | 221~222 |
| Layer | Unique OTUs | Shared OTUs | |||||
|---|---|---|---|---|---|---|---|
| X | L | Z | SL | ZL | CK | ||
| Upper | 2519 | 2374 | 2368 | 2299 | 2455 | 3410 | 420 |
| Lower | 2268 | 2266 | 2093 | 2008 | 2268 | 3234 | 432 |
| CV (%) | 7.72 | 3.29 | 8.72 | 9.56 | 5.60 | 3.75 | 1.99 |
| Sample | X | L | Z | SL | ZL | CK | |
|---|---|---|---|---|---|---|---|
| Unique OTUs | Upper | 1493 | 1430 | 1462 | 1696 | 1586 | 977 |
| Lower | 1597 | 1941 | 1422 | 1170 | 1403 | 696 | |
| Shared OTUs | 1847 | 1893 | 2016 | 1887 | 2342 | 2282 | |
| Total | 4937 | 5264 | 4900 | 4753 | 5331 | 3955 | |
| CV (%) | 4.76 | 21.44 | 1.96 | 25.96 | 8.66 | 23.75 | |
| Sample | Simpson | Chao1 | ACE | Shannon |
|---|---|---|---|---|
| X1 | 1.00 ± 0.00a | 2415.56 ± 313.95a | 2628.13 ± 394.88a | 9.62 ± 0.18a |
| L1 | 1.00 ± 0.00a | 2583.06 ± 264.11a | 2814.98 ± 320.18a | 9.80 ± 0.28a |
| Z1 | 1.00 ± 0.00a | 2395.76 ± 190.76a | 2573.73 ± 234.52a | 9.86 ± 0.19a |
| SL1 | 1.00 ± 0.00a | 2525.06 ± 433.90a | 2741.37 ± 518.70a | 9.84 ± 0.23a |
| ZL1 | 1.00 ± 0.00a | 2884.11 ± 138.48a | 3097.92 ± 248.01a | 10.02 ± 0.10a |
| CK1 | 1.00 ± 0.00a | 2495.38 ± 266.87a | 2633.57 ± 289.19a | 10.06 ± 0.07a |
| p-value | 0.22 | 0.38 | 0.50 | 0.13 |
| X2 | 1.00 ± 0.00a | 2300.21 ± 278.30a | 2497.11 ± 306.65ab | 9.74 ± 0.16a |
| L2 | 1.00 ± 0.00a | 2280.97 ± 268.93a | 2472.52 ± 310.77ab | 9.59 ± 0.28a |
| Z2 | 1.00 ± 0.00a | 2483.66 ± 168.47a | 2702.35 ± 195.05ab | 9.75 ± 0.17a |
| SL2 | 1.00 ± 0.00a | 2382.02 ± 427.27a | 2486.05 ± 342.33ab | 9.67 ± 0.17a |
| ZL2 | 1.00 ± 0.00a | 2673.35 ± 518.46a | 2947.23 ± 620.24a | 9.94 ± 0.21a |
| CK2 | 1.00 ± 0.00a | 2074.42 ± 297.47a | 2187.47 ± 395.47b | 9.84 ± 0.07a |
| p-value | 0.51 | 0.44 | 0.320 | 0.31 |
| Sample | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|
| X1 | 18.33 ± 0.58ab | 54.67 ± 2.89c | 71.00 ± 6.08b | 134.00 ± 6.93b | 180.67 ± 13.65 | 82.67 ± 8.02ab |
| L1 | 16.00 ± 2.00b | 57.00 ± 2.00bc | 76.00 ± 4.36b | 145.33 ± 6.35ab | 192.33 ± 8.02 | 90.33 ± 8.08a |
| Z1 | 17.33 ± 1.15ab | 57.67 ± 4.51bc | 73.33 ± 3.21b | 142.00 ± 7.00ab | 193.33 ± 11.59 | 86.67 ± 8.33ab |
| SL1 | 18.67 ± 2.08ab | 60.00 ± 1.73b | 73.67 ± 0.58b | 142.33 ± 8.51ab | 196.33 ± 13.05 | 83.33 ± 9.07ab |
| ZL1 | 17.67 ± 2.08ab | 60.33 ± 2.08b | 78.67 ± 3.21b | 149.33 ± 7.09a | 195.33 ± 4.93 | 89.33 ± 2.52ab |
| CK1 | 20.00 ± 1.73a | 68.67 ± 2.31a | 90.67 ± 4.51a | 150.33 ± 5.51a | 196.00 ± 5.57 | 76.33 ± 3.51b |
| p-value | 0.17 | 0.01 | 0.01 | 0.12 | 0.44 | 0.23 |
| X2 | 18.00 ± 1.73 | 57.00 ± 3.61b | 68.33 ± 6.81b | 129.00 ± 4.36c | 159.33 ± 4.93 | 76.67 ± 6.51ab |
| L2 | 17.33 ± 2.52 | 58.00 ± 2.00b | 69.67 ± 1.53b | 130.67 ± 11.15bc | 160.33 ± 18.90 | 72.67 ± 6.66ab |
| Z2 | 16.33 ± 0.58 | 58.00 ± 6.08b | 71.67 ± 5.03b | 140.67 ± 6.03abc | 183.67 ± 18.58 | 81.00 ± 9.17a |
| SL2 | 17.33 ± 1.53 | 61.67 ± 2.89b | 69.67 ± 5.03b | 129.67 ± 4.04bc | 162.00 ± 10.54 | 75.33 ± 6.43ab |
| ZL2 | 16.67 ± 1.15 | 62.00 ± 2.00b | 74.33 ± 4.73b | 145.33 ± 2.89a | 179.67 ± 5.51 | 77.33 ± 6.51ab |
| CK2 | 18.67 ± 1.15 | 68.00 ± 1.73a | 87.67 ± 4.73a | 143.67 ± 11.37ab | 165.67 ± 8.74 | 65.33 ± 4.62b |
| p-value | 0.51 | 0.02 | 0.00 | 0.05 | 0.13 | 0.18 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Xin, Y.; Zhao, Y. Diversity and Functional Potential of Soil Bacterial Communities in Different Types of Farmland Shelterbelts in Mid-Western Heilongjiang, China. Forests 2019, 10, 1115. https://doi.org/10.3390/f10121115
Zhang J, Xin Y, Zhao Y. Diversity and Functional Potential of Soil Bacterial Communities in Different Types of Farmland Shelterbelts in Mid-Western Heilongjiang, China. Forests. 2019; 10(12):1115. https://doi.org/10.3390/f10121115
Chicago/Turabian StyleZhang, Jun, Ying Xin, and Yusen Zhao. 2019. "Diversity and Functional Potential of Soil Bacterial Communities in Different Types of Farmland Shelterbelts in Mid-Western Heilongjiang, China" Forests 10, no. 12: 1115. https://doi.org/10.3390/f10121115
APA StyleZhang, J., Xin, Y., & Zhao, Y. (2019). Diversity and Functional Potential of Soil Bacterial Communities in Different Types of Farmland Shelterbelts in Mid-Western Heilongjiang, China. Forests, 10(12), 1115. https://doi.org/10.3390/f10121115





