The Development of Populus alba L. and Populus tremula L. Species Specific Molecular Markers Based on 5S rDNA Non-Transcribed Spacer Polymorphism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Isolation
2.2. Analysis of Sequences and Primer Design
2.3. PCR and Electrophoresis
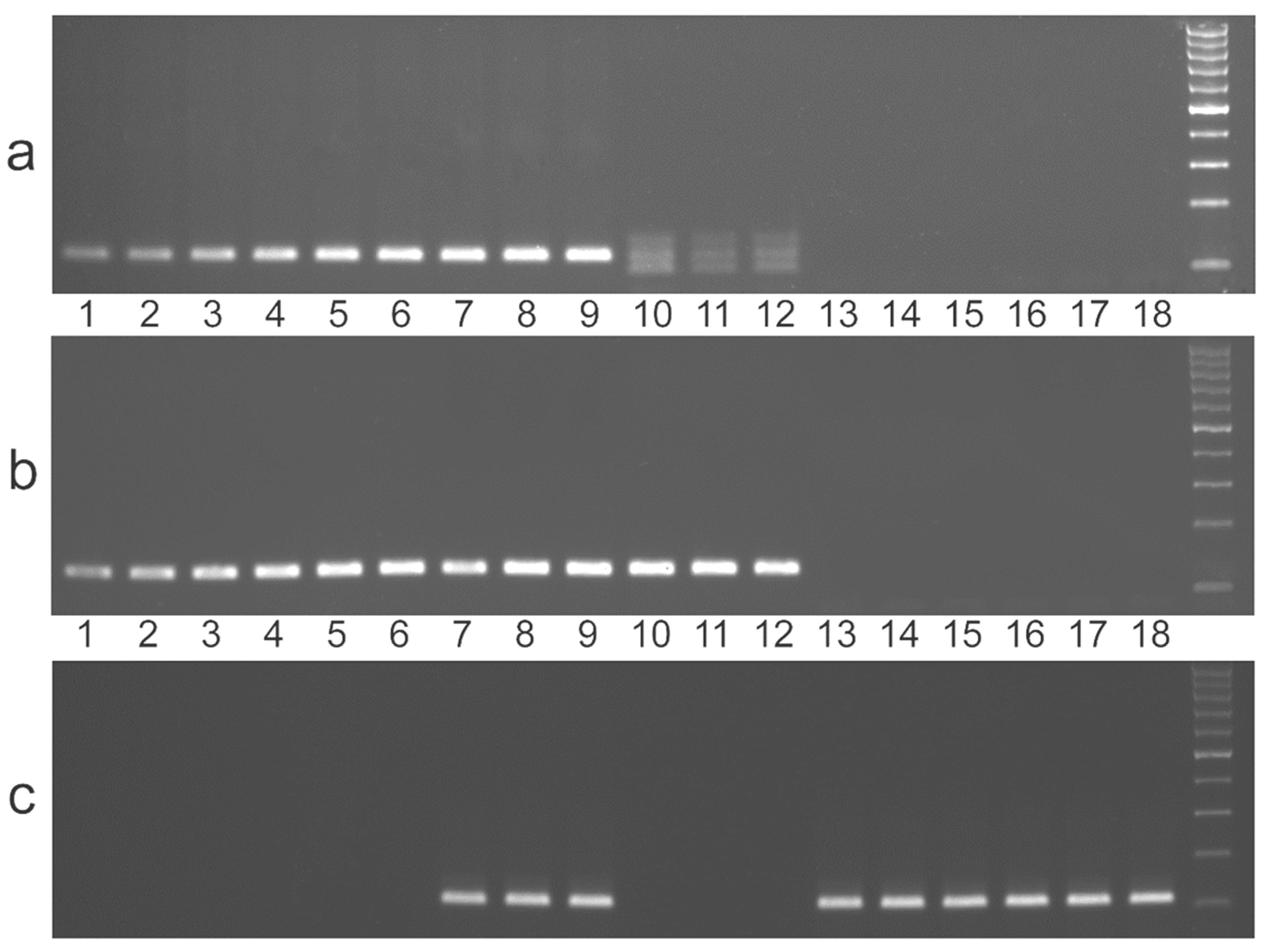
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Isebrands, J.G.; Richardson, J. Poplars and Willows: Trees for Society and the Environment; CABI: Boston, MA, USA; FAO: Rome, Italy, 2014; p. 14. [Google Scholar]
- Eckenwalder, J.E. Systematics and evolution of Populus. In Biology of Populus and its Implications for Management and Conservation; Part I; Stettler, R.F., Bradshaw, H.D., Jr., Heilman, P.E., Hinckley, T.M., Eds.; NRC Research Press: Ottawa, ON, Canada, 1996; pp. 7–32. [Google Scholar]
- Proshkin, B.V.; Klimov, A.V. Spontaneous hybridization of Populus × sibirica and Populus nigra in the city of Novokuznetsk (Kemerovo region). Turczaninowia 2017, 20, 206–218. [Google Scholar]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Schein, J. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed]
- Mader, M.; Le Paslier, M.C.; Bounon, R.; Bérard, A.; Rampant, P.F.; Fladung, M.; Leplé, J.C.; Kersten, B. Whole-genome draft assembly of Populus tremula × P. alba clone INRA 717-1B4. Silvae Genet. 2016, 65, 74–79. [Google Scholar] [CrossRef]
- Lin, Y.C.; Wang, J.; Delhomme, N.; Schiffthaler, B.; Sundström, G.; Zuccolo, A.; Hoeppner, M.P. Functional and evolutionary genomic inferences in Populus through genome and population sequencing of American and European aspen. Proc. Natl. Acad. Sci. USA 2018, 115, E10970–E10978. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Wang, J.; Zhou, G.; Yue, Z.; Hu, Q.; Chen, Y.; Wang, K. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2019, 4, 2797. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Wang, X.R.; Zeng, Q.Y. De novo assembly of white poplar genome and genetic diversity of white poplar population in Irtysh River basin in China. Sci. China Life Sci. 2019, 62, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Vanden Broeck, A.; Villar, M.; Van Bockstaele, E.; Van Slycken, J. Natural hybridization between cultivated poplars and their wild relatives: Evidence and consequences for native poplar populations. Ann. Forest Sci. 2005, 62, 601–613. [Google Scholar] [CrossRef]
- Vanden Broeck, A.; Cox, K.; Quataert, P.; Van Bockstaele, E.; Van Slycken, J. Flowering phenology of Populus nigra L., P. nigra cv. italica and P. × canadensis Moench. and the potential for natural hybridization in Belgium. Silvae Genet. 2003, 52, 280–283. [Google Scholar]
- Ma, H.; Dong, Y.; Chen, Z.; Liao, W.; Lei, B.; Gao, K.; Li, S.; An, X. Variation in the growth traits and wood properties of hybrid white poplar clones. Forests 2015, 6, 1107–1120. [Google Scholar] [CrossRef]
- Christe, C.S.; Bresadola, K.N.; Fussi, L.; Heinze, B.; Wegmann, B.; Lexer, D.C. Selection against recombinant hybrids maintains reproductive isolation in hybridizing Populus species despite F1 fertility and recurrent gene flow. Mol. Ecol. 2016, 11, 2482–2498. [Google Scholar] [CrossRef]
- Konovalov, N.A. New forms of the hybrid pyramidal crown poplars. Not. Sverdl. Dep. All-USSR Bot. Soc. 1964, 3, 129–132. (In Russian) [Google Scholar]
- Banayev, Y.V.; Shishkin, S.V.; Voronkova, M.S.; Belanova, A.P.; Tomoshevich, M.A. Morphological and biochemical features of Populus × canescens in natural populations of the Altai region. Bull. Alt. St. Agr. Univ. 2017, 8, 90–97. (In Russian) [Google Scholar]
- Allendorf, F.W.; Leary, R.F.; Spruell, P.; Wenburg, J.K. The problems with hybrids: Setting conservation guidelines. Trends Ecol. Evol. 2001, 16, 613–622. [Google Scholar] [CrossRef]
- Rajora, O.P.; Dancik, B.P. Genetic characterization and relationships of Populus alba, P. tremula, and P. × canescens, and their clones. Theor. Appl. Genet. 1992, 84, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Fossati, T.; Patrignani, G.; Zapelli, I.; Sabatti, M.; Sala, F.; Castiglione, S. Development of molecular markers to assess the level of introgression of Populus tremula into P. alba natural populations. Plant. Breed. 2004, 123, 382–385. [Google Scholar] [CrossRef]
- Schroeder, H.; Kersten, B.; Fladung, M. Development of multiplexed marker sets to identify the most relevant poplar species for breeding. Forests 2017, 8, 492. [Google Scholar] [CrossRef]
- Joseph, J.A.; Lexer, C. A set of novel DNA polymorphisms within candidate genes potentially involved in ecological divergence between Populus alba and P. tremula, two hybridizing European forest trees. Mol. Ecol. Res. 2008, 8, 188–192. [Google Scholar] [CrossRef]
- Tsarev, A.; Tsareva, R.; Tsarev, V.; Fladung, M.; von Wühlisch, G. Aspen hybridization: Parents’ compatibility and seedlings’ growth. Silvae Genet. 2018, 67, 12–19. [Google Scholar] [CrossRef]
- Fedulova, T.P.; Kondratyeva, A.M.; Evlakov, P.M.; Marchuk, I.I. Investigation of genetic diversity of poplar variety samples (Populus L.) based on SSR markers. Lesoteh. Žurnal [For. Eng. J.] 2016, 4, 105–111. (In Russian) [Google Scholar]
- Alexandrov, O.S.; Karlov, G.I. Development of 5S rDNA-based molecular markers for the identification of Populus deltoides Bartr. ex Marshall, Populus nigra L., and Their Hybrids. Forests 2018, 9, 604. [Google Scholar] [CrossRef]
- Apples, R.; Gerlach, W.L.; Dennis, E.S.; Swift, H.; Peacock, W.J. Molecular and chromosomal organization of DNA sequence coding for the ribosomal RNAs in cereals. Chromosoma 1980, 78, 293–311. [Google Scholar] [CrossRef]
- Pendás, A.M.; Morán, P.; Martínez, J.L.; Garcia-Vásquez, E. Applications of 5S rDNA in Atlantic salmon, brown trout, and in Atlantic salmon x brown trout hybrid identification. Mol. Ecol. 1995, 4, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Feodorova, T.A.; Aleksandrov, O.S. The molecular-phylogenetic study of Petrosimonia species of Chenopodiaceae Juss. family. Izv. TAA 2015, 5, 54–60. [Google Scholar]
- Negi, M.S.; Rajagopal, J.; Chauhan, N.; Cronn, R.; Lakshmikumaran, M. Length and sequence heterogeneity in 5S rDNA of Populus deltoides. Genome 2002, 45, 1181–1188. [Google Scholar] [CrossRef]
- Wilson, N. Genome Analysis of Populus Species: Assessment of Genetic Diversity of P. deltoides, Characterization of Wide Hybrids and Phylogenetic Analysis Using Molecular Markers. Ph.D. Thesis, Teri University, New Delhi, India, 2013. [Google Scholar]
- Alexandrov, O.S.; Karlov, G.I.; Sorokin, A.N.; Potapenko, N.C. Development of the molecular marker system for species identification of poplars and analysis of hybrids. In Proceedings of the III (V) All-Russia Youth Conference with Participation of Foreign Scientists “Prospects of Development and Problems of Modern Botany”, Novosibirsk, Russia, 10–14 November 2014; Asbaganov, S.V., Ed.; Akademizdat: Novosibirsk, Russia, 2014; pp. 123–124. [Google Scholar]
- Kisileva, K.V.; Mayorov, S.R.; Novikov, V.S. Flora of Central Russia: Atlas—Determinant; ZAO; “Phython+”: Moscow, Russia, 2010; pp. 1–544. [Google Scholar]
- Vinogradova, Y.K.; Mayorov, S.R.; Khorun, L.V. The Black Book of Flora of Central Russia: Alien Plant Species in the Ecosystems of Central Russia; GEOS: Moscow, Russia, 2010; pp. 427–436. [Google Scholar]
- Kostina, M.V.; Nasimovich, J.A. On the systematics of Populus L. II. importance of fruit characters for identification of cultivated and adventive species in Moscow region. Bull. Mosc. Soc. Naturalists. Biol. Ser. 2014, 119, 74–79. [Google Scholar]
- Mamaev, S.A. Key to Trees and Shrubs of the Urals; USSR Academy of Sciences, Ural Branch: Sverdlovsk/Moscow, Russia, 1965; pp. 44–69. [Google Scholar]
- Borodina, N.A.; Nekrasov, V.I.; Nekrasova, N.S.; Petrova, I.P.; Plotnikova, L.S.; Smirnova, N.G. Trees and Shrubs of the USSR; Mysl’: Moscow, Russia, 1966; pp. 268–276. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Razumova, O.V.; Alexandrov, O.S.; Divashuk, M.G.; Sukhorada, T.I.; Karlov, G.I. Molecular cytogenetic analysis of monoecious hemp (Cannabis sativa L.) cultivars reveals its karyotype variations and sex chromosomes constitution. Protoplasma 2016, 253, 895–901. [Google Scholar] [CrossRef]
- Falistocco, E.; Passeri, V.; Marconi, G. Investigations of 5S rDNA of Vitis vinifera L.: Sequence analysis and physical mapping. Genome 2007, 50, 927–938. [Google Scholar] [CrossRef]
- GeneDoc: Analysis and Visualization of Genetic Variation. Available online: http://www.nrbsc.org/gfx/genedoc/ebinet.htm (accessed on 16 October 2019).
- Welsh, S.L.; Atwood, N.D.; Goodrich, S.; Higgins, L.C. A Utah Flora, The Great Basin Naturalist Memoir No. 9; Brigham Young University: Provo, UT, USA, 1987; 894p. [Google Scholar]
- The Plant List: A Working List of All Plant Species. Populus bolleana Lauche. Available online: http://www.theplantlist.org/tpl1.1/record/kew-5004590 (accessed on 16 October 2019).
| Species | Parents of Hybrid | Sample Name | Co-ordinates |
|---|---|---|---|
| P. alba L. | - | tree#1 | 55°81′46.58″ 37°55′98.41″ |
| tree#2 | 44°22′78.85″ 38°89′55.87″ | ||
| tree#3 | 44°64′65.02″ 39°14′36.91″ | ||
| tree#4 | 45°29′43.82″ 36°42′96.96″ | ||
| tree#5 | 50°44′89.88″ 39°63′36.73″ | ||
| tree#6 | 55°83′62.65″ 37°56′70.35″ | ||
| P. tremula L. | - | tree#7 | 55°83′50.41″ 37°55′53.78″ |
| tree#8 | 56°23′92.56″ 38°11′17.26″ | ||
| tree#9 | 56°06′58.33″ 37°90′21.71″ | ||
| tree#10 | 55°90′05.39″ 37°56′67.05″ | ||
| tree#11 | 55°41′81.88″ 37°84′72.91″ | ||
| tree#12 | 67°08′07.96″ 32°86′88.48″ | ||
| P. × canescens (Aiton) Sm. | P. alba L. × P. tremula L. | tree#13 | 55°45′11.99″ 36°94′16.11″ |
| tree#14 | 55°72′38.02″ 37°56′49.92″ | ||
| tree#15 | 55°85′23.54″ 37°54′33.94″ | ||
| P. bolleana Lauche | tree#16 | 45°29′56.17″ 36°43′13.14″ | |
| tree#17 | 45°30′05.94″ 36°42′79.16″ | ||
| tree#18 | 45°30′04.41″ 36°42′81.74″ | ||
| P. nigra L. | tree#19 | 50°83′42.96″ 39°39′21.76″ | |
| P. deltoides Bartr. ex Marshall | - | tree#20 | 55°83′51.72″ 37°55′55.98″ |
| P. × canadensis Moench. | P. deltoides Bartr. ex Marshall × P. nigra L. | tree#21 | 55°82′83.67″ 37°57′47.36″ |
| P. trichocarpa Torr. et A. Gray | - | tree#22 | 55°83′48.51″ 37°55′55.97″ |
| P. maximowiczii Henry | - | tree#23 | 55°83′52.52″ 37°55′53.57″ |
| P. simonii Can. | - | tree#24 | 55°83′52.74″ 37°55′54.93″ |
| P. candicans Ait. | - | tree#25 | 55°83′53.31″ 37°55′52.29″ |
| P. × moskoviensis R. I. Schrod. | P. suaveolens Fish. (syn. P. maximowiczii Henry) × P. laurifolia Ldb. | tree#26 | 55°83′53.36″ 37°55′54.88″ |
| P. × berolinensis K. Koch. | P. laurifolia Ldb. × P. nigra L. | tree#27 | 55°83′51.80″ 37°55′55.15″ |
| Primer Name | Sequence | PCR Product Length, bp |
|---|---|---|
| alb2-f | 5′-TTTTGCCGTTTTCC-3′ | 110 |
| alb2-r | 5′-AATCGCCCGGGAAAGGAAA-3′ | |
| alb9-f | 5′-TCGGAGTAGCGATTCACAGC-3′ | 126 |
| alb9-r | 5′-GTTTGCGTCGGACCATAACA-3′ | |
| tremu1 | 5′-AGCCTCCCGCTGGG-3′ | 113 |
| 5Srev | 5′-CGCTTAACTGCGGAGT-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexandrov, O.S.; Karlov, G.I. The Development of Populus alba L. and Populus tremula L. Species Specific Molecular Markers Based on 5S rDNA Non-Transcribed Spacer Polymorphism. Forests 2019, 10, 1092. https://doi.org/10.3390/f10121092
Alexandrov OS, Karlov GI. The Development of Populus alba L. and Populus tremula L. Species Specific Molecular Markers Based on 5S rDNA Non-Transcribed Spacer Polymorphism. Forests. 2019; 10(12):1092. https://doi.org/10.3390/f10121092
Chicago/Turabian StyleAlexandrov, Oleg S., and Gennady I. Karlov. 2019. "The Development of Populus alba L. and Populus tremula L. Species Specific Molecular Markers Based on 5S rDNA Non-Transcribed Spacer Polymorphism" Forests 10, no. 12: 1092. https://doi.org/10.3390/f10121092
APA StyleAlexandrov, O. S., & Karlov, G. I. (2019). The Development of Populus alba L. and Populus tremula L. Species Specific Molecular Markers Based on 5S rDNA Non-Transcribed Spacer Polymorphism. Forests, 10(12), 1092. https://doi.org/10.3390/f10121092






