Phytophthora Species from Xinjiang Wild Apple Forests in China
Abstract
:1. Introduction
2. Material and Methods
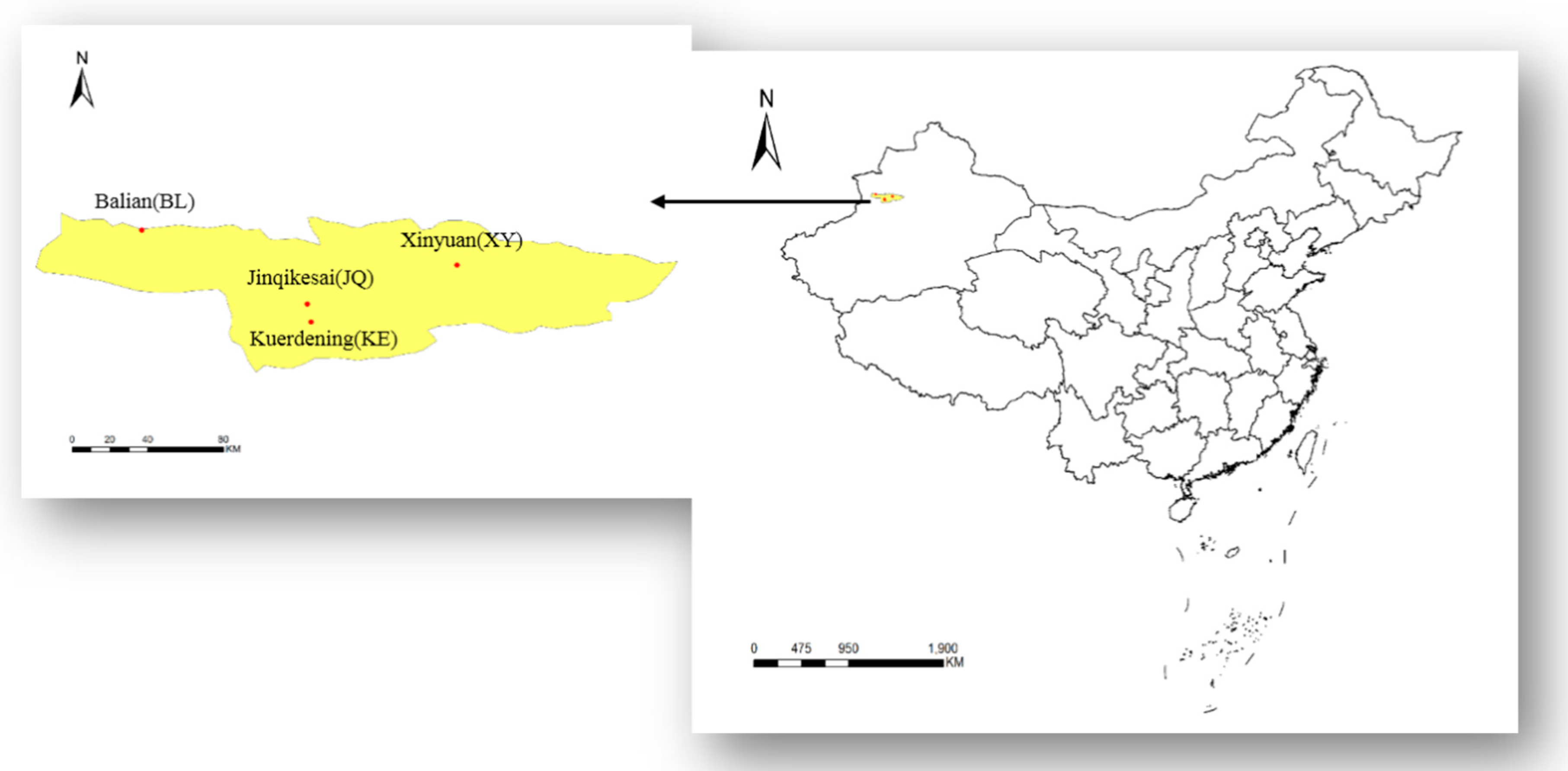
2.1. Study Area
2.2. Sampling and Phytophthora Isolation
2.3. Classical Identification of Isolates
2.4. Molecular Identification of Isolates
2.5. Phylogenetic Analysis
3. Results
3.1. Phytophthora Species Identification
3.2. Phylogenetic Analysis
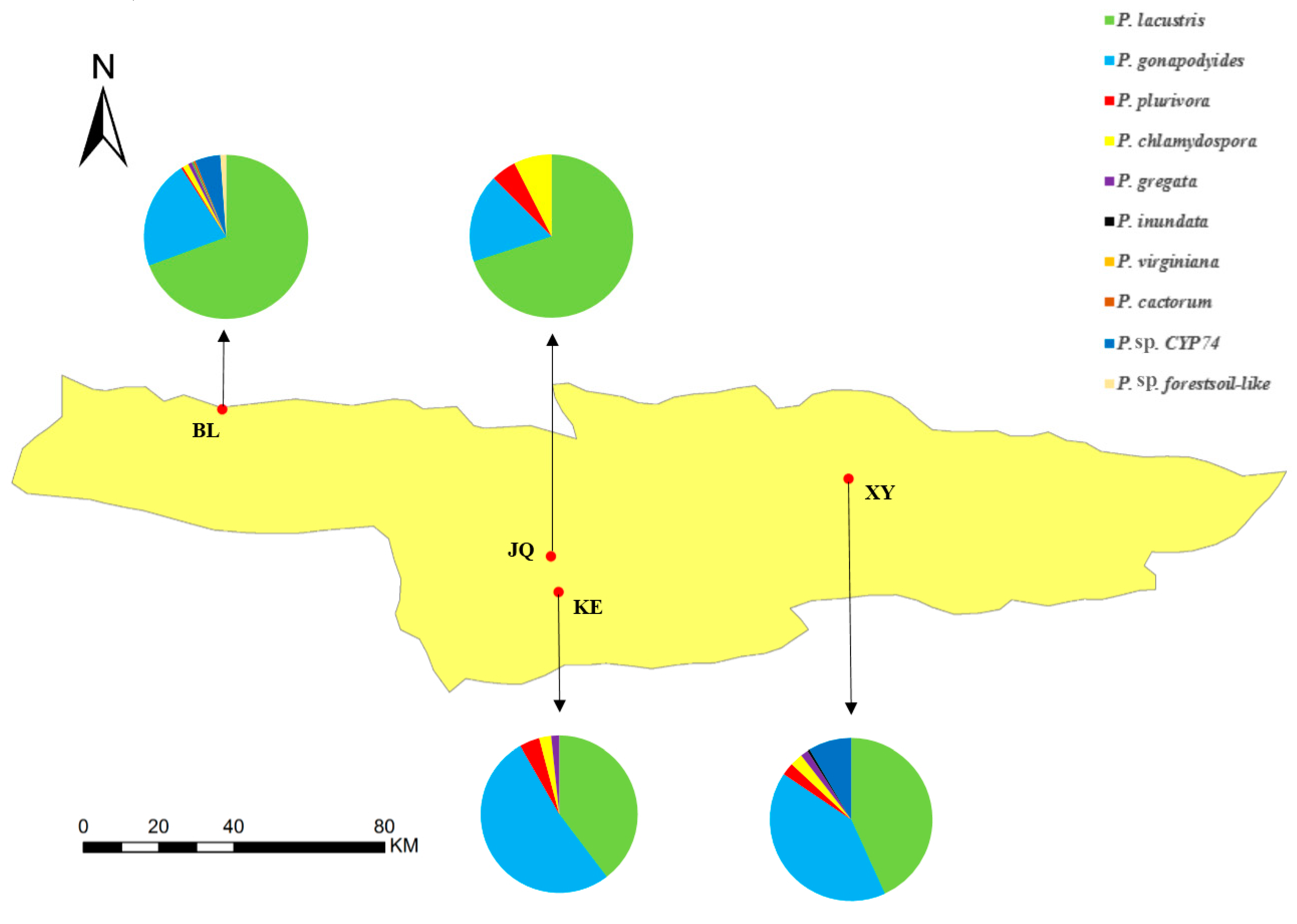
3.3. The Distribution of Phytophthora Species
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, X.S.; Lin, P.J.; Zhong, J.P. Analysis of habitat for wild fruit forests in Ili and discussion on its occurrence. Arid Zone Res. 1993, 3, 28–30. [Google Scholar]
- Yan, G.; Long, H.; Song, W.; Chen, R. Genetic polymorphism of Malus sieversii populations in Xinjiang, China. Genet. Resour. Crop Evol. 2008, 55, 171–181. [Google Scholar] [CrossRef]
- Zhang, H.X.; Zhang, M.L.; Wang, L.N. Genetic structure and historical demography of Malus sieversii in the Yili Valley and the western mountains of the Junggar Basin, Xinjiang, China. J. Arid Land 2015, 7, 264–271. [Google Scholar] [CrossRef]
- Zhang, X.S. On the eco-geographical characters and the problems of classification of the wild fruit-tree forest in the Ili Valley of Sinkiang. Acta Bot. Sin. 1973, 15, 239–253. [Google Scholar]
- Chen, L.Z. Present Situations of and Conservation Strategies for Biodiversity in China; Science China Press: Beijing, China, 1993. [Google Scholar]
- Wang, N.; Jiang, S.; Zhang, Z.; Fang, H.; Xu, H.; Wang, Y.; Chen, X. Malus sieversii: The origin, flavonoid synthesis mechanism, and breeding of red-skinned and red-fleshed apples. Hortic. Res. 2018, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Panyushkina, I.; Mukhamadiev, N.; Lynch, A.; Ashikbaev, N.; Arizpe, A.; O’Connor, C.; Sagitov, A. Wild Apple Growth and Climate Change in Southeast Kazakhstan. Forests 2017, 8, 406. [Google Scholar] [CrossRef]
- Zhang, P.; Lü, Z.Z.; Zhang, X.; Zhao, X.P.; Zhang, Y.G.; Gulzhanat, T.; Maisupova, B.; Adilbayeva, Z.; Cui, Z.J. Age Structure of Malus sieversii Population in Ili of Xinjiang and Kazakhstan. Arid Zone Res. 2019, 36, 844–853. [Google Scholar]
- Wang, Z.Y.; Yang, Z.Q.; Zhang, Y.L.; Wang, X.Y.; Tang, Y.L. Biological control of agrilus mali (coleoptera: Buprestidae) by applying four species of bethylid wasp (hymenoptera: Bethylidae) on malus sieversii in Xinjiang. Sci. Silvae Sin. 2014, 50, 97–101. [Google Scholar]
- Yan, G.; Zheng, X.U. Study on the wild fruit tree diseases of Tianshan mountains and their distribution in Xinjiang. Arid Zone Res. 2001, 18, 47–49. [Google Scholar]
- Liu, A.H.; Zhang, X.P.; Wen, J.B.; Yue, C.Y.; Alimu, J.; Zhang, S.P.; Kereman, J.W. Preliminary research on the composite damage of Agrilus mali matsumura and Valsa mali miyabe et yamada in wild apple trees in tianshan mountain. Xinjiang Agric. Sci. 2014, 51, 2240–2244. [Google Scholar]
- Niu, C.; Wang, J.; Zhu, X.; Chen, X.; Guo, L. Brown rot pathogens on stone and pome fruit trees in Xinjiang wild forest. Mycosystema 2016, 35, 1514–1525. [Google Scholar]
- Cheng, Y.; Zhao, W.; Lin, R.; Yao, Y.; Yu, S.; Zhou, Z.; Huai, W. Fusarium species in declining wild apple forests on the northern slope of the Tianshan Mountains in north-western China. For. Pathol. 2019. [Google Scholar] [CrossRef]
- Jung, T.; Pérez-Sierra, A.; Durán, A.; Horta, M.J.; Balci, Y.; Scanu, B. Canker and decline diseases caused by soil- and airborne Phytophthora species in forests and woodlands. Persoonia Mol. Phylogeny Evol. Fungi 2018, 40, 182–220. [Google Scholar] [CrossRef] [PubMed]
- Erwin, D.C.; Ribeiro, O.K. Phytophthora Diseases-Worldwide; APS Press: St. Paul, MN, USA, 1996. [Google Scholar]
- Jung, T.; Durßn, A.; Sanfuentes von Stowasser, E.; Schena, L.; Mosca, S.; Fajardo, S.; Tomšovskř, M. Diversity of Phytophthora species in Valdivian rainforests and association with severe dieback symptoms. For. Pathol. 2018, 48, e12443. [Google Scholar] [CrossRef]
- Liu, A.H.; Shang, J.; Zhang, J.W.; Kong, T.T.; Yue, Z.Y.; Wen, J.B. Canker and fine-root loss of Malus sieversii (Ldb.) Roem. caused by Phytophthora plurivora in Xinjiang Province in China. For. Pathol. 2018, 48, e12462. [Google Scholar] [CrossRef]
- Lin, P.Y.; Cui, N.R. Wild Fruit Forest Resources in Tianshan Mountains—Comprehensive Research on Wild Fruit Forests in Ili. Xinjiang, China; China Forestry Publishing House: Beijing, China, 2000. [Google Scholar]
- Hüberli, D.; Hardy, G.E.S.J.; White, D.; Williams, N.; Burgess, T.I. Fishing for Phytophthora from Western Australia’s waterways: A distribution and diversity survey. Australas. Plant Pathol. 2013, 42, 251–260. [Google Scholar] [CrossRef]
- Huai, W.X.; Tian, G.; Hansen, E.M.; Zhao, W.X.; Goheen, E.M.; Grünwald, N.J.; Cheng, C. Identification of Phytophthora species baited and isolated from forest soil and streams in northwestern Yunnan province, China. For. Pathol. 2013, 43, 87–103. [Google Scholar] [CrossRef]
- Reeser, P.; Sutton, W.; Hansen, E. Phytophthora species in tanoak trees, canopy-drip, soil, and streams in the sudden oak death epidemic plot of south-western Oregon, USA. N. Z. J. For. Sci. 2011, 41, S65–S73. [Google Scholar]
- Wingfield, B.D.; Oh, E.; Gryzenhout, M.; Burgess, T.I.; Wingfield, M.J. Surveys of soil and water reveal a goldmine of Phytophthora diversity in South African natural ecosystems. IMA Fungus 2013, 4, 123–131. [Google Scholar]
- Li, W.W.; Zhao, W.X.; Huai, W.X. Phytophthora pseudopolonica sp. Nov., a new species recovered from stream water in subtropical forests of China. Int. J. Syst. Evol. Microbiol. 2017, 67, 3666–3675. [Google Scholar] [CrossRef]
- Jung, T.; Chang, T.T.; Bakonyi, J.; Seress, D.; Pérez-Sierra, A.; Yang, X.; Maia, C. Diversity of Phytophthora species in natural ecosystems of Taiwan and association with disease symptoms. Plant Pathol. 2017, 66, 194–211. [Google Scholar] [CrossRef]
- Huai, W.X.; Guo, L.D.; He, W. Genetic diversity of an ectomycorrhizal fungus Tricholoma terreum in a Larix principis-rupprechtii stand assessed using random amplified polymorphic DNA. Mycorrhiza 2003, 13, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Cooke, D.E.L.; Drenth, A.; Duncan, J.M.; Wagels, G.; Brasier, C.M. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 2000, 30, 17–32. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal Rna Genes for Phylogenetics; Academic Press Inc.: Cambridge, MA, USA, 1990. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped-BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Yang, X.; Tyler, B.M.; Hong, C. An expanded phylogeny for the genus Phytophthora. IMA Fungus 2017, 8, 355–384. [Google Scholar] [CrossRef]
- Jung, T.; Horta Jung, M.; Cacciola, S.O.; Cech, T.; Bakonyi, J.; Seress, D.; Mosca, S.; Schena, L.; Seddaiu, S.; Pane, A.; et al. Multiple new cryptic pathogenic Phytophthora species from Fagaceae forests in Austria, Italy and Portugal. IMA Fungus 2017, 8, 219–244. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.C.; Ho, H.H.; Zheng, F.C. A survey of Phytophthora species on Hainan Island of South China. J. Phytopathol. 2009, 157, 33–39. [Google Scholar] [CrossRef]
- Sutton, W.; Adams, G.C.; Remigi, P.; Reeser, P.W.; Hansen, E.M. Phytophthora species in forest streams in Oregon and Alaska. Mycologia 2011, 103, 22–35. [Google Scholar]
- Akilli, S.; Ulubaş Serçe, Ç.; Katircioǧlu, Y.Z.; Maden, S. Phytophthora dieback on narrow leaved ash in the black sea region of turkey. For. Pathol. 2013, 43, 252–256. [Google Scholar] [CrossRef]
- Aday Kaya, A.G.; Lehtijärvi, A.; Şaşmaz, Y.; Nowakowska, J.A.; Oszako, T.; Doğmuş Lehtijärvi, H.T.; Woodward, S. Phytophthora species detected in the rhizosphere of Alnus glutinosa stands in the Floodplain Forests of Western Turkey. For. Pathol. 2018, 48, 11–14. [Google Scholar] [CrossRef]
- Tkaczyk, M.; Nowakowska, J.A.; Oszako, T. Phytophthora species isolated from ash stands in Białowieża Forest nature reserve. For. Pathol. 2016, 46, 660–662. [Google Scholar] [CrossRef]
- Kanoun-Boulé, M.; Vasconcelos, T.; Gaspar, J.; Vieira, S.; Dias-Ferreira, C.; Husson, C. Phytophthora ×alni and Phytophthora lacustris associated with common alder decline in Central Portugal. For. Pathol. 2016, 46, 174–176. [Google Scholar] [CrossRef]
- Nechwatal, J.; Bakonyi, J.; Cacciola, S.O.; Cooke, D.E.L.; Jung, T.; Nagy, Z.A.; Brasier, C.M. The morphology, behaviour and molecular phylogeny of Phytophthora taxon Salixsoil and its redesignation as Phytophthora lacustris sp. nov. Plant Pathol. 2012, 62, 355–369. [Google Scholar] [CrossRef]
- Jung, T.; La Spada, F.; Pane, A.; Aloi, F.; Evoli, M.; Horta Jung, M.; Magnano di San Lio, G. Diversity and Distribution of Phytophthora Species in Protected Natural Plots in Sicily. Forests 2019, 10, 259. [Google Scholar] [CrossRef]
- Stamler, R.A.; Sanogo, S.; Goldberg, N.P.; Randall, J.J. Phytophthora Species in Rivers and Streams of the Southwestern United States. Appl. Environ. Microbiol. 2016, 82, 4696–4704. [Google Scholar] [CrossRef]
- Ghimire, S.R.; Richardson, P.A.; Kong, P.; Hu, J.; Lea-Cox, J.D.; Ross, D.S.; Hong, C. Distribution and Diversity of Phytophthora species in Nursery Irrigation Reservoir Adopting Water Recycling System During Winter Months. J. Phytopathol. 2011, 159, 713–719. [Google Scholar] [CrossRef]
- Jung, T.; Blaschke, H.; Neumann, P. Isolation, identification and pathogenicity of Phytophthora species from declining oak stands. For. Pathol. 1996, 26, 253–272. [Google Scholar] [CrossRef]
- Balcì, Y.; Halmschlager, E. Phytophthora species in oak ecosystems in Turkey and their association with declining oak trees. Plant Pathol. 2003, 52, 694–702. [Google Scholar] [CrossRef]
- Orlikowski, L.B.; Ptaszek, M.; Rodziewicz, A.; Nechwatal, J.; Thinggaard, K.; Jung, T. Phytophthora root and collar rot of mature Fraxinus excelsior in forest stands in Poland and Denmark. For. Pathol. 2011, 41, 510–519. [Google Scholar] [CrossRef]
- Belhaj, R.; McComb, J.; Burgess, T.I.; Hardy, G.E.S.J. Pathogenicity of 21 newly described Phytophthora species against seven Western Australian native plant species. Plant Pathol. 2017, 67, 1140–1149. [Google Scholar] [CrossRef]
- Brasier, C.M.; Sanchez-Hernandez, E.; Kirk, S.A. Phytophthora inundata sp. nov., a part heterothallic pathogen of trees and shrubs in wet or flooded soils. Mycol. Res. 2003, 107, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Parkunan, V.; Johnson, C.S.; Bowman, B.C.; Hong, C.X. First report of Phytophthora inundata associated with a latent infection of tobacco (Nicotiana tabacum) in Virginia. Plant Pathol. 2010, 59, 1164. [Google Scholar] [CrossRef]
- Jung, T.; Orlikowski, L.; Henricot, B.; Abad-Campos, P.; Aday, A.G.; Aguín Casal, O.; Bakonyi, J.; Cacciola, S.O.; Cech, T.; Chavarriaga, D.; et al. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For. Pathol. 2016, 46, 134–163. [Google Scholar] [CrossRef]
- O’Hanlon, R.; Choiseul, J.; Corrigan, M.; Catarame, T.; Destefanis, M. Diversity and detections of Phytophthora species from trade and non-trade environments in Ireland. EPPO Bull. 2016, 46, 594–602. [Google Scholar] [CrossRef]
- Hansen, E.M.; Reeser, P.; Sutton, W.; Brasier, C.M. Redesignation of Phytophthora taxon Pgchlamydo as Phytophthora chlamydospora sp. nov. N. Am. Fungi 2015, 10, 1–14. [Google Scholar]
- Brasier, C.M.; Cooke, D.E.L.; Duncan, J.M.; Hansen, E.M. Multiple new phenotypic taxa from trees and riparian ecosystems in Phytophthora gonapodyides-P. megasperma ITS Clade 6, which tend to be high-temperature tolerant and either inbreeding or sterile. Mycol. Res. 2003, 107, 277–290. [Google Scholar] [CrossRef]
- Li, H. Identification of Phytophthora species infecting staple crops in Xinjiang. Acta Phytopathol. Sin. 1999, 29, 364–371. [Google Scholar]
- Jung, T.; Blaschke, M. Phytophthora root and collar rot of alders in Bavaria: Distribution, modes of spread and possible management strategies. Plant Pathol. 2004, 53, 197–208. [Google Scholar] [CrossRef]
- Kovács, J.; Lakatos, F.; Szabó, I. Occurrence and diversity of soil borne Phytophthoras in a declining black walnut stand in Hungary. Acta Silv. Lignaria Hung. 2015, 9, 57–69. [Google Scholar] [CrossRef]
- Ankowiak, R.; St, H.; Bila, P.; Kola, M. Occurrence of Phytophthora plurivora and other Phytophthora species in oak forests of southern Poland and their association with site conditions and the health status of trees. Folia Microbiol. 2014, 59, 531–542. [Google Scholar] [CrossRef]
- Zamora-Ballesteros, C.; Haque, M.M.U.; Diez, J.J.; Martín-García, J. Pathogenicity of Phytophthora alni complex and P. plurivora in Alnus glutinosa seedlings. For. Pathol. 2017, 47, e12299. [Google Scholar] [CrossRef]
- Mircetich, S.M. Phytophthora Root and Crown Rot of Apricot Trees. Acta Hortic. 2015, 121, 385–396. [Google Scholar] [CrossRef]
- Day, W.R. Root- rot of sweet chestnut and beech caused by species of Phytophthora. I. Cause and symptoms of disease: Its relation to soil conditions. Forestry 1938, 12, 101–116. [Google Scholar] [CrossRef]
- Mircetich, S.M.; Matheron, M.E. Phytophthora root and crown rot of walnut trees. Phytopathology 1983, 73, 1481–1488. [Google Scholar] [CrossRef]
- Wilcox, W.F.; Ellis, M.A. Phytophthora root and crown rots of peach trees in the eastern Great Lakes region. Plant Dis. 1989, 73, 794–798. [Google Scholar] [CrossRef]
- Yang, X.; Hong, C. Phytophthora virginiana sp. nov., a high-temperature tolerant species from irrigation water in Virginia. Mycotaxon 2014, 126, 167–176. [Google Scholar] [CrossRef]


| Clade | Species | Plot | Method | Number |
|---|---|---|---|---|
| clade1a | P. cactorum | BL | canopy drip | 1 |
| clade2c | P. plurivora | BL | soil | 1 |
| XY | soil | 2 | ||
| XY | stream | 3 | ||
| KE | stream | 5 | ||
| clade6b | P. lacustris | BL | stream | 175 |
| BL | canopy drip | 2 | ||
| BL | soil | 4 | ||
| XY | stream | 84 | ||
| XY | canopy drip | 1 | ||
| XY | soil | 1 | ||
| KE | stream | 48 | ||
| JQ | stream | 28 | ||
| P. gonapodyides | BL | stream | 56 | |
| XY | stream | 74 | ||
| XY | canopy drip | 5 | ||
| XY | soil | 3 | ||
| KE | stream | 63 | ||
| JQ | stream | 7 | ||
| P. chlamydospora | BL | stream | 3 | |
| XY | stream | 3 | ||
| P. gregata | BL | stream | 2 | |
| XY | stream | 4 | ||
| XY | soil | 1 | ||
| P. inundata | XY | stream | 1 | |
| P. sp. CYP74 | BL | stream | 13 | |
| XY | stream | 15 | ||
| XY | canopy drip | 2 | ||
| KE | stream | 3 | ||
| JQ | stream | 2 | ||
| P. sp. forestsoil-like | BL | stream | 3 | |
| KE | stream | 2 | ||
| JQ | stream | 3 | ||
| clade9a | P. virginiana | BL | stream | 1 |
| Species | Isolate No. | ITS Clade | Genbank Number |
|---|---|---|---|
| Phytophthora cactorum | 8BLL3 | 1 | MN175469 |
| Phytophthora plurivora | 1KEX3(6) | 2 | MN175458 |
| Phytophthora lacustris | 4GLX9(3) | 6 | MN175455 |
| 8KEX3(2) | 6 | MN175463 | |
| Phytophthora gregata | 9XYX6(5) | 6 | MN175456 |
| 7XYT6(2) | 6 | MN175459 | |
| Phytophthora gonapodyides | 9XYX7(2) | 6 | MN175457 |
| 2KEX5(5) | 6 | MN175462 | |
| Phytophthora sp. CYP74 | 1KEX9(1) | 6 | MN175460 |
| 15XYX2(1) | 6 | MN175465 | |
| Phytophthora chlamydospora | 9XYX5(4) | 6 | MN175461 |
| 5BLX9(2) | 6 | MN175466 | |
| Phytophthora sp. forestsoil-like | 1BLX1(7) | 6 | MN175464 |
| 8JQX4(1) | 6 | MN175468 | |
| Phytophthora inundata | 15XYX3(1) | 6 | MN209784 |
| Phytophthora virginiana | 1BLX1(3) | 9 | MN175467 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Huai, W.; Hamiti; Zhang, X.; Zhao, W. Phytophthora Species from Xinjiang Wild Apple Forests in China. Forests 2019, 10, 927. https://doi.org/10.3390/f10100927
Xu X, Huai W, Hamiti, Zhang X, Zhao W. Phytophthora Species from Xinjiang Wild Apple Forests in China. Forests. 2019; 10(10):927. https://doi.org/10.3390/f10100927
Chicago/Turabian StyleXu, Xiaoxue, Wenxia Huai, Hamiti, Xuechao Zhang, and Wenxia Zhao. 2019. "Phytophthora Species from Xinjiang Wild Apple Forests in China" Forests 10, no. 10: 927. https://doi.org/10.3390/f10100927
APA StyleXu, X., Huai, W., Hamiti, Zhang, X., & Zhao, W. (2019). Phytophthora Species from Xinjiang Wild Apple Forests in China. Forests, 10(10), 927. https://doi.org/10.3390/f10100927




