An Introduction to Clique Minimal Separator Decomposition
Abstract
:1. Introduction
- First, not every graph has a clique separator.
- Second, the subgraphs obtained are not disjoint, which means that if the overlap is large, little is to be gained by a Divide-and-Conquer approach.
- Section 2 gives some general graph notions, some particulars on minimal separation and minimal triangulation.
- Section 3 provides a precise definition of clique minimal separator decomposition, as well as examples; we also discuss how to find the clique minimal separators of a graph efficiently, and give some properties of this decomposition.
- Section 4 gives an efficient process for computing the clique minimal separator decomposition of a graph.
- Section 5 provides a brief history of clique decomposition, with the bibliographic background as well as high-level proofs of the results we present in this paper.
- We conclude in Section 6.
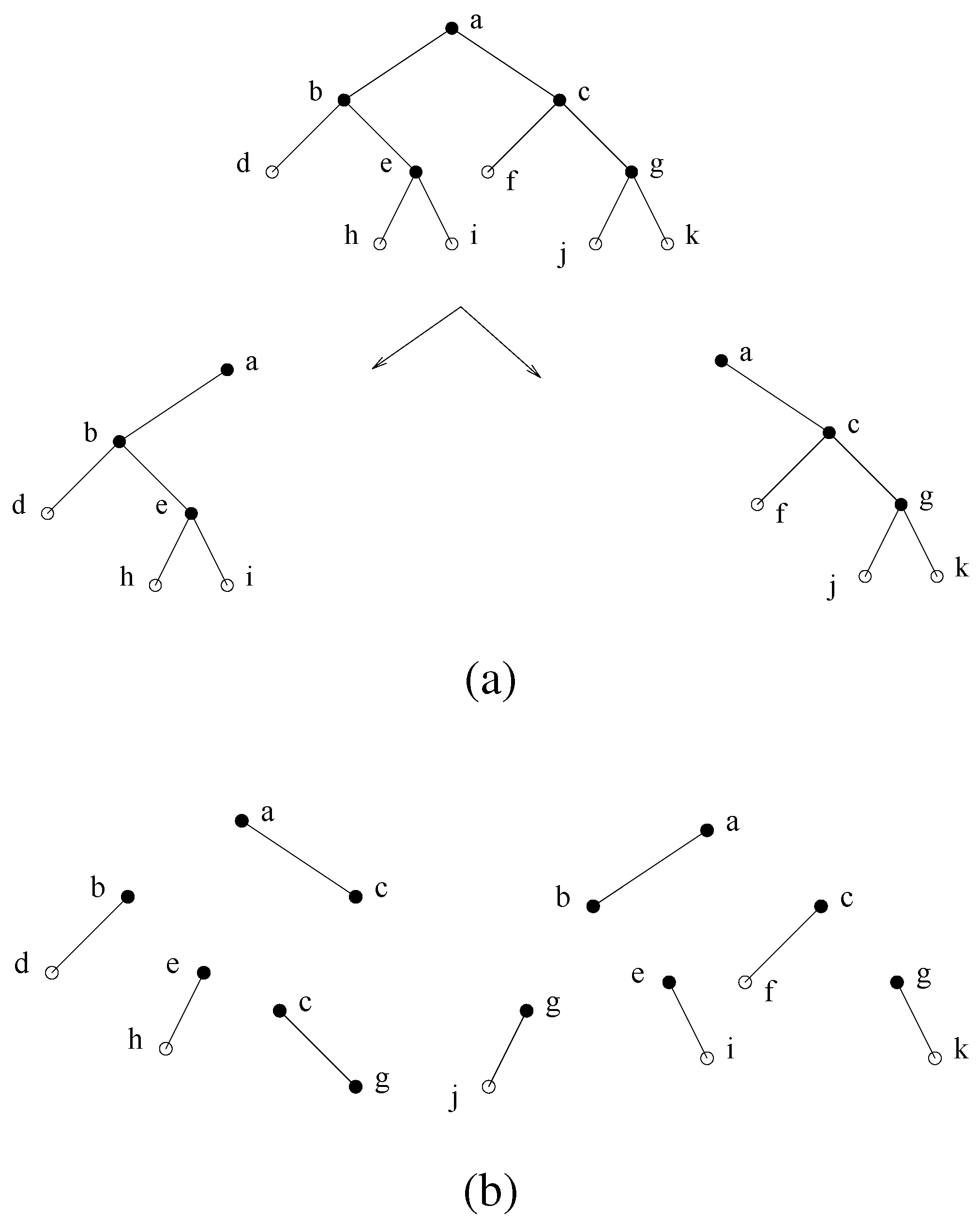
2. Graph Notions
2.1. General notions
2.2. Minimal separation, chordal graphs, and minimal triangulation
- A subset S of vertices of a connected graph G is called a separator (or sometimes a cutset) if is not connected.
- A separator S is called an -separator if a and b lie in different connected components of, a minimal -separator if S is an -separator and no proper subset of S is an -separator.
- A separator S is a minimal separator, if there is some pair such that S is a minimal -separator.


- minimal if for no proper subset of F, is chordal.
- minimum if no other minimal triangulation has less fill edges.

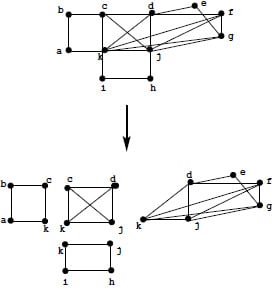
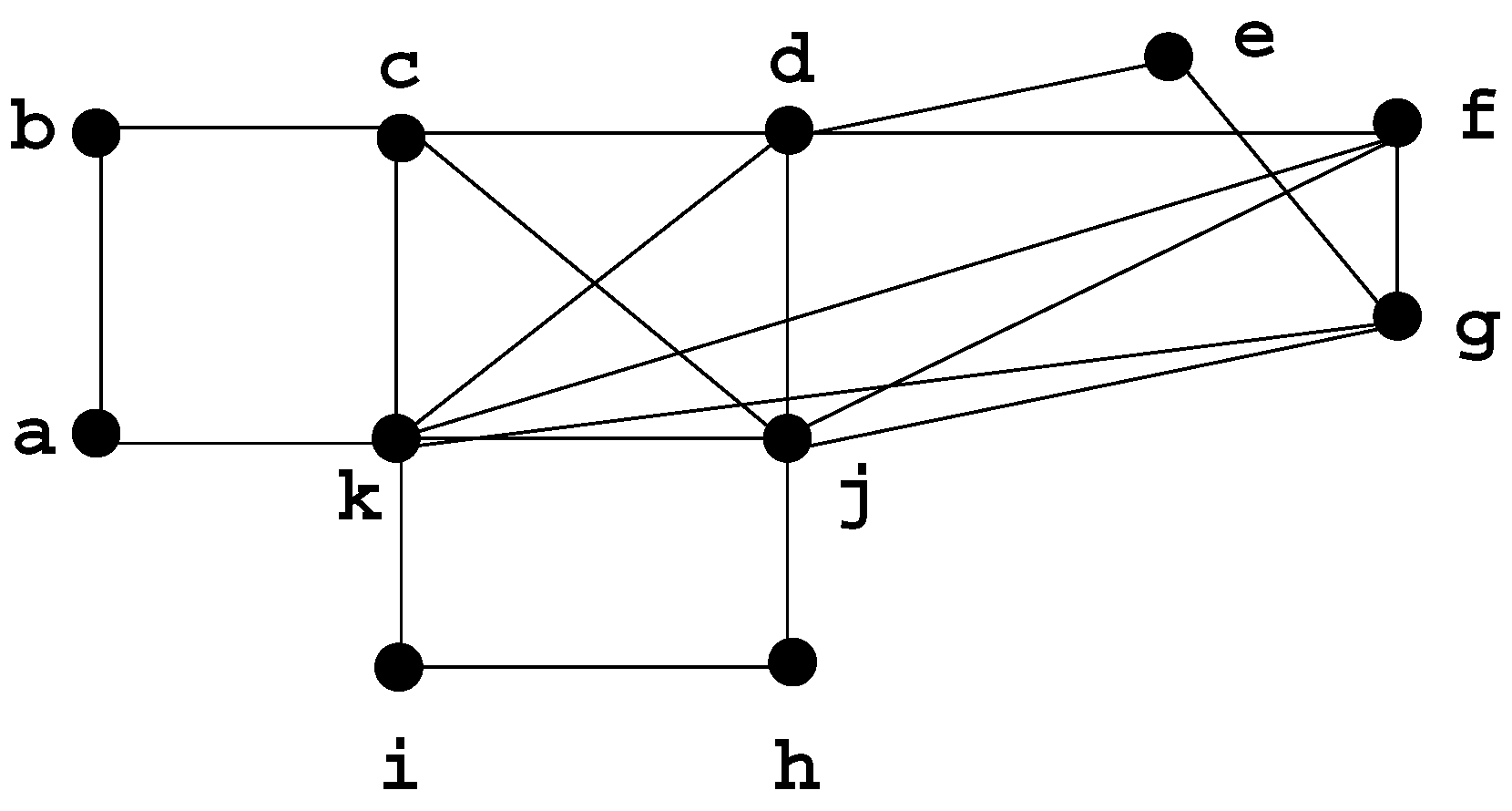
3. Defining clique minimal separator decomposition
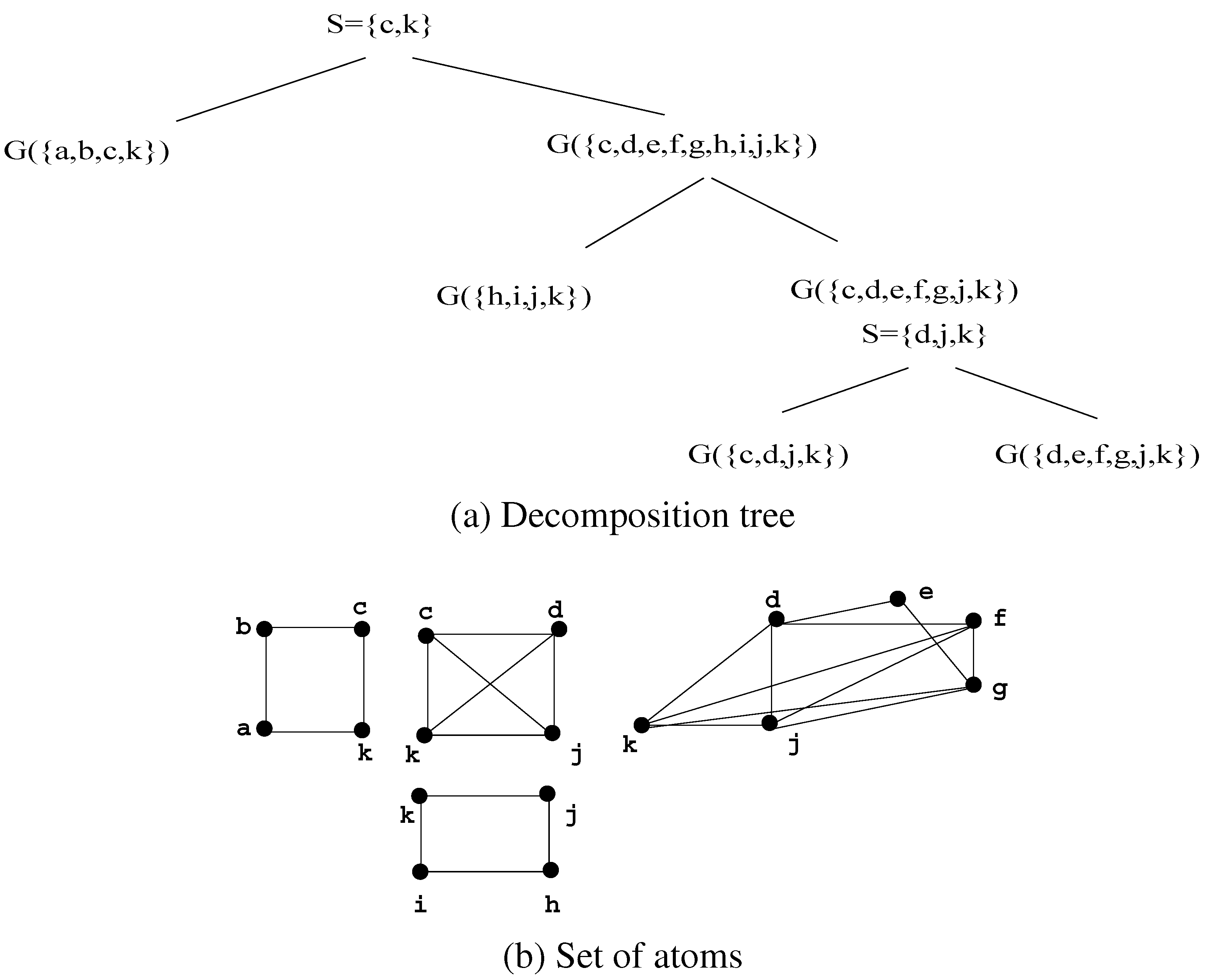
3.1. Definitions and examples
- Figure 6 gives the decomposition of a tree.
- On any chordal graph, this decomposition yields the set of maximal cliques of the input graph.
3.2. Properties of the atoms
- The number of atoms is at most n.
- The intersection between atoms is always a clique (which may be empty), and it is not necessarily a minimal separator (even when it is not empty).
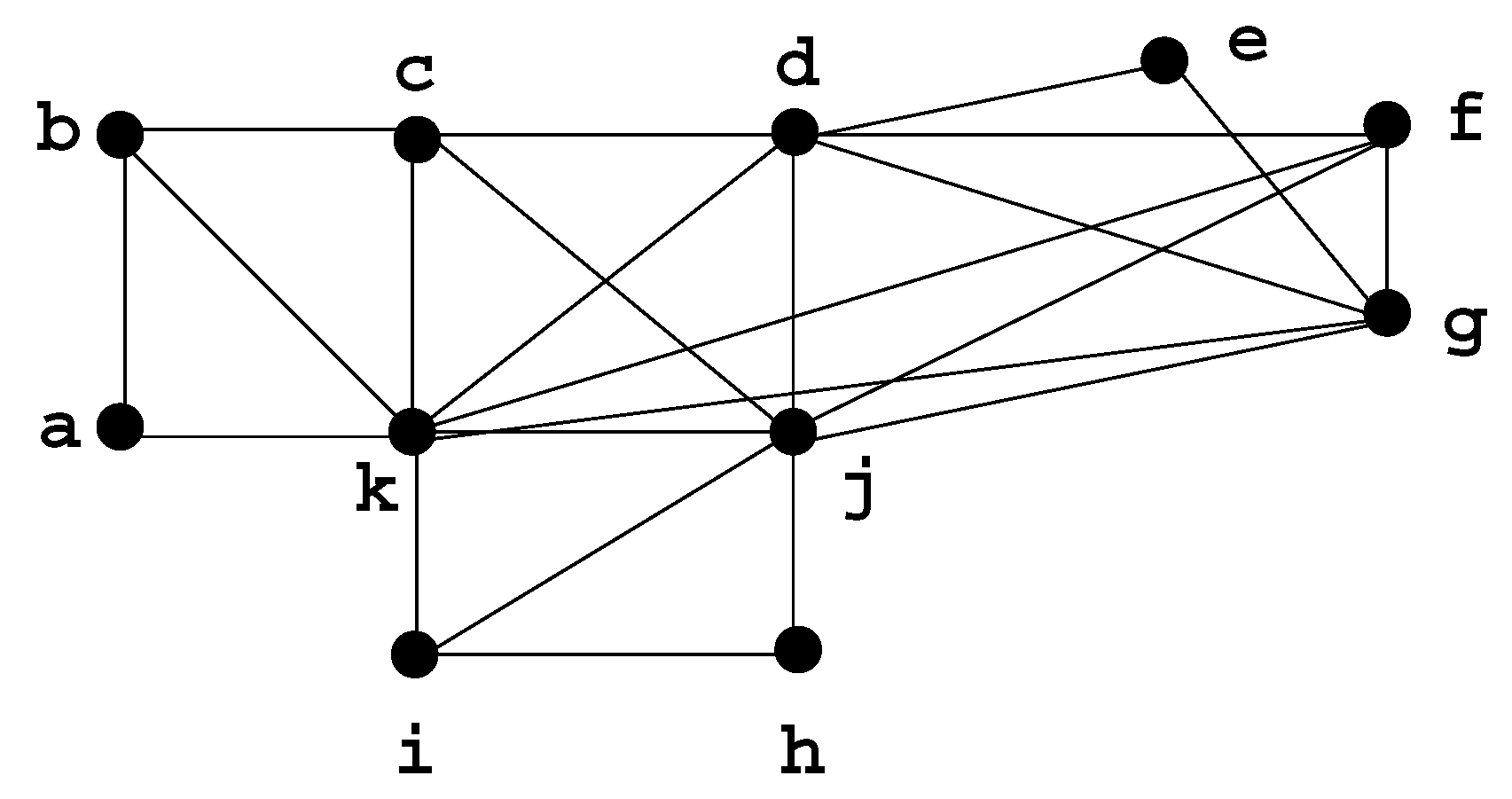
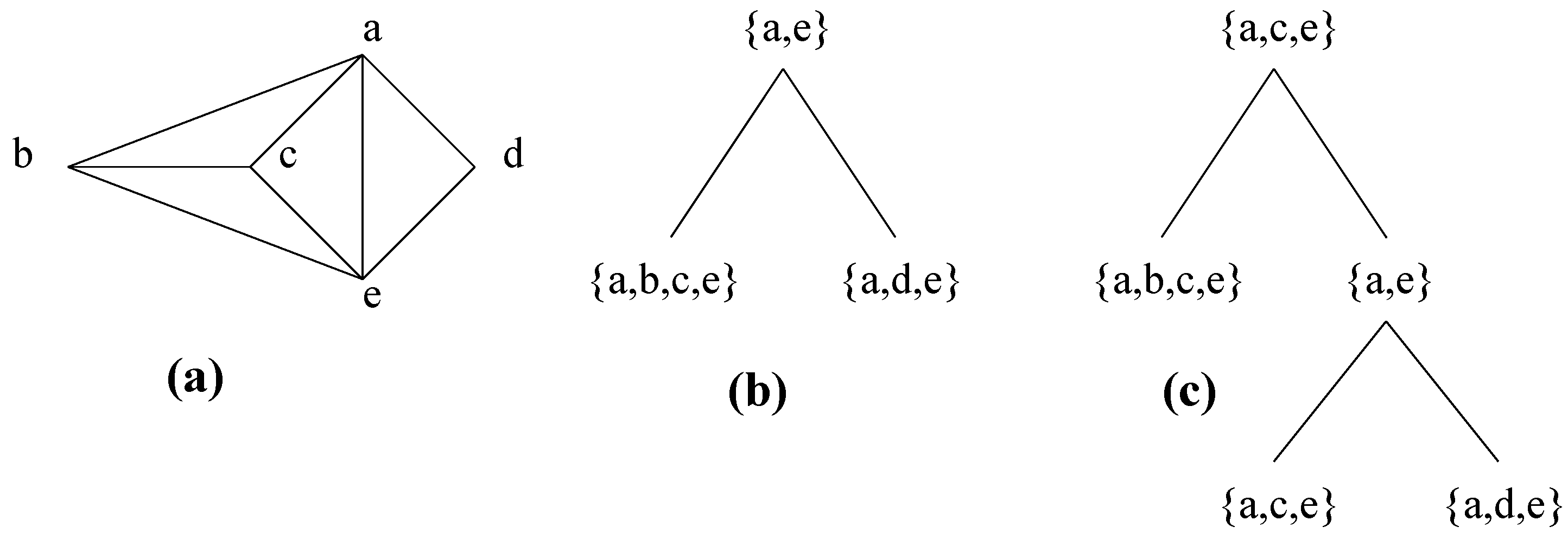
3.3. An equivalent process
3.4. How to compute the clique minimal separators

- compute a minimal triangulation H of G.
- compute the minimal separators of H.
- check each minimal separator of H to see whether it is a clique in G.
3.5. Some problems which can be solved using the atoms
- Minimal and minimum triangulation: if a minimal triangulation is computed for each of the atoms of the clique minimal separator decomposition of a graph, then the union of the set of fill edges obtained defines a minimal triangulation for G. Thus if for each atom a minimum-sized fill is computed, the resulting fill will be a minimum triangulation of G, since the sets of fill edges in the atoms are pairwise disjoint.
- Treewidth: the treewidth is obtained by taking the largest treewidth over all the atoms.
- Perfection: Any chordless cycle of length 4 or more is preserved by a decomposition step, as well as any antihole, so this decomposition preserves holes and antiholes. Thus a graph is perfect if and only if all its atoms are perfect.
- Coloring: An optimal coloring is obtained by merging optimal colorings of the atoms.
- Maximum clique: Any maximal clique of the graph is preserved after a decomposition step, so finding the size of the largest clique of each atom will yield the size of the largest clique of the original graph.
4. Algorithms and implementations



5. Theoretical and bibliographical background
5.1. A brief history of clique minimal separator decomposition
5.2. Computing the clique minimal separators of a graph

5.3. Decomposing a graph into atoms
5.4. The unicity of clique minimal separator decomposition
6. Conclusions
References
- Didi Biha, M.; Kaba, B.; Meurs, M.J.; SanJuan, E. Graph decomposition approaches for terminology graphs. Proc. MICAI 2007, 883–893. [Google Scholar]
- Kaba, B.; Pinet, N.; Lelandais, G.; Sigayret, A.; Berry, A. Clustering gene expression data using graph separators. In Silico Biol. 2007, 7, 433–452. [Google Scholar] [PubMed]
- Dirac, G.A. On rigid circuit graphs. Abh. Math. Sem. Univ. Hamburg 1961, 25, 71–76. [Google Scholar] [CrossRef]
- Rose, D.J. Triangulated graphs and the elimination process. J. Math. Anal. Appl. 1970, 32, 597–609. [Google Scholar] [CrossRef]
- Berry, A.; Pogorelcnik, R. A simple algorithm to generate the minimal separators of a chordal graph; Research Report LIMOS RR-10-04; LIMOS UMR CNRS: Aubière, France, 2010. [Google Scholar]
- Yannakakis, M. Computing the minimum fill-in is NP-complete. SIAM J. Algebr. Discrete Method 1981, 2, 77–79. [Google Scholar] [CrossRef]
- Kratsch, D.; Spinrad, J. Minimal fill in O(n2.69) time. Discrete Math. 2006, 306, 366–371. [Google Scholar] [CrossRef]
- Heggernes, P.; Telle, J.A.; Villanger, Y. Computing minimal triangulations in time O(nαlogn)=o(n2.376). SIAM J. Discrete Math. 2005, 19, 900–913. [Google Scholar] [CrossRef]
- Berry, A.; Blair, J.R.S.; Heggernes, P. Maximum cardinality search for computing minimal triangulations of graphs. Algorithmica 2004, 39, 287–298. [Google Scholar] [CrossRef]
- Berry, A.; Bordat, J.P.; Heggernes, P.; Simonet, G.; Villanger, Y. A wide-range algorithm for minimal triangulation from an arbitrary ordering. J. Algor. 2006, 58, 33–66. [Google Scholar] [CrossRef]
- Berry, A.; Heggernes, P.; Villanger, Y. A vertex incremental approach for maintaining chordality. Discrete Math. 2006, 306, 318–336. [Google Scholar] [CrossRef]
- Heggernes, P. Minimal triangulations of graphs: A survey. Discrete Math. 2006, 306, 297–317. [Google Scholar] [CrossRef]
- Rose, D.J.; Tarjan, R.E.; Lueker, G.S. Algorithmic aspects of vertex elimination on graphs. SIAM J. Comput. 1976, 5, 266–283. [Google Scholar] [CrossRef]
- Berry, A.; Krueger, R.; Simonet, G. Maximal label search algorithms to compute perfect and minimal elimination orderings. SIAM J. Discrete Math. 2009, 23, 428–446. [Google Scholar] [CrossRef]
- Tarjan, R.E. Decomposition by clique separators. Discrete Math. 1985, 55, 221–232. [Google Scholar] [CrossRef]
- Gavril, F. Algorithms on clique separable graphs. Discrete Math. 1977, 19, 159–165. [Google Scholar] [CrossRef]
- Whitesides, S. An algorithm for finding clique cutsets. Inf. Process. Lett. 1981, 12, 31–32. [Google Scholar] [CrossRef]
- Dahlhaus, E.; Karpinski, M.; Novick, M.B. Fast parallel algorithms for the clique separator decomposition. In Proceedings of the First Annual ACM-SIAM Symposium on Discrete Algorithms: SODA ’90, Philadelphia, PA, USA, January 1990; pp. 244–251.
- Leimer, H.G. Optimal decomposition by clique separators. Discrete Math. 1993, 113, 99–123. [Google Scholar] [CrossRef]
- Olesen, K.G.; Madsen, A.L. Maximal prime subgraph decomposition of Bayesian networks. IEEE Trans. Syst. Man Cybernet. B 2002, 32, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.R.S.; Peyton, B.W. An introduction to chordal graphs and clique trees. Graph Theory Sparse Matrix Comput. 1993, 84, 1–29. [Google Scholar]
- Brandstädt, A.; Hoàng, C.T. On clique separators, nearly chordal graphs, and the Maximum Weight Stable Set Problem. Theor. Comput. Sci. 2007, 389, 295–306. [Google Scholar] [CrossRef]
- Bodlaender, H.L.; Koster, A.M.C.A. Safe separators for treewidth. Discret Math. 2006, 306, 337–350. [Google Scholar] [CrossRef]
© 2010 by the authors. Licensee MDPI, Basel, Switzerland. This article is an Open Access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Berry, A.; Pogorelcnik, R.; Simonet, G. An Introduction to Clique Minimal Separator Decomposition. Algorithms 2010, 3, 197-215. https://doi.org/10.3390/a3020197
Berry A, Pogorelcnik R, Simonet G. An Introduction to Clique Minimal Separator Decomposition. Algorithms. 2010; 3(2):197-215. https://doi.org/10.3390/a3020197
Chicago/Turabian StyleBerry, Anne, Romain Pogorelcnik, and Geneviève Simonet. 2010. "An Introduction to Clique Minimal Separator Decomposition" Algorithms 3, no. 2: 197-215. https://doi.org/10.3390/a3020197
APA StyleBerry, A., Pogorelcnik, R., & Simonet, G. (2010). An Introduction to Clique Minimal Separator Decomposition. Algorithms, 3(2), 197-215. https://doi.org/10.3390/a3020197