HPANet: Hierarchical Path Aggregation Network with Pyramid Vision Transformers for Colorectal Polyp Segmentation
Abstract
1. Introduction
- Noise Interference: when the polyp image is disturbed by noise such as bubbles, it is easy to cause discontinuity and blurred segmentation edges.
- Large Differences in Shape and Size: the polyp scale is large, and the shape characteristics are also different, making it easy to generate incorrect segmentation when dealing with small polyps.
- Semantic Information Loss: through continuous sampling operations during the segmentation process, the semantic information contained in the polyp context is easily gradually lost.
2. Related Works
2.1. Polyp Segmentation
2.2. Vision Transformer
3. Materials and Methods
3.1. Overall Architecture
3.2. Encoder
3.3. Backward Multi-Scale Feature Fusion Module (BMFM)
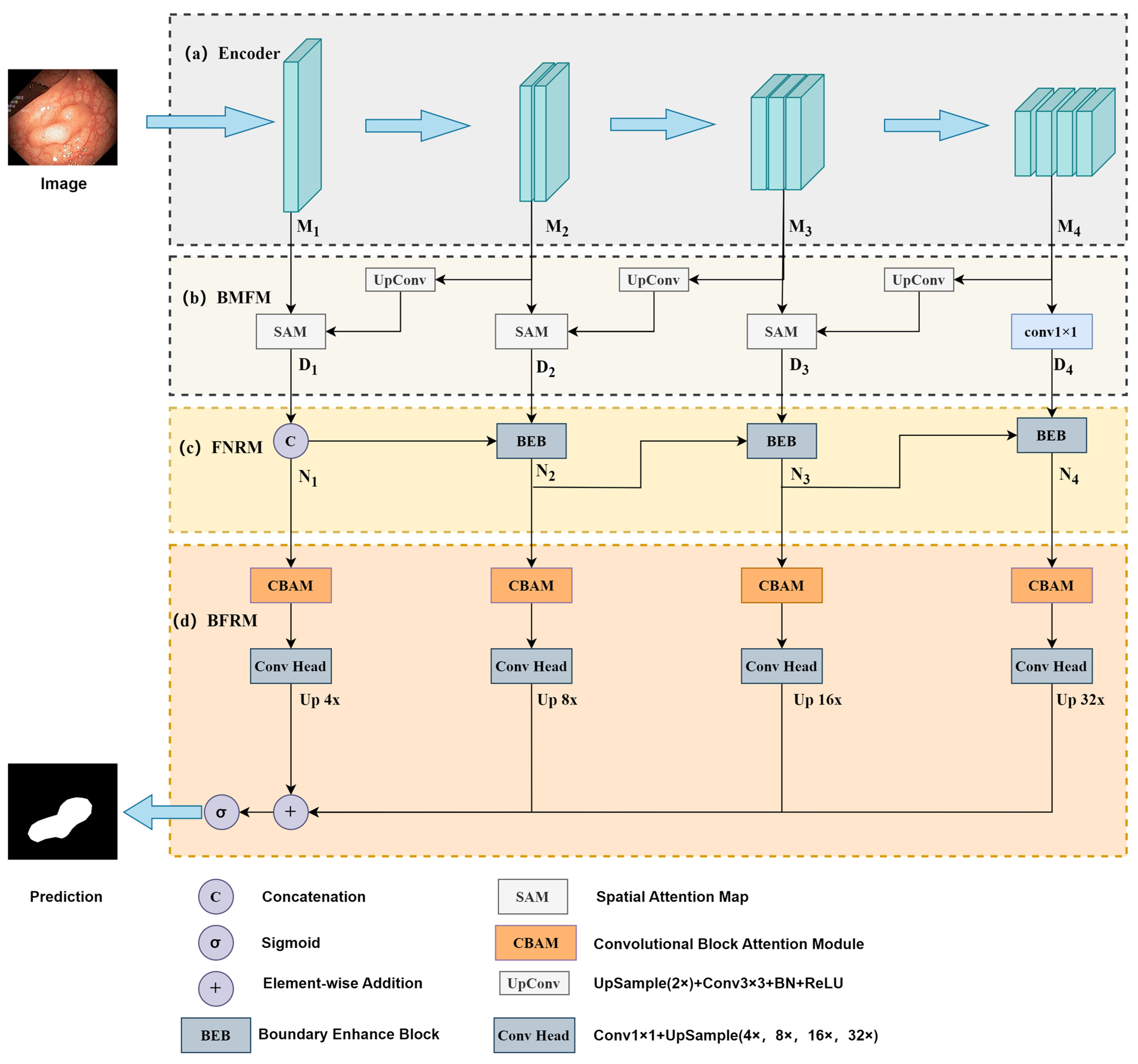
3.4. Forward Noise Reduction Module (FNRM)
3.5. Boundary Feature Refinement Module (BFRM)

3.6. Loss Function
4. Experiments and Results
4.1. Datasets and Training Settings
4.2. Learning Capacity
4.3. Generalization Capability
4.4. Comparative Analysis of Polyp Segmentation Performance Across Diverse Imaging Scenarios
4.5. Ablation Study
4.6. Comparison of Inference Time and Model Complexity
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CRC | Colorectal Cancer |
| OC | Optical Colonoscopy |
| CNN | Convolutional Neural Network |
| PVTv2 | Pyramid Vision Transformer v2 |
| ViT | Vision Transformer |
| MLP | Multi-Layer Perceptron |
| GAP | Global Average Pooling |
| CBAM | Convolutional Block Attention Module |
| SA | Spatial Attention |
| CA | Channel Attention |
| IoU | Intersection over Union |
| BCE | Binary Cross-Entropy |
| mDice | Mean Dice Coefficient |
| PPD | Parallel Partial Decoder |
| SAM | Spatial Attention Map |
| BEB | Boundary Enhance Block |
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.H.; Schrag, D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021, 325, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Fang, Y.; Zhu, D.; Yao, J.; Yuan, Y.; Tong, K. ABC-Net: Area-Boundary Constraint Network with Dynamical Feature Selection for Colorectal Polyp Segmentation. IEEE Sens. J. 2020, 21, 11799–11809. [Google Scholar] [CrossRef]
- Chen, F.; Ma, H.; Zhang, W. SegT: Separated Edge-Guidance Transformer Network for Polyp Segmentation. Math. Biosci. Eng 2023, 20, 17803–17821. [Google Scholar] [CrossRef]
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. PraNet: Parallel Reverse Attention Network for Polyp Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2020; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; Volume 12266, pp. 263–273. ISBN 978-3-030-59724-5. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Zhang, Z.; Zhang, W. Pyramid Medical Transformer for Medical Image Segmentation. arXiv 2022, arXiv:2104.14702. [Google Scholar]
- Chowdary, G.J.; Yin, Z. Med-Former: A Transformer Based Architecture for Medical Image Classification. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2024; Linguraru, M.G., Dou, Q., Feragen, A., Giannarou, S., Glocker, B., Lekadir, K., Schnabel, J.A., Eds.; Lecture Notes in Computer Science; Springer Nature Switzerland: Cham, Switzerland, 2024; Volume 15011, pp. 448–457. ISBN 978-3-031-72119-9. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.-P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. PVT v2: Improved Baselines with Pyramid Vision Transformer. Comput. Vis. Media 2022, 8, 415–424. [Google Scholar] [CrossRef]
- Gao, S.H.; Cheng, M.M.; Zhao, K.; Zhang, X.Y.; Yang, M.H.; Torr, P. Res2Net: A New Multi-Scale Backbone Architecture. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 652–662. [Google Scholar] [CrossRef]
- Wei, J.; Hu, Y.; Zhang, R.; Li, Z.; Zhou, S.K.; Cui, S. Shallow Attention Network for Polyp Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; De Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2021; Volume 12901, pp. 699–708. ISBN 978-3-030-87192-5. [Google Scholar]
- Tomar, N.K.; Jha, D.; Bagci, U.; Ali, S. TGANet: Text-Guided Attention for Improved Polyp Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2022; Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S., Eds.; Lecture Notes in Computer Science; Springer Nature Switzerland: Cham, Switzerland, 2022; Volume 13433, pp. 151–160. ISBN 978-3-031-16436-1. [Google Scholar]
- Oukdach, Y.; Garbaz, A.; Kerkaou, Z.; El Ansari, M.; Koutti, L.; El Ouafdi, A.F.; Salihoun, M. UViT-Seg: An Efficient ViT and U-Net-Based Framework for Accurate Colorectal Polyp Segmentation in Colonoscopy and WCE Images. J. Imaging Inform. Med. 2024, 37, 2354–2374. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Z.; Yu, J.; Gao, Y.; Liu, M. Multi-Scale Nested UNet with Transformer for Colorectal Polyp Segmentation. J. Appl. Clin. Med. Phys. 2024, 25, e14351. [Google Scholar] [CrossRef]
- Zhang, W.; Fu, C.; Zheng, Y.; Zhang, F.; Zhao, Y.; Sham, C.-W. HSNet: A Hybrid Semantic Network for Polyp Segmentation. Comput. Biol. Med. 2022, 150, 106173. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tian, S.; Yu, L.; Zhou, Z.; Wang, F.; Wang, Y. HIGF-Net: Hierarchical Information-Guided Fusion Network for Polyp Segmentation Based on Transformer and Convolution Feature Learning. Comput. Biol. Med. 2023, 161, 107038. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Wang, W.; Fan, D.P.; Li, J.; Fu, H.; Shao, L. Polyp-PVT: Polyp Segmentation with Pyramid Vision Transformers. CAAI Artif. Intell. Res. 2023, 2, 9150015. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Han, K.; Xiao, A.; Wu, E.; Guo, J.; Xu, C.; Wang, Y. Transformer in Transformer. Adv. Neural Inf. Process. Syst. 2021, 34, 15908–15919. [Google Scholar]
- Touvron, H.; Cord, M.; Sablayrolles, A.; Synnaeve, G.; Jégou, H. Going Deeper with Image Transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021; pp. 32–42. [Google Scholar]
- Zhou, D.; Kang, B.; Jin, X.; Yang, L.; Lian, X.; Jiang, Z.; Hou, Q.; Feng, J. DeepViT: Towards Deeper Vision Transformer. arXiv 2021, arXiv:2103.11886. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.-P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid Vision Transformer: A Versatile Backbone for Dense Prediction without Convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018. [Google Scholar]
- Bhojanapalli, S.; Chakrabarti, A.; Glasner, D.; Li, D.; Unterthiner, T.; Veit, A. Understanding Robustness of Transformers for Image Classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021. [Google Scholar]
- Khanh, T.L.B.; Dao, D.-P.; Ho, N.-H.; Yang, H.-J.; Baek, E.-T.; Lee, G.; Kim, S.-H.; Yoo, S.B. Enhancing U-Net with Spatial-Channel Attention Gate for Abnormal Tissue Segmentation in Medical Imaging. Appl. Sci. 2020, 10, 5729. [Google Scholar] [CrossRef]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA Maps for Accurate Polyp Highlighting in Colonoscopy: Validation vs. Saliency Maps from Physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; de Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-SEG: A Segmented Polyp Dataset. In Proceedings of the MultiMedia Modeling, Daejeon, Republic of Korea, 5–8 January 2020; Ro, Y.M., Cheng, W.-H., Kim, J., Chu, W.T., Cui, P., Choi, J.W., Hu, M.C., De Neve, W., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 451–462. [Google Scholar]
- Vázquez, D.; Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; López, A.M.; Romero, A.; Drozdzal, M.; Courville, A. A Benchmark for Endoluminal Scene Segmentation of Colonoscopy Images. J. Healthc. Eng. 2017, 2017, 4037190. [Google Scholar] [CrossRef]
- Geetha, K.; Rajan, C. Automatic Colorectal Polyp Detection in Colonoscopy Video Frames. Asian Pac. J. Cancer Prev. 2016, 17, 4869–4873. [Google Scholar] [CrossRef]
- Silva, J.; Histace, A.; Romain, O.; Dray, X.; Granado, B. Toward Embedded Detection of Polyps in WCE Images for Early Diagnosis of Colorectal Cancer. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C. ResUNet-a: A Deep Learning Framework for Semantic Segmentation of Remotely Sensed Data. ISPRS J. Photogramm. Remote Sens. 2020, 162, 94–114. [Google Scholar] [CrossRef]
- Lou, A.; Guan, S.; Loew, M. CaraNet: Context Axial Reverse Attention Network for Segmentation of Small Medical Objects. J. Med. Imaging 2023, 10, 014005. [Google Scholar] [CrossRef] [PubMed]




| Model | mIoU | mDice | Recall | Precision | F2 |
|---|---|---|---|---|---|
| U-Net [3] | 0.6497 | 0.742 | 0.6959 | 0.902 | 0.7094 |
| ResUnet [34] | 0.7637 | 0.8417 | 0.8372 | 0.8875 | 0.8344 |
| PraNet [7] | 0.8155 | 0.8802 | 0.8718 | 0.9239 | 0.8714 |
| CaraNet [35] | 0.8135 | 0.8758 | 0.8978 | 0.8952 | 0.8819 |
| Polyp-PVT [19] | 0.8595 | 0.9144 | 0.929 | 0.9201 | 0.9204 |
| HPANet | 0.8655 | 0.9204 | 0.9323 | 0.927 | 0.9251 |
| Datasets | Model | mIoU | mDice | Recall | Precision | F2 |
|---|---|---|---|---|---|---|
| CVC-ClinicDB | U-Net [3] | 0.4051 | 0.4902 | 0.4534 | 0.7521 | 0.4619 |
| ResUnet [34] | 0.5558 | 0.6314 | 0.6059 | 0.7899 | 0.6128 | |
| PraNet [7] | 0.7382 | 0.8029 | 0.8218 | 0.8228 | 0.8082 | |
| CaraNet [35] | 0.5578 | 0.6213 | 0.7612 | 0.6262 | 0.6658 | |
| Polyp-PVT [19] | 0.7273 | 0.8045 | 0.8301 | 0.836 | 0.8154 | |
| HPANet | 0.7539 | 0.8264 | 0.8632 | 0.8586 | 0.8397 | |
| CVC-300 | U-Net [3] | 0.2224 | 0.3025 | 0.2389 | 0.7127 | 0.2584 |
| ResUnet [34] | 0.7278 | 0.8041 | 0.8207 | 0.822 | 0.8122 | |
| PraNet [7] | 0.7879 | 0.8621 | 0.9423 | 0.8182 | 0.9007 | |
| CaraNet [35] | 0.7269 | 0.7962 | 0.8622 | 0.8178 | 0.8217 | |
| Polyp-PVT [19] | 0.7799 | 0.8587 | 0.9404 | 0.8232 | 0.8978 | |
| HPANet | 0.827 | 0.8965 | 0.9613 | 0.8563 | 0.9302 | |
| CVC-ColonDB | U-Net [3] | 0.1855 | 0.252 | 0.2301 | 0.5649 | 0.2273 |
| ResUnet [34] | 0.4806 | 0.5507 | 0.5226 | 0.7056 | 0.5307 | |
| PraNet [7] | 0.556 | 0.6327 | 0.7078 | 0.6435 | 0.6486 | |
| CaraNet [35] | 0.4987 | 0.5635 | 0.6045 | 0.6488 | 0.5714 | |
| Polyp-PVT [19] | 0.6913 | 0.7773 | 0.7995 | 0.817 | 0.7842 | |
| HPANet | 0.7044 | 0.7867 | 0.815 | 0.8099 | 0.7953 | |
| ETIS-LaribPolypDB | U-Net [3] | 0.257 | 0.3053 | 0.312 | 0.4734 | 0.2977 |
| ResUnet [34] | 0.4656 | 0.5327 | 0.5519 | 0.6082 | 0.5386 | |
| PraNet [7] | 0.4626 | 0.5239 | 0.5881 | 0.5204 | 0.5461 | |
| CaraNet [35] | 0.4026 | 0.4782 | 0.4778 | 0.6013 | 0.4676 | |
| Polyp-PVT [19] | 0.606 | 0.6862 | 0.7436 | 0.7361 | 0.7131 | |
| HPANet | 0.6268 | 0.6965 | 0.7458 | 0.7061 | 0.7198 |
| BMFM | BFRM | FNRM | mIoU | mDice | Recall | Precision | F2 |
|---|---|---|---|---|---|---|---|
| × | × | × | 0.8392 | 0.8955 | 0.9061 | 0.9189 | 0.8934 |
| √ | × | × | 0.8455 | 0.9004 | 0.9023 | 0.9276 | 0.8961 |
| √ | √ | × | 0.8623 | 0.9129 | 0.9207 | 0.9292 | 0.9127 |
| √ | √ | √ | 0.8655 | 0.9204 | 0.9323 | 0.927 | 0.9251 |
| Model | Params ↓ (M) | Flops ↓ (G) | Inference Time ↑ (FPS) |
|---|---|---|---|
| U-Net [3] | 17.26 | 227.53 | 32 |
| ResUnet [34] | 32.63 | 156.3 | 35 |
| PraNet [7] | 30.5 | 13.15 | 46 |
| CaraNet [35] | 44.59 | 21.75 | 38 |
| Polyp-PVT [19] | 28.11 | 17.02 | 40 |
| HPANet | 26.3 | 15.4 | 42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ying, Y.; Li, H.; Zhong, Y.; Lin, M. HPANet: Hierarchical Path Aggregation Network with Pyramid Vision Transformers for Colorectal Polyp Segmentation. Algorithms 2025, 18, 281. https://doi.org/10.3390/a18050281
Ying Y, Li H, Zhong Y, Lin M. HPANet: Hierarchical Path Aggregation Network with Pyramid Vision Transformers for Colorectal Polyp Segmentation. Algorithms. 2025; 18(5):281. https://doi.org/10.3390/a18050281
Chicago/Turabian StyleYing, Yuhong, Haoyuan Li, Yiwen Zhong, and Min Lin. 2025. "HPANet: Hierarchical Path Aggregation Network with Pyramid Vision Transformers for Colorectal Polyp Segmentation" Algorithms 18, no. 5: 281. https://doi.org/10.3390/a18050281
APA StyleYing, Y., Li, H., Zhong, Y., & Lin, M. (2025). HPANet: Hierarchical Path Aggregation Network with Pyramid Vision Transformers for Colorectal Polyp Segmentation. Algorithms, 18(5), 281. https://doi.org/10.3390/a18050281







