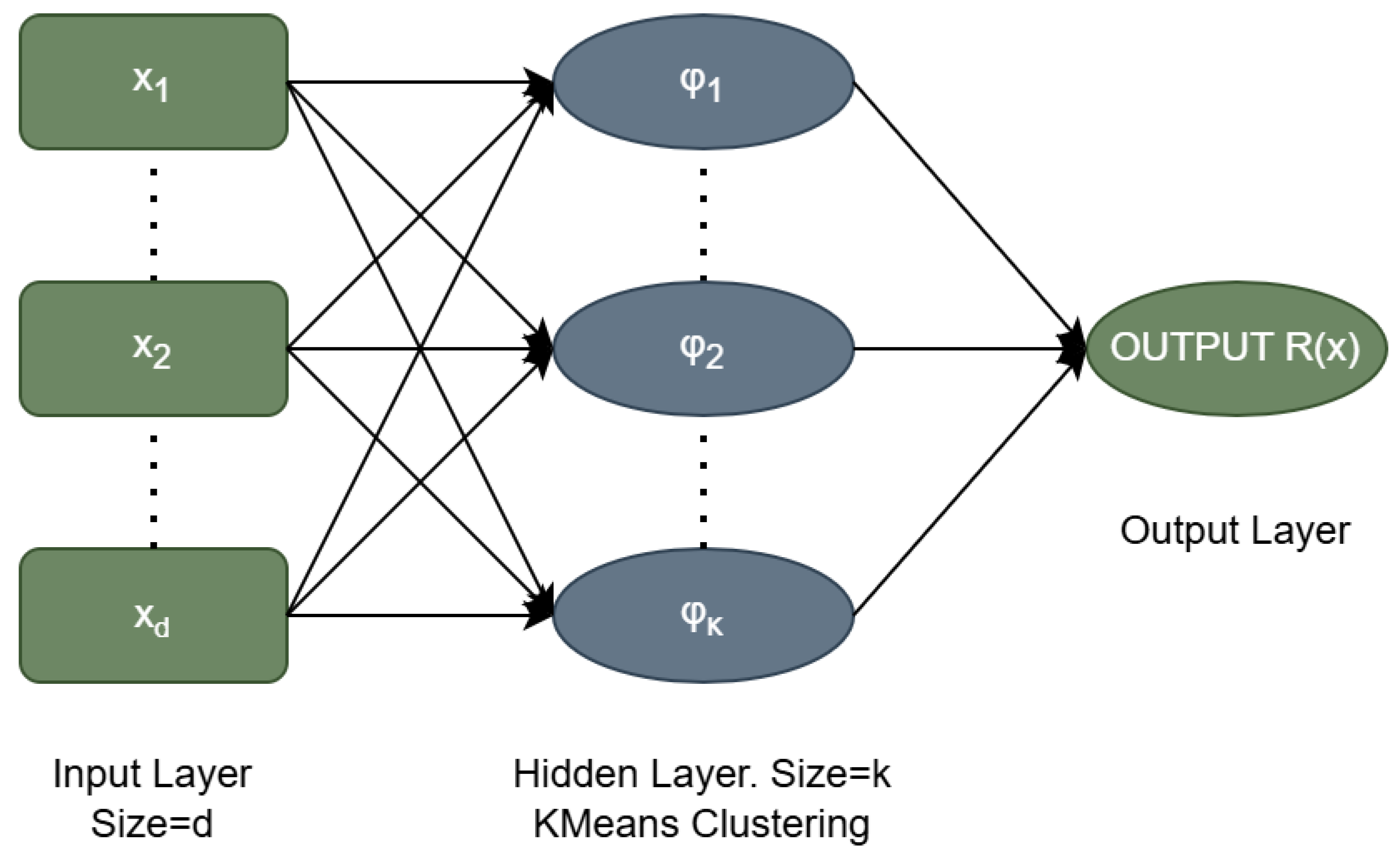
A series of classification and regression datasets from various websites was incorporated to test the proposed method and to measure its reliability. The datasets used in the conducted experiments were downloaded from the following online databases:
3.2. Experimental Results
The code used in the experiments was implemented in the C++ programming language and a machine equipped with 128 GB RAM running Debian Linux was utilized in the conducted experiments. The code was written with the assistance of the freely available GlobalOptimus optimization environment, that can be downloaded from
https://github.com/itsoulos/GlobalOptimus (accessed on 12 April 2025). Each experiment was executed 30 times and the average classification error was measured for the classification datasets and the average regression error was measured for the regression datasets. The classification error was computed using the following equation:
where the set
stands for the test set of the current problem and
is the RBF model. The regression error was calculated through the following equation:
The values for the parameters of the proposed method are mentioned in
Table 1. The selection of values for the experimental parameters was performed in such a way that there was a compromise between the speed and reliability of the proposed methodology. In the following tables, that describe the experimental results, the following notation is used:
The column Dataset represents the name of the objective problem.
The column BFGS denotes the application of the BFGS optimization method [
80] in the training of a neural network [
81,
82] with 10 processing nodes.
The column ADAM stands for the incorporation of the ADAM optimizer [
83] to train an artificial neural network with 10 processing nodes.
The column NEAT represents the usage of the NEAT method (NeuroEvolution of Augmenting Topologies) [
84].
The column RBF-KMEANS stands for the usage of the original two-phase method to train an RBF network with 10 processing nodes.
The column GENRBF represents the incorporation of the method proposed in [
85] to train an RBF network with 10 processing nodes.
The column Proposed denotes the usage of the proposed method to train an RBF network with 10 processing nodes.
The row Average represents the average classification or regression error.
The row W-average denotes the average classification error for all datasets and for each method. In this average, each individual classification error was multiplied by the number of patterns for the corresponding dataset.
The results from the application of the previously mentioned machine learning methods to the classification datasets are depicted in
Table 2, and for the regression datasets the results are presented in
Table 3.
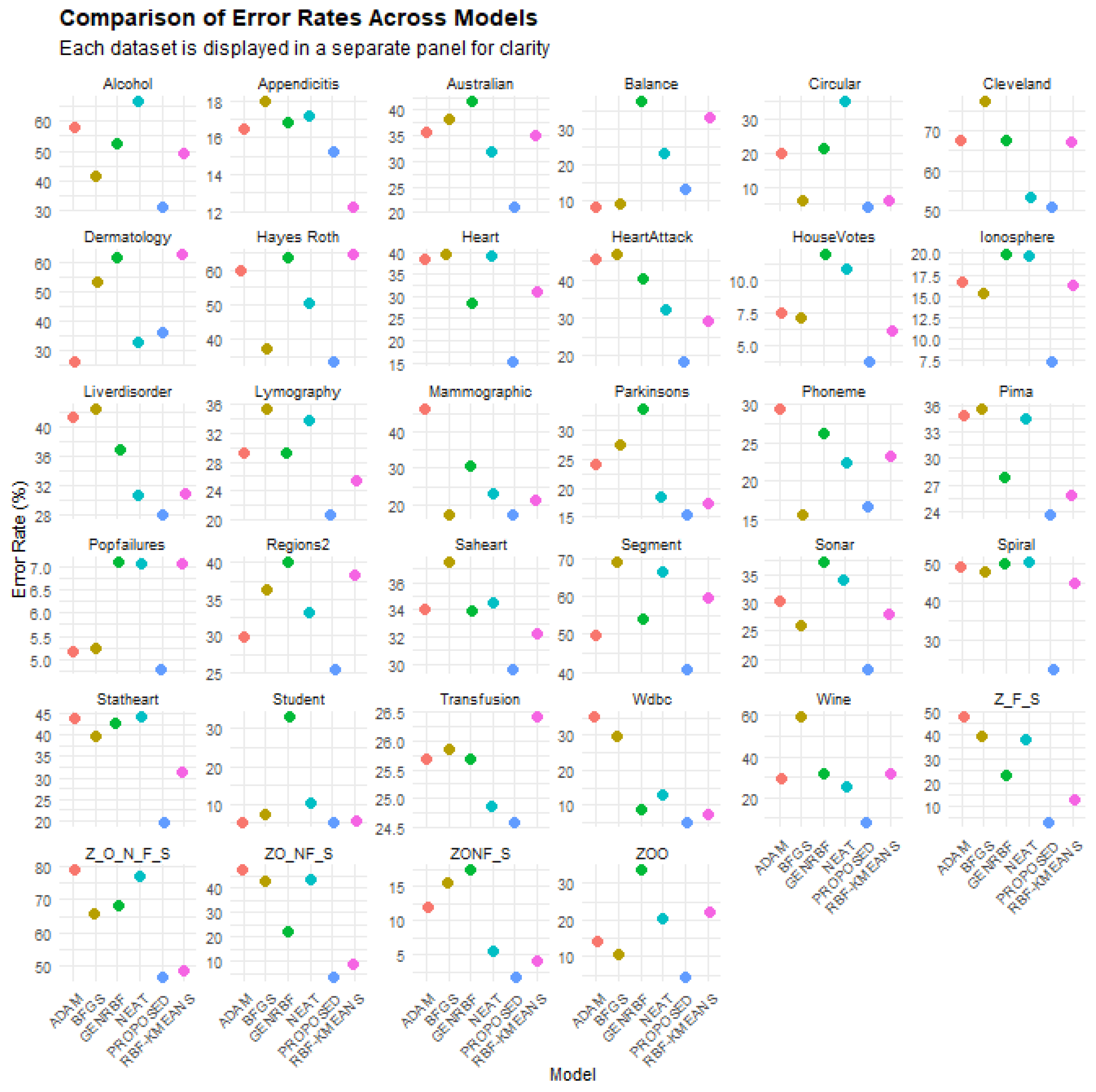
Table 2 presents the error rates of the various machine learning models (BFGS, ADAM, NEAT, RBF-KMEANS, GENRBF, proposed) on the different classification datasets. Each row corresponds to a dataset, while each column represents the error rate of a specific model. These values indicate the percentage of incorrect predictions, with lower values reflecting better performance. The last row of the table includes the average error rates for each model. Statistical analysis of the data reveals significant insights. The proposed model exhibits the lowest average error rate (18.67%) compared to the other models, establishing it as the optimal choice based on the table. Conversely, the other models demonstrate higher average error rates, with GENRBF showing the highest average error (34.64%). Additionally, significant variations in error rates across datasets are observed. For instance, on the “Circular” and “ZONF_S” datasets, the proposed model outperforms others, with very low error rates (4.19% and 1.79%, respectively). Conversely, on datasets like “Cleveland”, the NEAT model shows a lower error rate (53.44%) compared to the proposed model (50.82%). Notably, in certain datasets, the performance of the proposed model is significantly inferior to other models. For example, on the “Alcohol” and “Z_F_S” datasets, the proposed model exhibits much higher error rates compared to other models. This indicates that while the proposed model generally has the lowest average error rate, its performance may not be consistent across all datasets. In conclusion, the proposed model emerges as the best general choice for minimizing error rates, though its evaluation depends on the characteristics of each dataset. The performance differences among models highlight the need for careful model selection depending on the application.
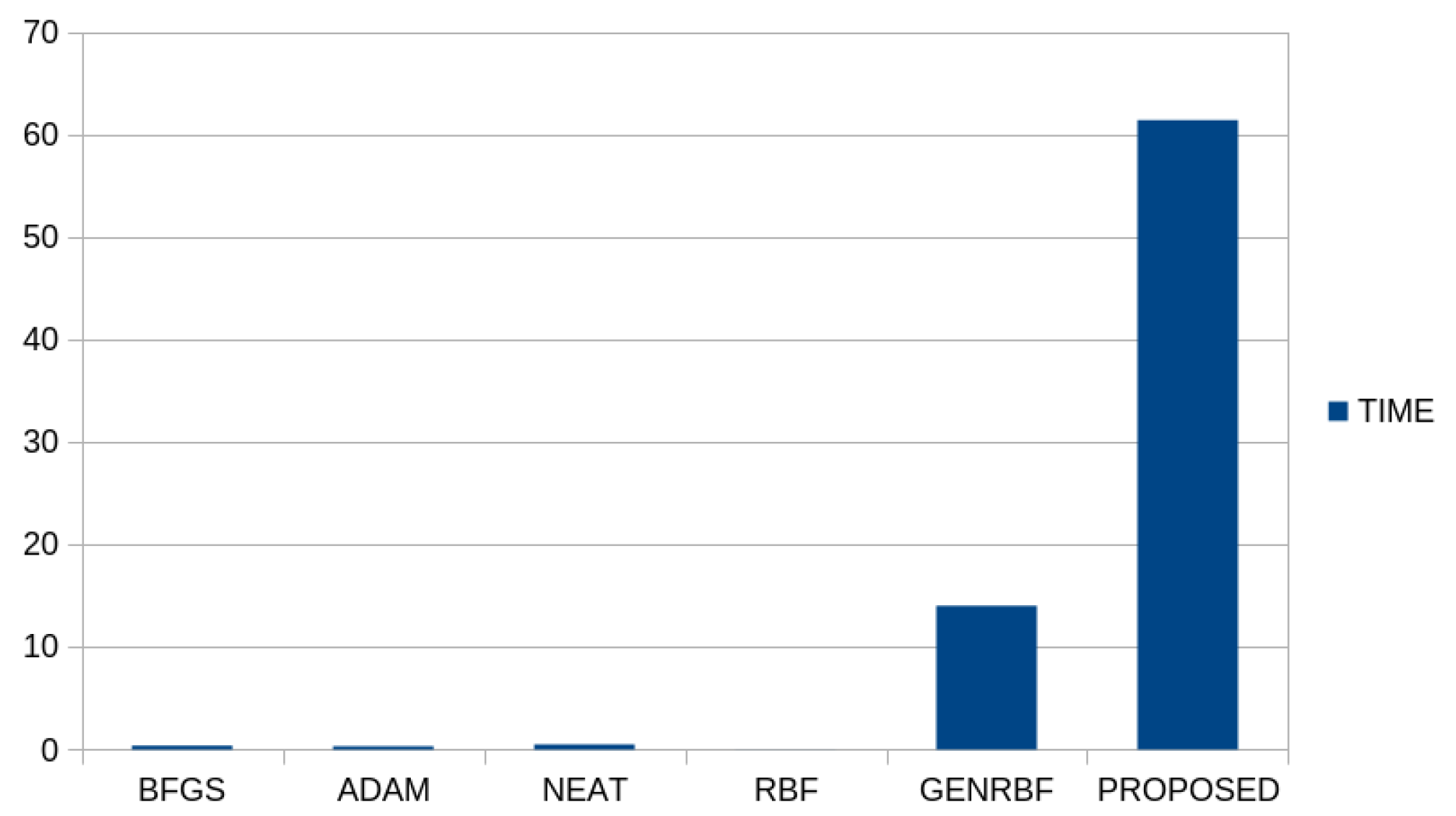
Also, the average execution time for each machine learning technique that was applied to the classification datasets is depicted in
Figure 4.
As expected, the proposed technique requires significantly more execution time than all the other techniques in the set, since it consists of the serial execution of global optimization techniques. Moreover, the method entitled GENRBF also required a significant amount of time with respect to other simpler methods in the set. The additional time required by the proposed technique can of course be significantly reduced by the use of parallel processing techniques in its various stages, such as, for example, the use of parallel Simulated Annealing techniques [
86]. Moreover,
Figure 5 depicts a comparison of the error rates across the models for all classification datasets involved in the conducted experiments.
Table 3 displays the absolute error values resulting from the application of the various machine learning models (BFGS, ADAM, NEAT, RBF-KMEANS, GENRBF, proposed) on the regression datasets. Each row corresponds to a dataset, while each column shows the error of a specific model. The last row records the average error for each model. Lower error values indicate better model performance. The analysis shows that the proposed model has the lowest average error (5.48), making it the most efficient choice among the available models. The second-best model is RBF-KMEANS, with an average error of 9.19, while other models, such as BFGS (26.43) and ADAM (19.62), exhibit significantly higher error values. The performance of the proposed model is particularly impressive on datasets such as BL, where its error is nearly negligible (0.0002), and Mortgage, where it has a very low error (0.14) compared to other models. On datasets like Stock and Plastic, where errors are high across all models, the proposed model still outperforms the other models, with error values of 1.53 and 2.29, respectively. However, there are instances where the performance difference of the proposed model relative to others is small or even unfavorable. For example, on the Laser dataset, the ADAM model has an error of 0.03, slightly higher than the proposed model’s 0.003, while on the HO dataset, the proposed model performs better (0.01), but the RBF-KMEANS model is comparably close (0.03). In summary, the proposed model achieves the lowest average error and the most consistent performance across most datasets, making it an ideal choice for regression problems. Nonetheless, certain models, such as RBF-KMEANS, may demonstrate competitive performance in specific cases, suggesting that model selection depends on the unique characteristics of each dataset.
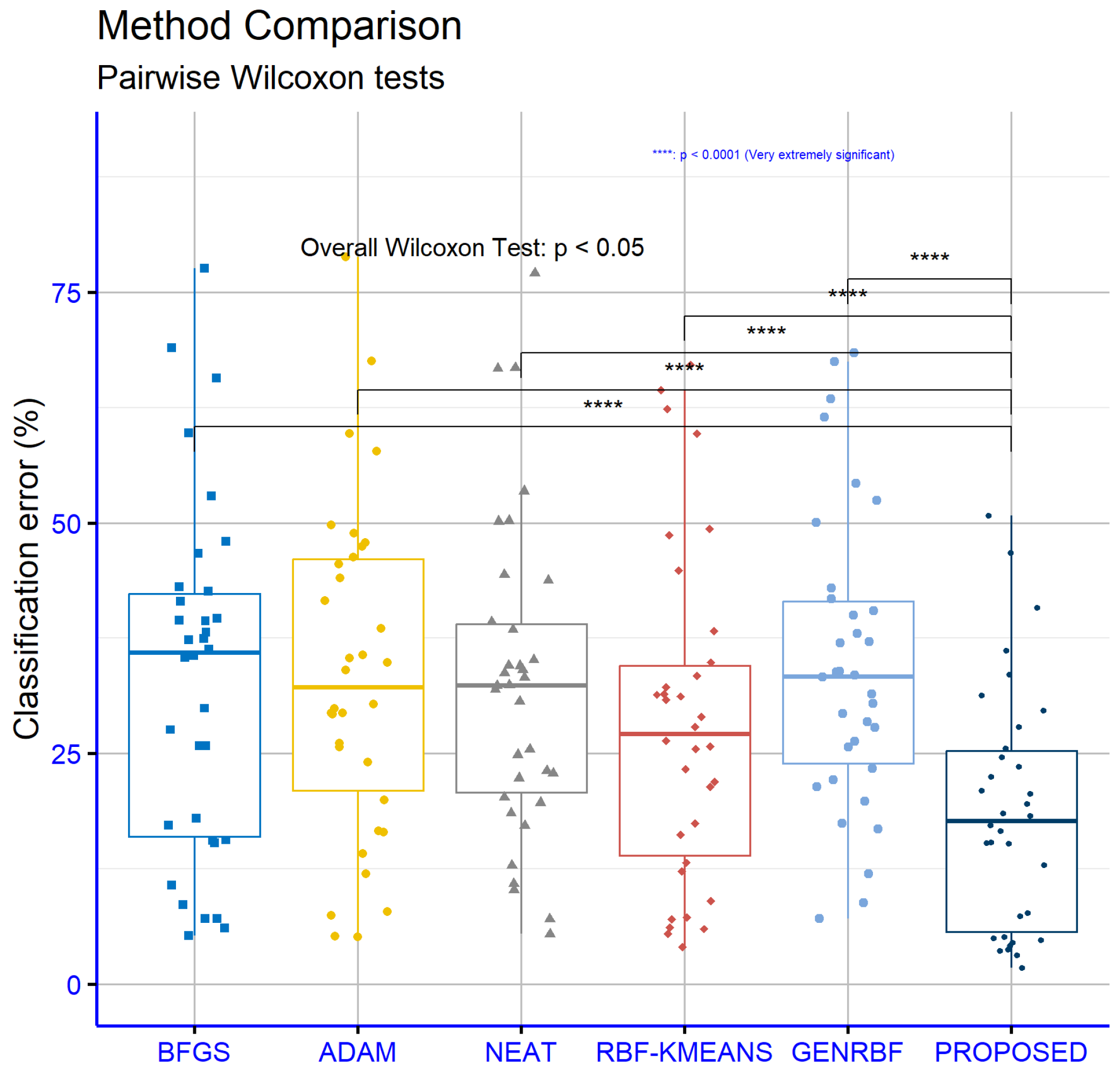
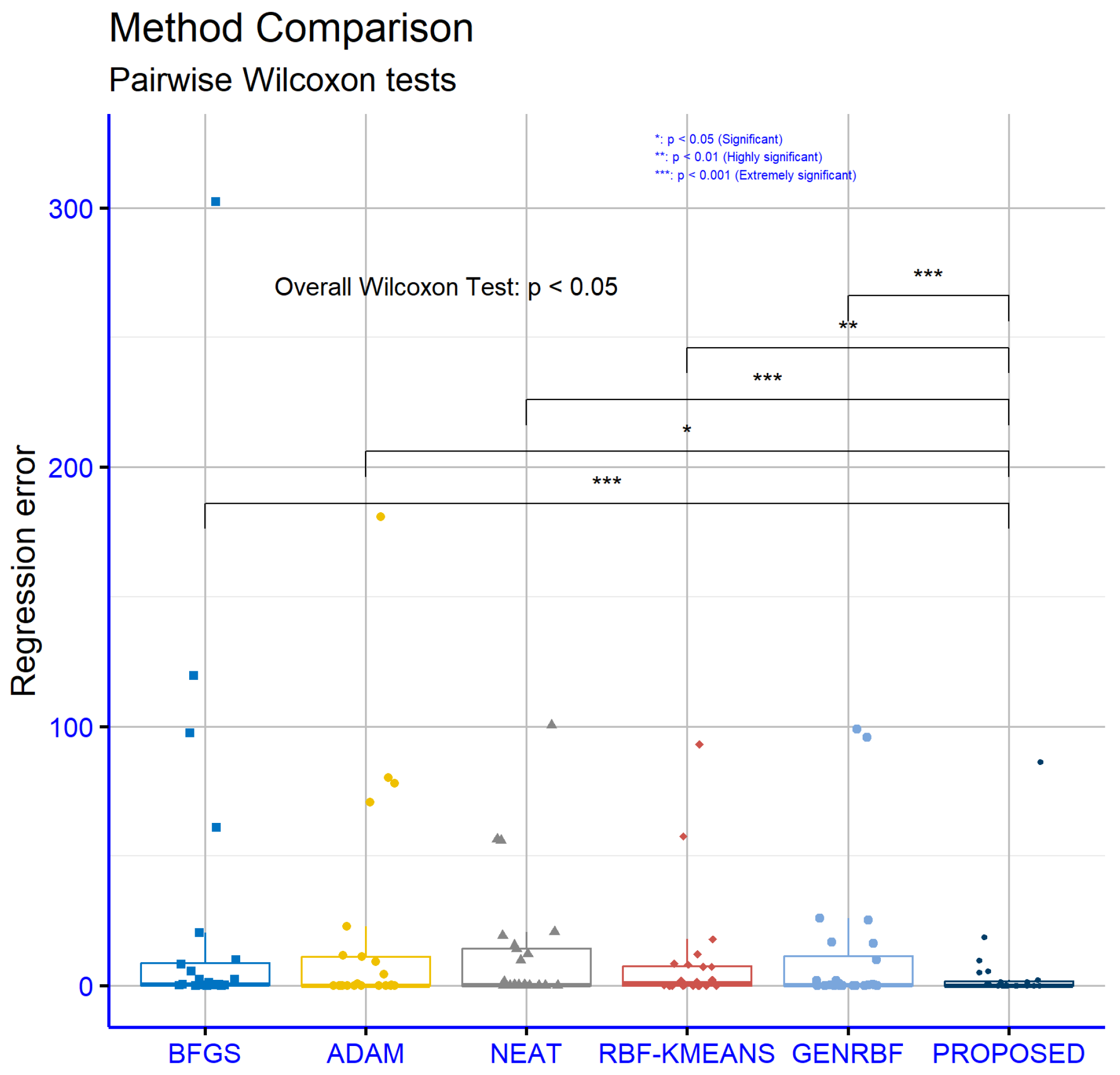
An analysis of significance levels for the classification datasets, as illustrated in
Figure 6, reveals that the proposed model statistically significantly outperforms all other models in every comparison pair. Specifically, the
p-values indicate strong statistical differences: Proposed vs. BFGS
, proposed vs. ADAM
, proposed vs. NEAT
, proposed vs. RBF-KMEANS
, and proposed vs. GENRBF
. These values suggest that the proposed model is significantly better than the others with high reliability.
In
Figure 7, which concerns the regression datasets, a similar pattern is observed, though the
p-values are generally higher compared to the classification datasets. The proposed model demonstrates statistically significant superiority over the other models in all comparison pairs: Proposed vs. BFGS (
), proposed vs. ADAM (
), proposed vs. NEAT (
), proposed vs RBF-KMEANS (
), and proposed vs. GENRBF (
). Although the significance is not as strong as in the classification datasets, the proposed model’s superiority remains clear.
3.3. Experiments on the Perturbation Factor a
In order to determine the stability of the proposed technique, another experiment was performed in which the perturbation factor
a, presented in the second stage of the proposed technique, took a series of different values.
Table 4 presents the error rates of the proposed machine learning model for three different values of the perturbation factor
a (0.001, 0.005, 0.01) across various classification datasets. Each row represents a dataset, and the values indicate the model’s error rate for each value of
a. The last row includes the average error rate for each value of
a. Analysis of the data shows that the smallest value of
a (0.001) achieves the lowest average error rate (18.67%), while the largest value (0.01) results in the highest average (19.06%). This suggests that the model generally performs better with smaller values of
a, although the difference in averages is minimal. At the dataset level, there are cases where the model’s performance is significantly affected by changes in the parameter. For instance, on the “Lymography” dataset, increasing
a from 0.001 to 0.01 leads to a significant increase in the error rate, from 20.64% to 30.33%. A similar trend is observed on the “ZOO” dataset, where the error rate rises from 4.50% to 6.87% for
, but decreases again to 4.60% for
. On the other hand, on datasets like “ZO_NF_S”, the error remains unchanged at 3.63%, regardless of changes in
a. Datasets such as “Z_F_S” and “ZONF_S” exhibit nonlinear behavior. On “Z_F_S”, the error rate significantly decreases from 3.16% to 2.79% as
a increases from 0.001 to 0.01, while on “ZONF_S”, a similar decrease is observed from 1.79% to 1.74%. In conclusion, the analysis indicates that the perturbation factor
a has a notable impact on the performance of the proposed model. Smaller values of
a are generally associated with better performance; however, the optimal value may depend on the characteristics of each dataset. Instances where error rates increase or decrease nonlinearly with changes in “a” suggest the need for further investigation into the tuning of “a” for specific applications.
Table 5 displays the absolute error values of the proposed machine learning model across various regression datasets for three different values of the perturbation factor
a (0.001, 0.005, 0.01). Data analysis reveals that the parameter
yields the lowest average error (4.92), while the values
and
result in slightly higher averages (5.48 and 5.13, respectively). This difference indicates that 0.005 is generally the most suitable value for the model, ensuring better performance in most cases. At the dataset level, the impact of
a varies. Some datasets, such as “Airfoil”, “Concrete”, “Dee”, “HO”, “Laser”, “NT”, “PL”, “Plastic”, and “Quake”, show no change in error with variations in
a as the error values remain constant. In contrast, other datasets exhibit significant variations. For example, on the “Baseball” dataset, the error decreases from 86.19 for
to 77.46 for
then increases again to 81.97 for
. Similarly, on the “MB” dataset, the error drastically decreases from 5.49 for
to 0.56 for
and further to 0.48 for
. On datasets like “FA” and “FY”, the error increases as
a changes from 0.001 to 0.005, then decreases again for
. On the “Treasury” dataset, the error shows a slight decline as
a increases. In conclusion, the parameter
a has a significant impact on the model’s performance on certain datasets, while on others, its effect is negligible. The lowest average error observed for
suggests that this value is generally optimal for the model, though further tuning may be required for specific datasets. Cases with high variability in errors highlight the need for deeper analysis and optimization of the
a parameter based on the characteristics of each dataset.
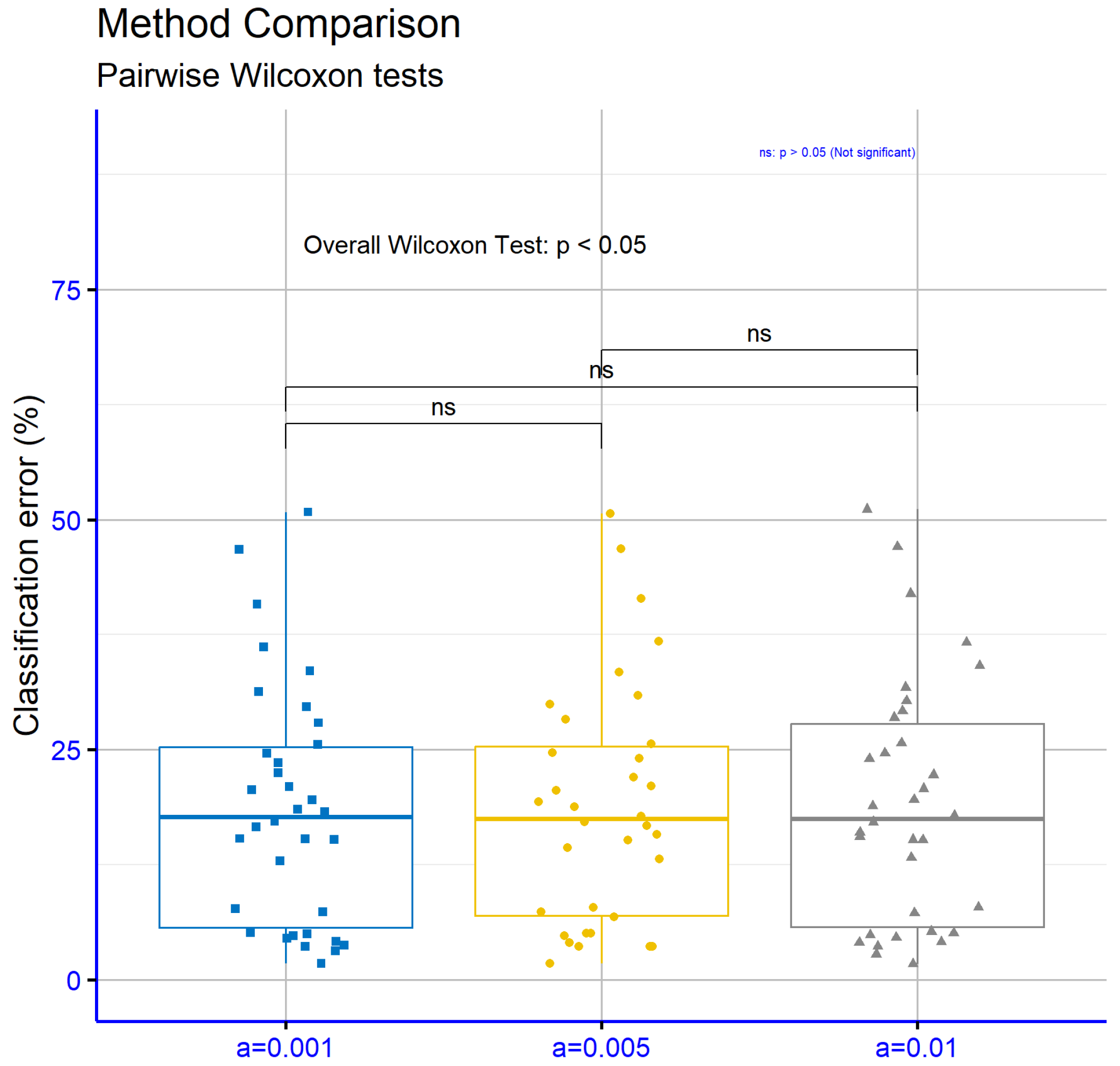
Figure 8 compares different values of the parameter
a for the classification datasets. The
p-values for the comparisons
vs.
(
),
vs.
(
), and
vs.
(
) indicate that the differences between the parameter values are not statistically significant. This suggests that varying the parameter
a within this range does not substantially affect the model’s performance on these datasets.
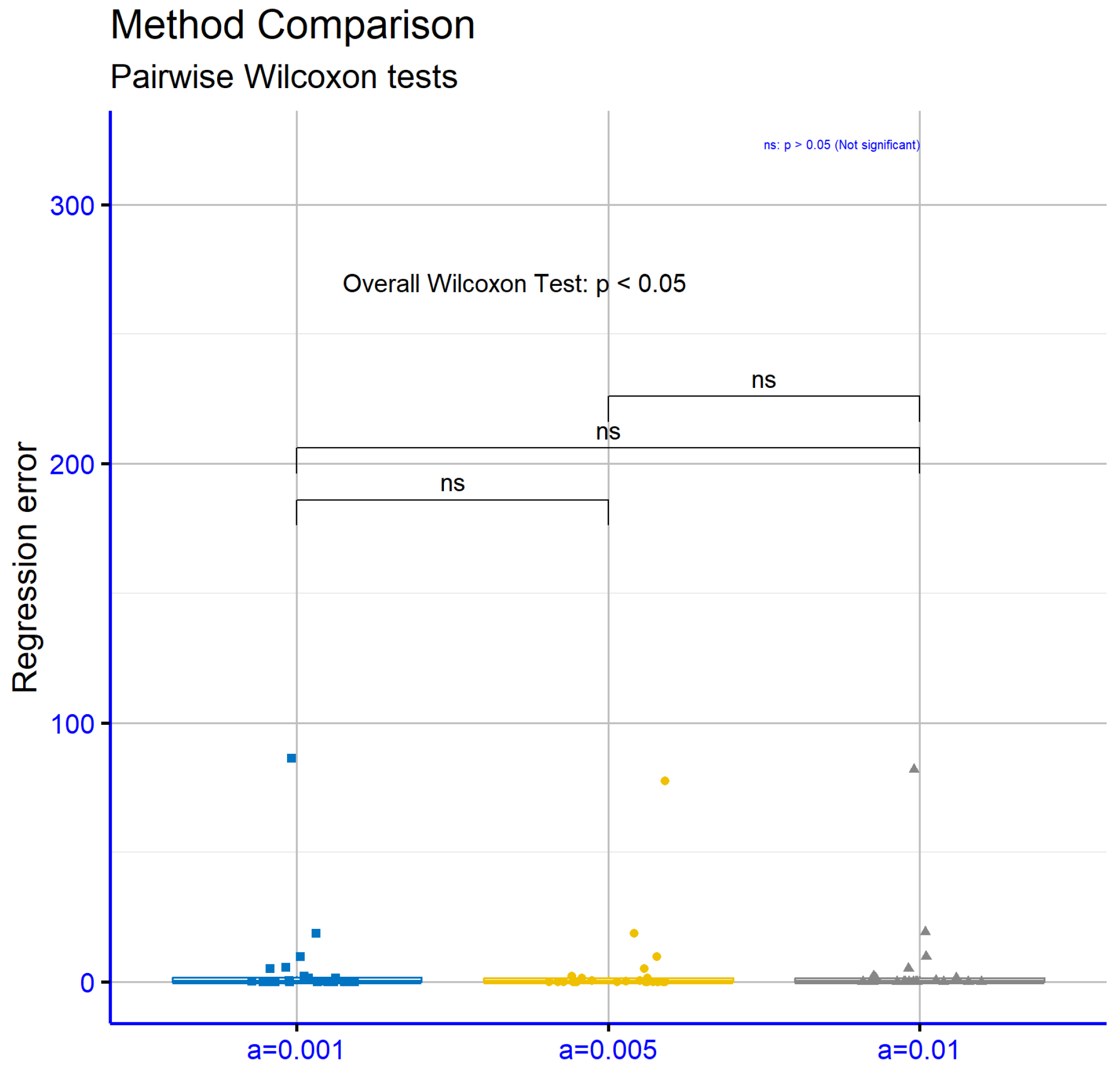
Figure 9 presents corresponding comparisons for the regression datasets, where a similar result is observed. The
p-values for the comparisons
vs.
(
),
vs.
(
), and
vs.
(
) indicate the absence of statistically significant differences. This shows that the choice of parameter
a does not significantly influence the model’s performance on the regression datasets.
3.4. Experiments on the Parameter F
Another experiment was conducted using the initialization factor
F.
Table 6 presents the percentage error rates of the proposed machine learning model across various classification datasets for four different values of the parameter
F (1.5, 3.0, 5.0, 10.0). Analyzing the data reveals that the parameter
F influences the model’s performance, but this effect varies by dataset. The lowest average error rate is observed for
(18.58%), indicating that this value is generally optimal. For the other values, slightly higher average error rates are noted: 18.88% for
; 18.67% for
; and the highest rate, 20.32%, for
. Examining individual datasets, it is evident that for many of them, increasing
F improves performance, as reflected in reduced error rates. Examples include the “Ionosphere”, “Wine”, and “ZONF_S” datasets, where error rates decrease as
F increases. On “Ionosphere”, the error rate drops from 12.92% for
to 7.39% for
. On “Wine”, the error rate decreases from 10.90% for
to 7.71% for
. Similarly, on “ZONF_S”, the error rate steadily decreases from 2.59% for
to 1.79% for
. However, there are cases where increasing
F does not lead to improvement or results in higher error rates. For example, on the “Segment” dataset, the error rate rises from 35.81% for
to 40.83% for
. On the “Spiral” dataset, the error rate consistently increases from 13.28% for
to 22.52% for
. A similar trend is observed on the “Z_O_N_F_S” dataset, where the error rate rises from 46.00% for
to 46.77% for
. Overall, the parameter
F significantly affects the model’s performance, and the optimal value appears to be
, as evidenced by the lowest average error rate. However, the exact impact depends on the characteristics of each dataset, emphasizing the need to fine-tune the parameter value for specific datasets to achieve optimal performance.
Table 7 provides the absolute error values of the proposed machine learning model across various regression datasets for four different values of the parameter
F (1.5, 3.0, 5.0, 10.0). The data analysis shows that the parameter
F affects the model’s performance differently depending on the dataset. The average errors indicate that
yields the lowest overall error (5.22), followed by
, with an average of 5.25. Higher averages are observed for
(5.52) and
(5.48), suggesting that deviating from
tends to increase error in some cases. Examining the datasets, it is evident that in several cases, increasing
F improves performance, reducing error rates. For example, on the “Abalone” dataset, the error decreases from 6.39 for
to 5.10 for
. Similarly, on the “Friedman” dataset, the error significantly decreases from 6.59 for
to 1.45 for
. On the “Laser” dataset, the error decreases progressively from 0.022 for
to 0.003 for
. Conversely, there are datasets where the effect of
F is nonlinear or increases the error rate. For instance, on the “Housing” dataset, the error rises from 16.75 for
to 18.70 for
. On the “MB” dataset, there is a sharp increase in error from 0.116 for
to 5.49 for
, indicating that
F significantly impacts model performance for this dataset. In summary, the parameter
F has varying effects on the model’s performance across different datasets. While the average indicates that
is the optimal choice, precise optimization of the parameter should be dataset-specific. Additionally, extreme parameter values may lead to significant performance degradation in certain datasets, as seen in examples like “MB” and “Housing”.
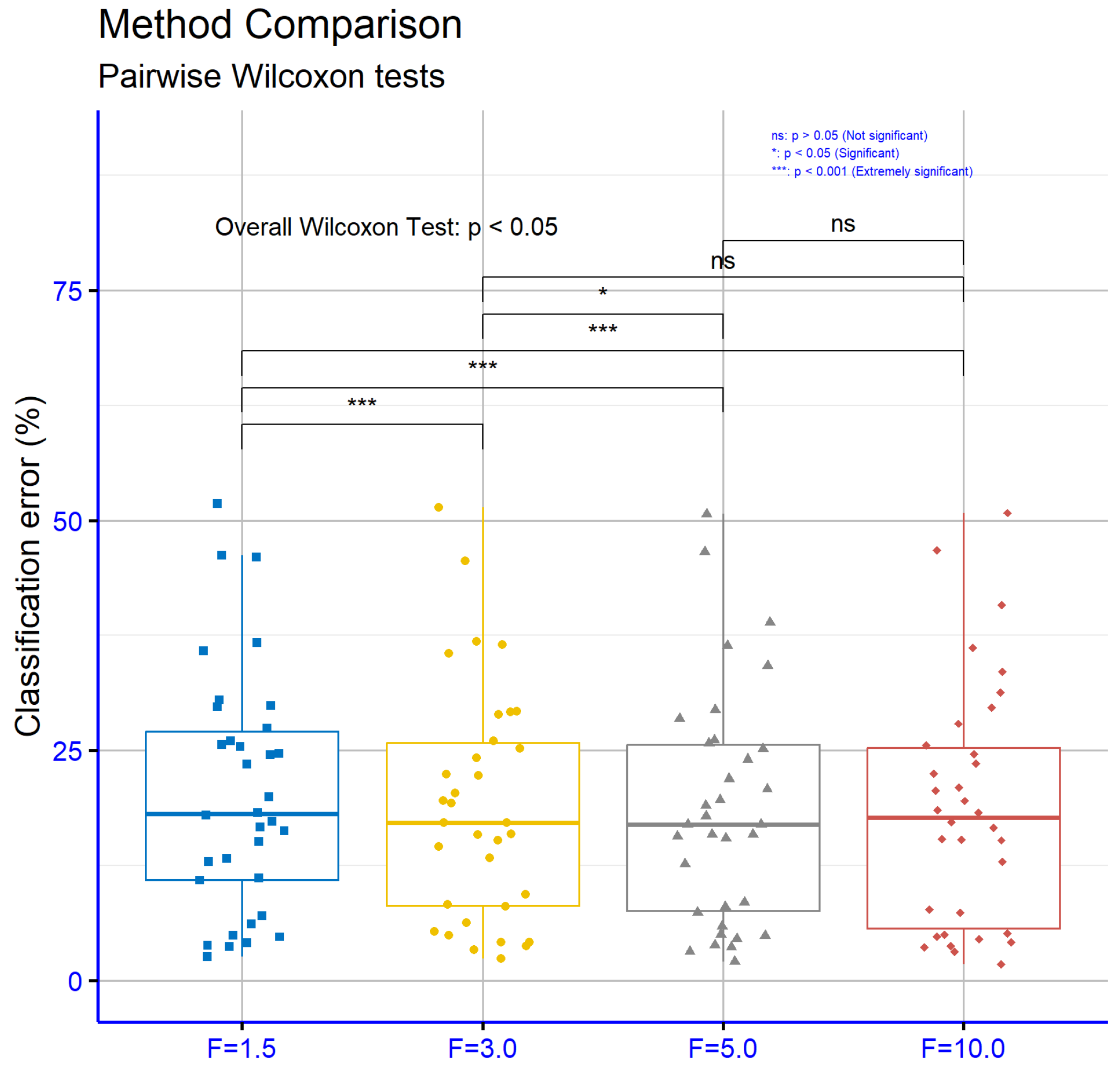
In
Figure 10, which compares different values of the parameter
F for the classification datasets, several statistically significant differences are observed. The
p-values for the comparisons
vs.
(
),
vs.
(
), and
vs.
(
) indicate a strong difference in the model’s performance. In contrast, the values for the comparisons
vs.
(
),
vs.
(
), and
vs.
(
) show that the differences between larger values of the parameter
F are less significant.
Figure 11 examines comparisons of the parameter
F for the regression datasets and shows no statistically significant differences. The
p-values for the comparisons
vs.
(
),
vs.
(
),
vs.
(
),
vs.
(
),
vs.
(
), and
vs.
(
) indicate that variations in the value of the parameter
F do not significantly affect the model’s performance on the regression datasets. This may suggest greater stability of the model to changes in this parameter compared to the classification datasets.
3.5. Experiments on Number of Generations
In order to evaluate the convergence of the genetic algorithm, an additional experiment was conducted where the number of generations was altered from 25 to 200. The experimental results using the proposed method for the classification datasets are depicted in
Table 8 and for the regression datasets in
Table 9.
In
Table 8, the experimental results indicate a general trend of decreasing error rates as the number of generations
increases from 25 to 200. This reduction suggests that increasing
improves the performance of the genetic algorithm, as it allows the model to better approximate the optimal solution. For most datasets, the lowest error rate is observed when
. For instance, on the “Alcohol” dataset, the error rate decreased from 38.19% to 31.28%, while a significant reduction was also observed in the “Sonar” dataset, from 21.77% to 18.25%. Similar reductions were noted on several other datasets, such as “Spiral” (31.82% to 22.52%) and “ZO_NF_S” (4.19% to 3.63%). However, there are datasets like “Mammographic” and “Lymography” where the changes are minor and not consistently positive, indicating that increasing the number of generations does not always have a dramatic impact on performance. The overall average error rate across all datasets steadily decreases, from 19.99% when
to 18.67% when
. This confirms the general trend of the genetic algorithm converging towards improved solutions with more generations. The statistical analysis of the results demonstrates that increasing the number of generations is effective in the majority of cases, enhancing the accuracy of the proposed method on classification datasets. Nonetheless, the performance improvement appears to also depend on the specific characteristics of each dataset as well as the inherent properties of the method.
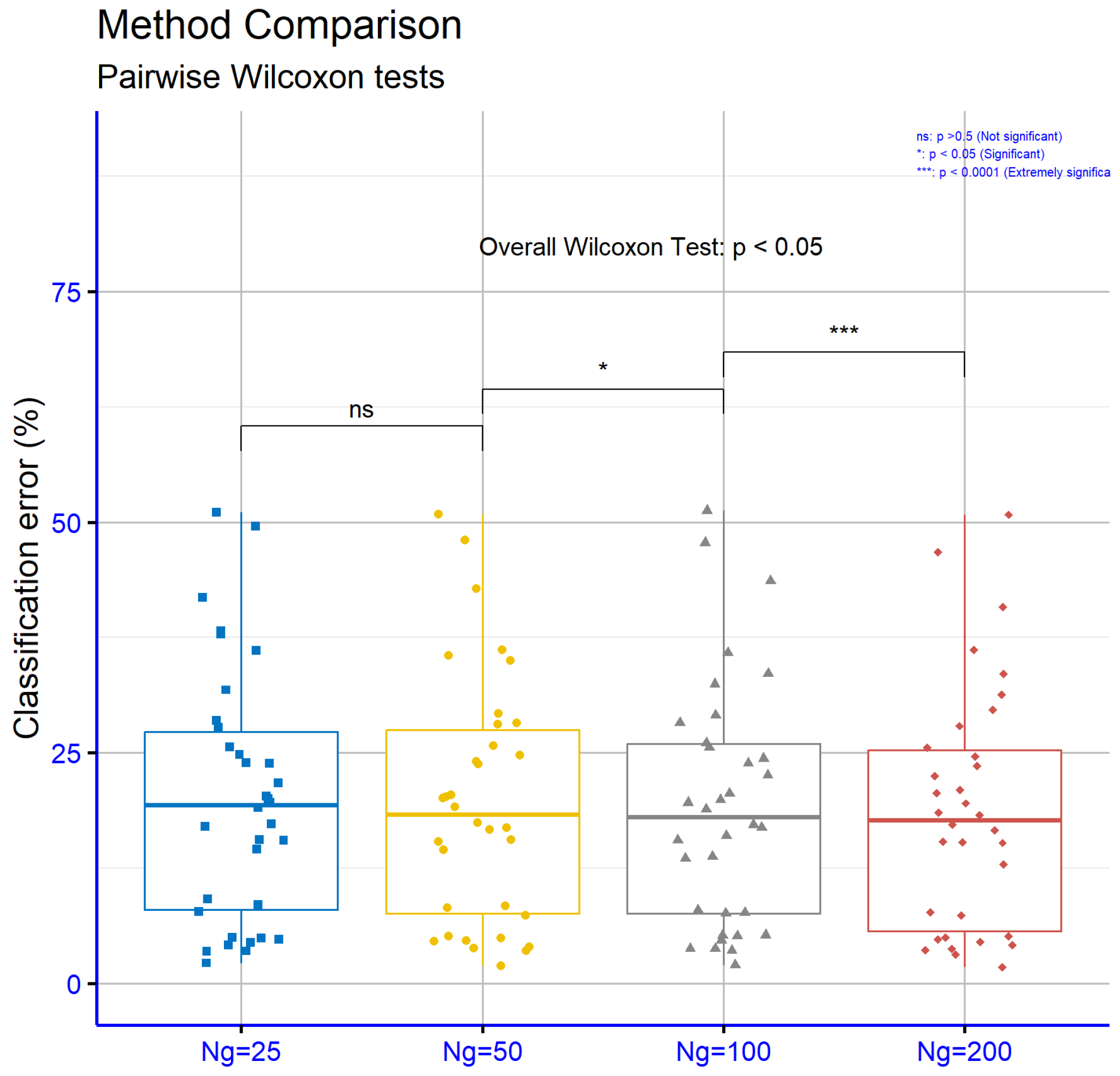
Figure 12 presents the significance levels p for the experiments conducted on the classification datasets with various models. The results indicate that the difference between
and
is not statistically significant, as
p is 0.12. In contrast, the transition from
to
is marginally significant, with
p = 0.048, suggesting that the increase in generations begins to influence model performance. Finally, the difference between
and
shows high statistical significance, with
p = 0.00083, confirming that the increase in the number of generations significantly contributes to performance improvement.
In
Table 9, the experimental findings suggest that the convergence behavior of the genetic algorithm on regression datasets shows varied patterns as the number of generations
increases from 25 to 200. Specifically, a reduction in error values is observed on several datasets, while on others, errors either increase or remain stable. For instance, on the “Abalone” dataset, the error decreases steadily from 6.08 at
to 5.10 at
, indicating improved performance. On the “Friedman” dataset, there is a significant reduction from 3.22 to 1.45, demonstrating a clear trend of convergence toward optimal solutions. Similar reductions are observed on “Stock” (2.29 to 1.53) and “Treasury” (0.82 to 0.51). However, there are datasets such as “Auto”, where the reduction is negligible, and the error slightly increases from 9.60 at
to 9.68 at
. Furthermore, on the “Housing” dataset, a gradual increase in error is observed, from 17.72 to 18.70, suggesting that the increased number of generations did not improve performance. On the “MB” dataset, the error rises significantly, from 0.15 at
= 25 to 5.49 at
, indicating potential instability of the method on this specific dataset. The average across all datasets shows a nonlinear pattern, with values fluctuating. The average error decreases from 5.54 at
= 25 to 5.13 at
, but then increases to 5.48 at
. This indicates that increasing the number of generations does not always lead to consistent improvement in performance across regression datasets. In conclusion, the statistical analysis reveals that increasing the number of generations
can improve the performance of the genetic algorithm on certain datasets; however, its impact is not always consistent. The outcome depends on the specific characteristics of each dataset as well as the inherent properties of the method.
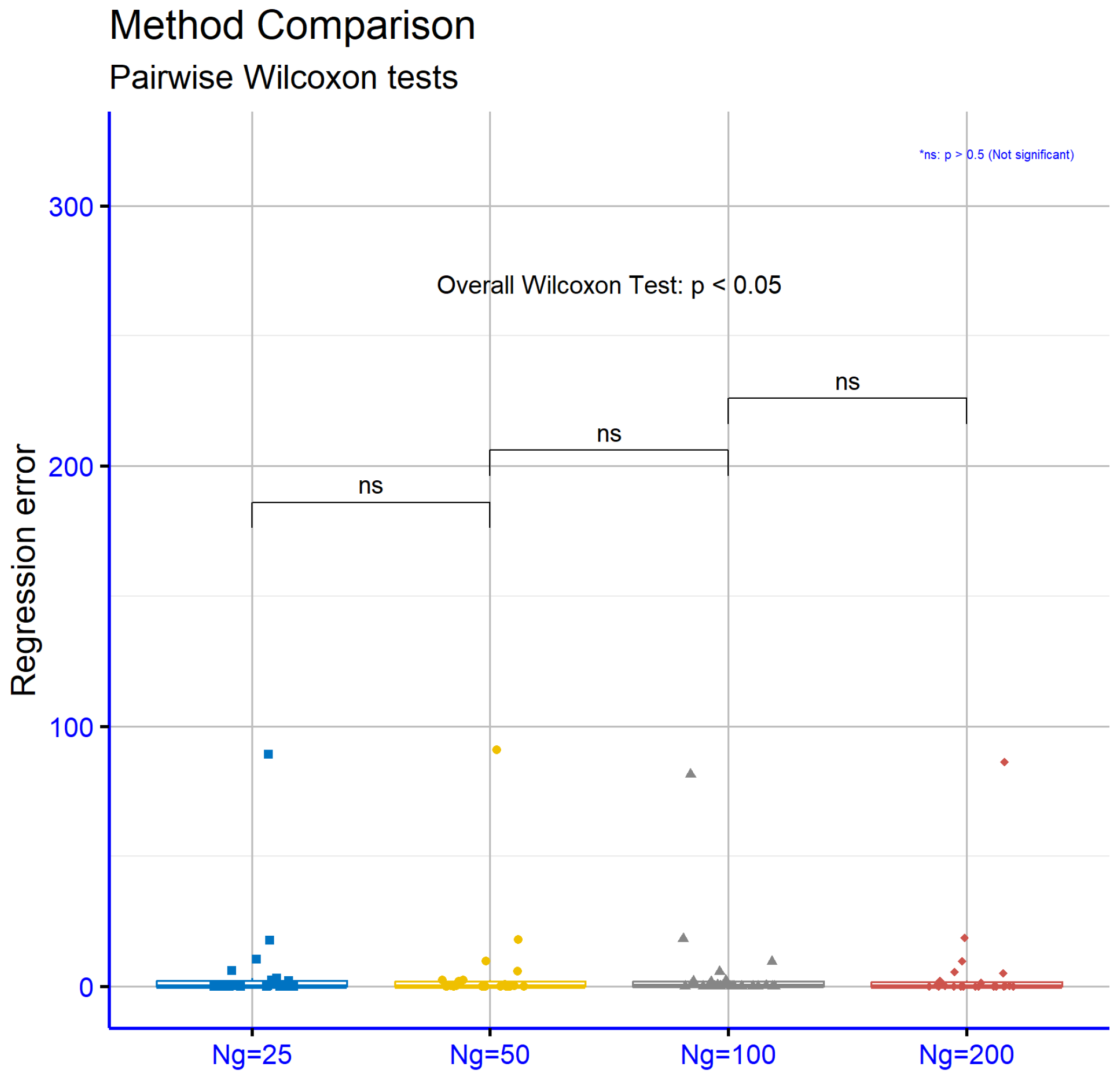
Figure 13 pertains to the regression datasets and shows that the performance differences between consecutive
values are not statistically significant. Specifically, for
versus
,
p is 0.46, for
versus
,
p is 0.27, and for
versus
,
p is 0.9. These results suggest that increasing the number of generations in regression datasets does not lead to significant performance changes, indicating that the characteristics of these datasets may limit the effectiveness of the approach.