An Alternative Estimator for Poisson–Inverse-Gaussian Regression: The Modified Kibria–Lukman Estimator
Abstract
1. Introduction
2. Methodology
2.1. Poisson–Inverse-Gaussian Regression Model (PIGM)
2.2. Biased Estimators
2.3. Suggested Biased Estimators
3. Theoretical Comparisons Between Estimators
4. Simulation Study
- -
- Set the sample size .
- -
- Define the total number of replications of the simulation, .
- -
- Specify the number of predictors .
- -
- Define the true regression coefficients .
- -
- Increment the replication counter: r = r + 1.
- -
- If r = 1000, stop the simulation; otherwise, return to Step 3.
5. Real Data Application
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zha, L.; Lord, D.; Zou, Y. The Poisson inverse Gaussian (PIG) generalized linear regression model for analyzing motor vehicle crash data. J. Transp. Saf. Secur. 2015, 8, 18–35. [Google Scholar] [CrossRef]
- Månsson, K. On ridge estimators for the negative binomial regression model. Econ. Model. 2012, 29, 178–184. [Google Scholar] [CrossRef]
- Abonazel, M.R.; Saber, A.A.; Awwad, F.A. Kibria–Lukman estimator for the Conway–Maxwell Poisson regression model: Simulation and applications. Sci. Afr. 2023, 19, e01553. [Google Scholar] [CrossRef]
- Ashraf, B.; Amin, M.; Mahmood, T.; Faisa, M. Performance of Alternative Estimators in the Poisson-Inverse Gaussian Regression Model: Simulation and Application. Appl. Math. Nonlinear Sci. 2024, 9, 1–18. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, P.; Wang, Y. Robust heavy-tailed versions of generalized linear models with applications in actuarial science. Comput. Stat. Data Anal. 2024, 194, 107920. [Google Scholar] [CrossRef]
- Hardin, J.; Hilbe, J. Generalized Linear Models and Extensions, 2nd ed.; Stata Press: College Station, TX, USA, 2002; pp. 13–26. [Google Scholar]
- Dean, C.; Lawless, J.; Willmot, G. A mixed Poisson Inverse-Gaussian regression model. Can. J. Stat. 1989, 17, 171–181. [Google Scholar] [CrossRef]
- Putri, G.; Nurrohmah, S.; Fithriani, I. Comparing Poisson-Inverse Gaussian Model and Negative Binomial Model on case study: Horseshoe crabs data. J. Phys. Conf. Ser. 2017, 1442, 012028. [Google Scholar] [CrossRef]
- Rintara, P.; Ahmed, S.; Lisawadi, S. Post Improved Estimation and Prediction in the Gamma Regression Model. Thail. Stat. 2023, 21, 580–606. [Google Scholar]
- Batool, A.; Amin, M.; Elhassanein, A. On the performance of some new ridge parameter estimators in the Poisson-inverse Gaussian ridge regression. Alex. Eng. J. 2023, 70, 231–245. [Google Scholar] [CrossRef]
- Hocking, E.; Speed, M.; Lynn, J. A class of biased estimators in linear regression. Technometrics 1976, 18, 55–67. [Google Scholar] [CrossRef]
- James, W.; Stein, C. Estimation with quadratic loss. Proc. Fourth Berkely Symp. Math. Stat. Probab. 1961, 1, 361–379. [Google Scholar]
- Hoerl, A.; Kennard, W. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics 1970, 12, 55–67. [Google Scholar] [CrossRef]
- Hoerl, A.; Kennard, W. Ridge regression: Applications to nonorthogonal problems. Technometrics 1970, 12, 69–82. [Google Scholar] [CrossRef]
- Liu, K. A new class of biased estimate in linear regression. Commun. Stat.-Theory Methods 1993, 22, 393–402. [Google Scholar]
- Liu, K. Using Liu-type estimator to combat collinearity. Commun. Stat.-Theory Methods 2003, 32, 1009–2003. [Google Scholar] [CrossRef]
- Liu, K. More on Liu-type estimator in linear regression. Commun. Stat.-Theory Methods 2004, 33, 2723–2733. [Google Scholar] [CrossRef]
- Kibria, B.G.; Lukman, A.F. A new ridge-type estimator for the linear regression model: Simulations and applications. Scientifica 2020, 2020, 9758378. [Google Scholar] [CrossRef]
- Mandal, S.; Belaghi, R.; Mahmoudi, A.; Aminnejad, M. Stein-type shrinkage estimators in gamma regression model with application to prostate cancer data. Stat. Med. 2019, 38, 4310–4322. [Google Scholar] [CrossRef]
- Akram, M.; Amin, M.; Faisal, M. On the generalized biased estimators for the gamma regression model: Methods and applications. Commun. Stat.-Simul. Comput. 2023, 52, 4087–4100. [Google Scholar] [CrossRef]
- Akram, M.; Amin, M.; Qasim, M. A new biased estimator for the gamma regression model: Some applications in medical sciences. Commun. Stat.-Theory Methods 2023, 52, 3612–3632. [Google Scholar] [CrossRef]
- Lukman, A.; Ayinde, K.; Kibria, G.; Adewuyi, E. Modified ridge-type estimator for the gamma regression model. Commun. Stat.-Simul. Comput. 2022, 51, 5009–5023. [Google Scholar] [CrossRef]
- Aladeitan, B.; Adebimpe, O.; Lukman, A.; Oludoun, O.; Abiodun, O. Modified Kibria-Lukman (MKL) estimator for the Poisson Regression Model: Application and simulation. F1000Research 2021, 10, 548. [Google Scholar] [CrossRef] [PubMed]
- Holla, M. On a Poisson-inverse gaussian distribution. Metr. Int. J. Theor. Appl. Stat. 1967, 11, 115–121. [Google Scholar] [CrossRef]
- Abramowitz, M.; Stegun, I.A. (Eds.) Handbook of Mathematical Functions with Formulas, Graphs, and Mathematical Tables; US Government Printing Office: Washington, DC, USA, 1968; Volume 55.
- Nelder, J.; Wedderburn, R. Generalized Linear Models. J. R. Stat. Soc. Ser. A 1972, 135, 370–384. [Google Scholar] [CrossRef]
- Lukman, A.F.; Ayinde, K. Review and classifications of the ridge parameter estimation techniques. Hacet. J. Math. Stat. 2017, 46, 953–967. [Google Scholar] [CrossRef]
- Lukman, A.; Kibria, G.; Ayinde, K.; Jegede, S. Modified One-Parameter Liu Estimator for the Linear Regression Model. In Modelling and Simulation in Engineering; Wiley Online Library: New York, NY, USA, 2020; p. 9574304. [Google Scholar]
- Özkale, M.; KaçIranlar, S. The restricted and unrestricted two-parameter estimators. Commun. Stat.-Theory Methods 2007, 36, 2707–2725. [Google Scholar] [CrossRef]
- Akram, N.; Amin, M.; Qasim, M. A new liu-type estimator for the inverse gaussian regression model. J. Stat. Comput. Simul. 2020, 90, 1153–1172. [Google Scholar] [CrossRef]
- Alrweili, H. Liu-Type estimator for the Poisson-Inverse Gaussian regression model: Simulation and practical applications. Stat. Optim. Inf. Comput. 2024, 12, 982–1003. [Google Scholar] [CrossRef]
- Farebrother, R. Further Results on the Mean Square Error of Ridge Regression. J. R. Stat. Soc. 1976, 38, 248–250. [Google Scholar] [CrossRef]
- Trenkler, G.; Toutenburg, H. Mean squared error matrix comparisons between biased estimators—An overview of recent results. Stat. Pap. 1990, 31, 165–179. [Google Scholar] [CrossRef]
- Lukman, A.F.; Albalawi, O.; Arashi, M.; Allohibi, J.; Alharbi, A.A.; Farghali, R.A. Robust Negative Binomial Regression via the Kibria–Lukman Strategy: Methodology and Application. Mathematics 2024, 12, 2929. [Google Scholar] [CrossRef]
- Barreto-Souza, W.; Simas, A.B. General mixed Poisson regression models with varying dispersion. Stat. Comput. 2016, 26, 1263–1280. [Google Scholar] [CrossRef]
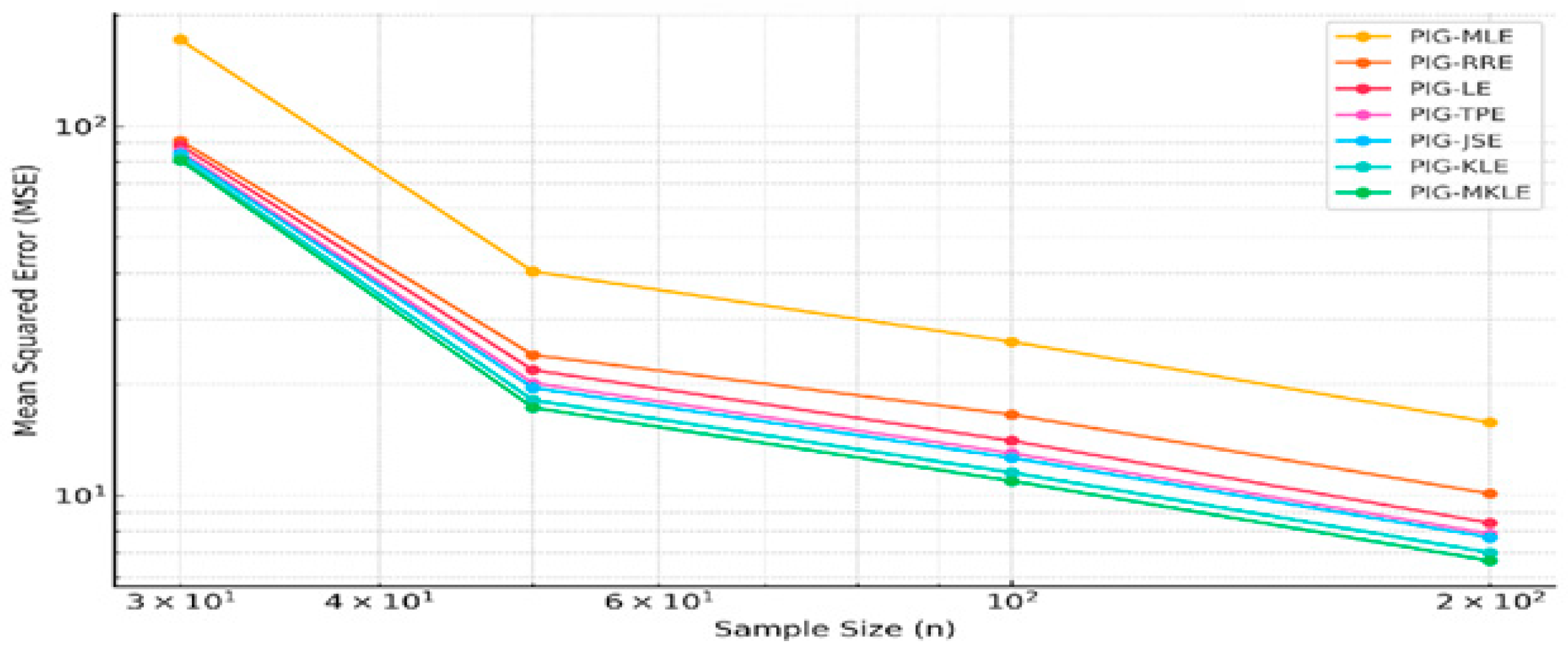
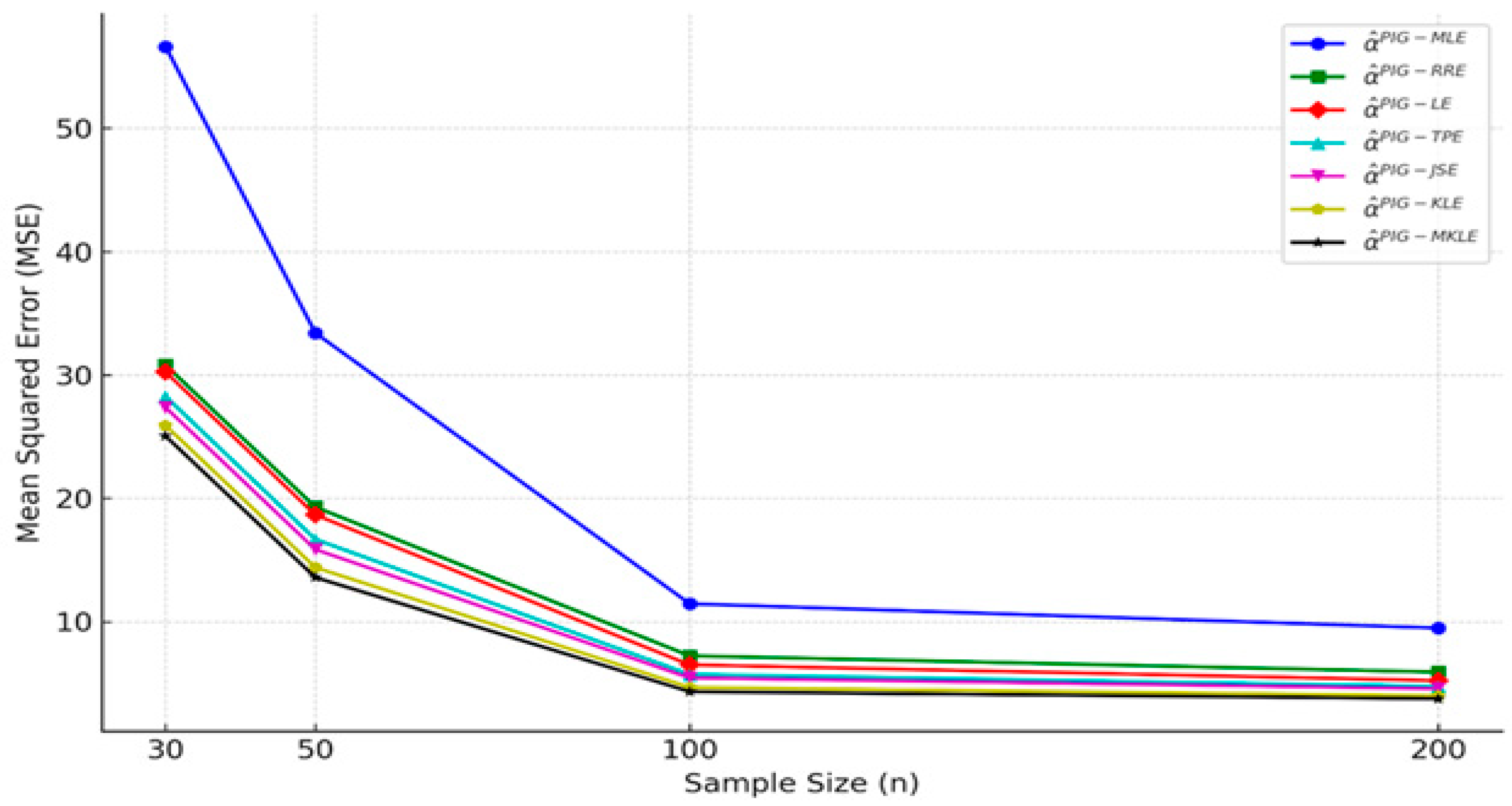
| n | 30 | 50 | 100 | 200 | 30 | 50 | 100 | 200 | 30 | 50 | 100 | 200 |
| 2.563 | 1.956 | 1.892 | 1.814 | 2.656 | 1.984 | 1.960 | 1.906 | 2.704 | 2.007 | 1.993 | 1.982 | |
| 1.347 | 0.965 | 0.925 | 0.919 | 1.531 | 0.981 | 0.957 | 0.939 | 1.680 | 1.000 | 0.981 | 0.972 | |
| 1.328 | 0.975 | 0.935 | 0.904 | 1.524 | 0.988 | 0.957 | 0.925 | 1.899 | 1.000 | 0.990 | 0.964 | |
| 1.281 | 0.958 | 0.957 | 0.946 | 1.327 | 0.962 | 0.963 | 0.960 | 1.451 | 1.004 | 0.986 | 0.976 | |
| 1.267 | 0.982 | 0.914 | 0.902 | 1.294 | 0.995 | 0.986 | 0.929 | 1.309 | 1.006 | 1.000 | 0.974 | |
| 1.126 | 0.952 | 0.919 | 0.906 | 1.251 | 0.976 | 0.947 | 0.914 | 1.008 | 1.000 | 0.963 | 0.955 | |
| 1.096 | 0.931 | 0.908 | 0.901 | 1.103 | 0.948 | 0.947 | 0.918 | 1.201 | 0.965 | 0.952 | 0.949 | |
| 3.933 | 2.229 | 1.952 | 1.895 | 4.830 | 2.253 | 1.993 | 1.937 | 4.993 | 2.714 | 2.022 | 2.019 | |
| 2.268 | 1.140 | 0.952 | 0.921 | 2.927 | 1.197 | 0.977 | 0.940 | 3.061 | 1.289 | 0.999 | 0.986 | |
| 2.209 | 1.150 | 0.958 | 0.920 | 2.900 | 1.212 | 0.986 | 0.938 | 3.036 | 1.305 | 1.003 | 0.988 | |
| 1.964 | 1.134 | 0.976 | 0.948 | 2.411 | 1.156 | 0.996 | 0.968 | 2.492 | 1.171 | 1.011 | 1.009 | |
| 1.914 | 1.136 | 0.956 | 0.953 | 2.298 | 1.169 | 1.002 | 0.976 | 2.369 | 1.179 | 1.015 | 1.015 | |
| 1.628 | 1.024 | 0.956 | 0.906 | 1.913 | 1.105 | 0.972 | 0.940 | 1.992 | 1.120 | 1.003 | 0.990 | |
| 1.549 | 0.952 | 0.935 | 0.902 | 1.784 | 1.008 | 0.945 | 0.928 | 1.897 | 1.699 | 0.971 | 0.950 | |
| 7.203 | 3.286 | 2.193 | 2.130 | 9.763 | 4.081 | 2.380 | 2.172 | 10.407 | 4.932 | 2.645 | 2.220 | |
| 4.251 | 1.819 | 1.072 | 1.038 | 5.757 | 2.454 | 1.104 | 1.041 | 6.187 | 3.045 | 1.184 | 1.140 | |
| 4.133 | 1.805 | 1.072 | 1.032 | 5.603 | 2.381 | 1.127 | 1.041 | 6.006 | 2.935 | 1.205 | 1.162 | |
| 3.596 | 1.642 | 1.096 | 1.065 | 4.873 | 2.038 | 1.110 | 1.061 | 5.195 | 2.462 | 1.210 | 1.122 | |
| 3.436 | 1.614 | 1.103 | 1.071 | 4.650 | 1.967 | 1.115 | 1.098 | 5.960 | 2.354 | 1.125 | 1.117 | |
| 2.911 | 1.367 | 1.042 | 1.036 | 4.118 | 1.622 | 1.016 | 1.075 | 5.330 | 1.919 | 1.113 | 1.100 | |
| 2.732 | 1.305 | 1.034 | 1.032 | 3.876 | 1.519 | 1.100 | 1.047 | 4.071 | 1.779 | 1.105 | 1.076 | |
| 45.884 | 20.772 | 7.035 | 5.806 | 53.706 | 25.398 | 9.405 | 7.780 | 56.620 | 33.407 | 11.446 | 9.476 | |
| 25.701 | 12.607 | 4.332 | 3.484 | 29.055 | 15.021 | 6.001 | 4.843 | 30.797 | 19.288 | 7.225 | 5.906 | |
| 24.798 | 11.718 | 3.866 | 3.093 | 28.652 | 14.309 | 5.382 | 4.212 | 30.233 | 18.652 | 6.495 | 5.215 | |
| 22.897 | 10.367 | 3.512 | 2.899 | 26.800 | 12.675 | 4.695 | 3.884 | 28.254 | 16.671 | 5.713 | 4.730 | |
| 22.063 | 9.816 | 3.396 | 2.834 | 25.978 | 11.995 | 4.461 | 3.766 | 27.361 | 15.848 | 5.433 | 4.560 | |
| 20.575 | 8.621 | 2.925 | 2.509 | 24.672 | 10.689 | 3.771 | 3.310 | 25.896 | 14.385 | 4.662 | 3.987 | |
| 19.771 | 8.045 | 2.738 | 2.381 | 23.869 | 10.005 | 3.485 | 3.113 | 25.036 | 13.575 | 4.317 | 3.740 | |
| n | 30 | 50 | 100 | 200 | 30 | 50 | 100 | 200 | 30 | 50 | 100 | 200 |
| 6.478 | 2.036 | 1.964 | 1.866 | 8.074 | 2.092 | 1.982 | 1.918 | 9.105 | 2.277 | 2.010 | 1.995 | |
| 3.460 | 1.012 | 0.914 | 0.899 | 4.491 | 1.105 | 0.970 | 0.910 | 5.004 | 1.185 | 1.000 | 0.972 | |
| 3.473 | 1.054 | 0.947 | 0.927 | 4.392 | 1.081 | 0.966 | 0.983 | 4.908 | 1.249 | 0.989 | 0.985 | |
| 3.234 | 1.018 | 0.943 | 0.931 | 4.030 | 1.046 | 0.982 | 0.957 | 4.545 | 1.137 | 1.005 | 0.999 | |
| 3.206 | 1.024 | 0.953 | 0.936 | 3.966 | 1.055 | 0.986 | 0.969 | 4.492 | 1.141 | 1.013 | 1.002 | |
| 2.984 | 0.977 | 0.924 | 0.903 | 3.708 | 1.002 | 0.940 | 0.897 | 4.218 | 1.030 | 0.996 | 0.983 | |
| 2.889 | 0.956 | 0.918 | 0.902 | 3.587 | 0.980 | 0.964 | 0.959 | 4.092 | 0.996 | 0.985 | 0.982 | |
| 16.613 | 2.844 | 2.419 | 2.050 | 16.705 | 3.951 | 2.891 | 2.086 | 20.355 | 4.371 | 3.504 | 2.093 | |
| 9.046 | 1.404 | 1.144 | 0.996 | 9.316 | 2.132 | 1.436 | 0.997 | 11.408 | 2.515 | 1.818 | 0.998 | |
| 8.804 | 1.501 | 1.238 | 1.013 | 8.978 | 2.168 | 1.531 | 1.039 | 11.002 | 2.473 | 1.907 | 1.050 | |
| 8.291 | 1.421 | 1.209 | 1.025 | 8.337 | 1.973 | 1.444 | 1.043 | 10.158 | 2.182 | 1.750 | 1.046 | |
| 8.191 | 1.430 | 1.221 | 1.031 | 8.191 | 1.963 | 1.452 | 1.052 | 9.961 | 2.159 | 1.749 | 1.058 | |
| 7.732 | 1.304 | 1.128 | 1.011 | 7.788 | 1.761 | 1.307 | 1.017 | 9.444 | 1.920 | 1.544 | 1.110 | |
| 6.495 | 1.266 | 1.104 | 0.990 | 7.590 | 1.687 | 1.264 | 1.007 | 9.165 | 1.832 | 1.478 | 1.011 | |
| 28.105 | 5.684 | 4.395 | 2.131 | 42.217 | 9.106 | 6.266 | 2.429 | 47.142 | 10.507 | 8.554 | 2.752 | |
| 15.747 | 3.182 | 2.341 | 1.036 | 23.076 | 5.348 | 3.593 | 1.154 | 25.580 | 6.252 | 5.118 | 1.307 | |
| 14.869 | 3.158 | 2.349 | 1.049 | 22.415 | 5.082 | 3.438 | 1.226 | 24.810 | 5.924 | 4.741 | 1.423 | |
| 14.026 | 2.838 | 2.195 | 1.065 | 21.067 | 4.545 | 3.128 | 1.214 | 23.524 | 5.244 | 4.270 | 1.375 | |
| 13.800 | 2.803 | 2.186 | 1.073 | 20.672 | 4.458 | 3.088 | 1.224 | 23.169 | 5.111 | 4.190 | 1.386 | |
| 13.139 | 2.473 | 1.951 | 1.015 | 19.837 | 3.996 | 2.699 | 1.160 | 22.349 | 4.593 | 3.678 | 1.274 | |
| 12.786 | 2.353 | 1.867 | 1.006 | 19.334 | 3.799 | 2.550 | 1.143 | 21.874 | 4.361 | 3.468 | 1.242 | |
| 171.420 | 40.414 | 26.096 | 15.786 | 225.455 | 58.279 | 39.836 | 20.008 | 247.230 | 71.848 | 56.252 | 21.255 | |
| 91.340 | 24.024 | 16.571 | 10.114 | 117.311 | 33.219 | 24.659 | 11.340 | 127.790 | 42.226 | 33.631 | 13.572 | |
| 88.722 | 21.862 | 14.071 | 8.431 | 115.666 | 31.413 | 21.324 | 10.849 | 126.440 | 38.562 | 30.151 | 11.425 | |
| 85.541 | 20.168 | 13.024 | 7.879 | 112.503 | 29.082 | 19.880 | 9.985 | 123.360 | 35.854 | 28.071 | 10.608 | |
| 84.340 | 19.572 | 12.662 | 7.710 | 111.383 | 27.246 | 19.310 | 9.671 | 122.330 | 34.748 | 28.246 | 10.338 | |
| 82.218 | 18.115 | 11.545 | 7.010 | 109.448 | 25.429 | 17.864 | 9.072 | 120.550 | 32.492 | 26.535 | 9.401 | |
| 80.789 | 17.299 | 10.949 | 6.661 | 108.081 | 24.335 | 17.036 | 8.704 | 119.240 | 31.090 | 25.499 | 8.912 | |
| n | 30 | 50 | 100 | 200 | 30 | 50 | 100 | 200 | 30 | 50 | 100 | 200 |
| 18.197 | 4.901 | 2.011 | 1.871 | 22.662 | 10.091 | 2.027 | 1.904 | 29.077 | 11.399 | 2.300 | 1.985 | |
| 9.630 | 2.537 | 0.956 | 0.915 | 12.012 | 5.592 | 0.977 | 0.937 | 15.433 | 6.476 | 1.057 | 0.995 | |
| 9.433 | 2.626 | 1.008 | 0.927 | 11.796 | 5.456 | 1.037 | 0.941 | 15.057 | 6.189 | 1.201 | 0.981 | |
| 9.080 | 2.447 | 1.005 | 0.935 | 11.309 | 5.037 | 1.013 | 0.952 | 14.510 | 5.689 | 1.149 | 0.992 | |
| 9.008 | 2.446 | 1.014 | 0.940 | 11.212 | 4.989 | 1.025 | 0.955 | 14.402 | 5.627 | 1.162 | 0.995 | |
| 8.819 | 2.276 | 0.972 | 0.929 | 10.910 | 4.684 | 0.993 | 0.947 | 14.108 | 5.287 | 1.080 | 0.980 | |
| 8.713 | 2.212 | 0.937 | 0.925 | 10.755 | 4.545 | 0.986 | 0.936 | 13.942 | 5.123 | 1.055 | 0.950 | |
| 36.880 | 10.744 | 2.778 | 1.998 | 38.850 | 22.156 | 3.691 | 2.015 | 39.860 | 27.797 | 5.009 | 2.025 | |
| 19.263 | 6.032 | 1.285 | 0.971 | 20.521 | 12.565 | 1.809 | 0.977 | 21.356 | 15.929 | 2.601 | 0.996 | |
| 19.008 | 5.809 | 1.437 | 0.998 | 20.117 | 11.935 | 1.960 | 1.000 | 20.788 | 15.050 | 2.696 | 1.018 | |
| 18.403 | 5.362 | 1.388 | 0.999 | 19.386 | 11.057 | 1.843 | 1.007 | 19.890 | 13.871 | 2.501 | 1.012 | |
| 18.265 | 5.304 | 1.401 | 1.008 | 19.206 | 10.882 | 1.853 | 1.013 | 19.680 | 13.643 | 2.504 | 1.023 | |
| 17.975 | 4.907 | 1.307 | 0.981 | 18.810 | 10.248 | 1.689 | 0.986 | 19.163 | 12.921 | 2.263 | 0.997 | |
| 17.574 | 4.729 | 1.277 | 0.945 | 17.799 | 9.924 | 1.631 | 0.948 | 18.879 | 12.544 | 2.173 | 0.975 | |
| 66.641 | 30.079 | 6.278 | 2.287 | 85.710 | 46.629 | 9.247 | 2.635 | 89.977 | 57.280 | 13.434 | 3.068 | |
| 34.588 | 17.375 | 3.445 | 1.089 | 45.226 | 26.555 | 5.347 | 1.292 | 46.842 | 32.207 | 7.943 | 1.396 | |
| 34.213 | 16.021 | 3.363 | 1.155 | 44.240 | 24.953 | 5.020 | 1.357 | 46.258 | 30.496 | 7.269 | 1.599 | |
| 33.254 | 15.010 | 3.134 | 1.143 | 42.769 | 23.269 | 4.616 | 1.317 | 44.898 | 28.583 | 6.705 | 1.533 | |
| 32.980 | 14.776 | 3.118 | 1.155 | 42.363 | 22.838 | 4.559 | 1.333 | 44.534 | 28.072 | 6.601 | 1.550 | |
| 32.492 | 13.972 | 2.822 | 1.101 | 41.555 | 21.743 | 4.105 | 1.238 | 43.850 | 26.860 | 6.023 | 1.423 | |
| 32.165 | 13.546 | 2.703 | 1.046 | 41.053 | 21.117 | 3.906 | 1.111 | 43.409 | 26.149 | 5.752 | 1.384 | |
| 306.507 | 183.213 | 56.727 | 13.293 | 375.705 | 254.194 | 57.365 | 19.084 | 383.607 | 323.779 | 111.401 | 24.977 | |
| 154.971 | 100.452 | 34.761 | 8.242 | 190.652 | 135.672 | 35.063 | 12.210 | 197.565 | 170.258 | 64.316 | 15.799 | |
| 154.611 | 95.371 | 30.083 | 7.086 | 189.952 | 131.931 | 30.675 | 10.177 | 194.846 | 167.306 | 58.826 | 13.420 | |
| 152.947 | 91.424 | 28.308 | 6.635 | 187.477 | 126.844 | 28.627 | 9.525 | 191.421 | 161.567 | 55.590 | 12.465 | |
| 152.599 | 89.952 | 27.650 | 6.534 | 186.913 | 125.012 | 27.897 | 9.361 | 190.396 | 159.480 | 54.318 | 12.212 | |
| 151.808 | 87.232 | 25.980 | 5.933 | 185.773 | 121.917 | 26.073 | 8.570 | 188.554 | 156.134 | 51.715 | 11.151 | |
| 151.189 | 85.386 | 24.991 | 5.648 | 184.901 | 119.712 | 25.000 | 8.179 | 187.200 | 153.668 | 50.016 | 10.617 | |
| Metrics | |||
|---|---|---|---|
| Residual Variance | 661.2 | 661.2 | 0.5498 |
| AIC | 873.1 | 397.9 | 406.0 |
| Coef. | |||||||
|---|---|---|---|---|---|---|---|
| 0.5436 | 0.0001 | 0.1434 | 0.0010 | −0.0296 | 0.0430 | −0.0001 | |
| 0.4398 | 0.0002 | 0.1440 | 0.0008 | 0.2138 | 0.0348 | −0.0002 | |
| −0.3814 | −0.0002 | −0.1318 | −0.0007 | −0.2544 | −0.0302 | 0.0002 | |
| 0.8503 | 0.0001 | 0.2116 | 0.0016 | −0.1302 | 0.0672 | −0.0001 | |
| SMSE | 987.4608 | 18.0435 | 17.5653 | 1.3558 | 171.9145 | 7.3136 | 1.3578 |
| PMSE | 453.3143 | 446.1027 | 448.5152 | 446.1133 | 450.5399 | 446.6592 | 446.0973 |
| PMAE | 16.1498 | 15.9001 | 15.9847 | 15.9005 | 16.0511 | 15.9198 | 15.8999 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farghali, R.A.; Lukman, A.F.; Algamal, Z.; Genc, M.; Attia, H. An Alternative Estimator for Poisson–Inverse-Gaussian Regression: The Modified Kibria–Lukman Estimator. Algorithms 2025, 18, 169. https://doi.org/10.3390/a18030169
Farghali RA, Lukman AF, Algamal Z, Genc M, Attia H. An Alternative Estimator for Poisson–Inverse-Gaussian Regression: The Modified Kibria–Lukman Estimator. Algorithms. 2025; 18(3):169. https://doi.org/10.3390/a18030169
Chicago/Turabian StyleFarghali, Rasha A., Adewale F. Lukman, Zakariya Algamal, Murat Genc, and Hend Attia. 2025. "An Alternative Estimator for Poisson–Inverse-Gaussian Regression: The Modified Kibria–Lukman Estimator" Algorithms 18, no. 3: 169. https://doi.org/10.3390/a18030169
APA StyleFarghali, R. A., Lukman, A. F., Algamal, Z., Genc, M., & Attia, H. (2025). An Alternative Estimator for Poisson–Inverse-Gaussian Regression: The Modified Kibria–Lukman Estimator. Algorithms, 18(3), 169. https://doi.org/10.3390/a18030169







