OIKAN: A Hybrid AI Framework Combining Symbolic Inference and Deep Learning for Interpretable Information Retrieval Models
Abstract
1. Introduction
2. Related Studies
2.1. Trustworthy and Interpretable AI
2.2. Kolmogorov–Arnold Networks (KANs) and Variants
2.3. KANs in Scientific and Physics-Informed Learning
2.4. Symbolic Regression Approaches
| Method | Use Case | Speed | Interpretability | Accuracy (Complex Data) | Tools/ Libraries |
|---|---|---|---|---|---|
| Genetic Programming (GP) | Non-linear symbolic modeling | Low (slow, resource-heavy) | High | High | Operon, gplearn, PySR [31] |
| Least Squares | Hybrid symbolic fitting | High | Medium (model-dependent) | Medium | Operon, PySINDy [32] |
| Sparse Regression (LASSO, STLSQ) | Sparse interpretable models | Medium (with regularization) | High | Medium (needs basis functions) | Lasso, PySINDy [33] |
| Bayesian SR | Uncertainty-aware modeling | Low (compute-intensive) | High (probabilistic) | Medium | Custom implementations [29] |
| NN-Based (AI Feynman) | Symbolic distillation from neural networks | Low (slow training) | Medium (postprocessed) | High (low-noise data) | AI Feynman [34] |
2.5. Neuro-Symbolic Frameworks
2.6. Positioning of OIKAN
2.7. Research Gaps
| Source | Tool | Algorithm Type | Time & Memory Efficiency | Tabular Data Focus | Multi- Obj. | Neural Integration | Interpretability |
|---|---|---|---|---|---|---|---|
| [45] | PySR | Genetic Programming | Medium (efficient Julia backend, moderate memory) | Partial (can integrate with ML pipelines) | Yes | Yes | High |
| [46] | Operon | Genetic Programming | High (fast C++ implementation, low memory) | No | Yes | Yes | High |
| [47] | GPLearn | Genetic Programming | Low (Python-based, slower) | No | No | No | High |
| [48] | EQL/ DeepSymReg | Neural Network | Medium (GPU- accelerated, moderate memory) | Yes | No | No | Medium |
| [49] | PyKAN | Mathematical Net | Medium | Partial (mathematical nets) | No | No | Medium |
| [50] | DSR | Reinforcement Learning | Low (computationally intensive, slower convergence) | Yes | No | No | Medium |
| [51] | QLattice | Graph-based | Medium | No | No | No | High |
| [52] | SR-Transformer | Transformer-based | Medium to High (depends on model size, GPU heavy) | Yes | No | No | Low |
| [53] | Eureqa | Evolutionary | Medium | No | Yes | Yes | High |
| Proposed | OIKAN | Neuro-Symbolic | High (lightweight MLPs) | Yes (MLPs) | Adapt. | Adapt. | High |
3. Materials and Methods
3.1. High-Level Overview and Main Contributions
3.2. Mathematical Foundation of the OIKAN
3.2.1. Kolmogorov–Arnold Representation Theorem (KART)
3.2.2. Neural Network Function Approximation (TabularNet)
- and are the weight matrix and bias vector for the layer .
- is the activation function used at layer .
- is the number of layers, with sizes defined by input_size, hidden_sizes, and output_size.
- For regression, Mean Squared Error (MSE):
- For classification, Cross-Entropy Loss:
3.2.3. Gaussian Noise-Based Data Augmentation
3.2.4. Two-Stage Sparse Symbolic Regression
- are basis functions (e.g., polynomials, logarithms, exponentials, sines).
- are coefficients estimated by ElasticNet.
- is the number of basis functions.
- is the vector of model coefficients.
- is the j-th feature (or basis function) evaluated at input .
- is the number of training samples.
- is the regularization strength (alpha parameter, ).
- is the number of polynomial features.
- : mixing parameter (default value: 0.5):
- ○
- : Lasso (L1) penalty only.
- ○
- : Ridge (L2) penalty only.
- ○
- : ElasticNet (mixture of both).
- : L1 norm, promoting sparsity.
- : L2 norm, promoting small coefficient magnitudes.
3.2.5. Symbolic Expression Generation
| Algorithm 1 Unified Symbolic Modeling Using OIKANClassifier or OIKANRegressor |
| Require: Labeled dataset (X,y); OIKANClassifier hyperparameters/OIKANRegressor hyperparameters; test set proportion t∈(0,1) Ensure: Trained symbolic model; extracted symbolic formulas; evaluation metrics (accuracy or R2); feature importance metrics 1: Load the dataset (e.g., Iris for classification, California Housing for regression) 2: Split data into training and testing subsets using proportion t 3: Initialize the OIKANClassifier or OIKANRegressor with the specified hyperparameters 4: Train the model on Xtrain,ytrain, including neural network evaluation and symbolic regression 5: Predict class labels or target values for the test set using model.predict(X_test) 6: For classification: compute and print accuracy using accuracy_score(y_test, y_pred; For regression: evaluate performance using the R2 score 7: Generate and print a classification report using classification_report(y_test, y_pred) 8: Retrieve and print symbolic formulas for each class using model.get_formula() 9: Retrieve and print feature importance values using model.feature_importances() 10: Save the trained model to disk using model.save(path) 11: Reload the model using model.load(path) 12: Verify the consistency of symbolic formulas in the original format from the loaded model 13: Retrieve and print symbolic formulas in SymPy format using get_formula(type = ‘sympy’) 14: Retrieve and print symbolic formulas in LaTeX format using get_formula(type = ‘latex’) 15: return Trained model, symbolic formula representations, accuracy, and feature importances |
3.3. Architecture and Base Class Parameters for the OIKAN
3.4. Datasets and Experimental Setup
4. Results
4.1. Performance of OIKAN Classifier on Benchmark Datasets
- Median accuracy: 0.886
- Average accuracy on top 10 datasets: 0.987
- Datasets with accuracy ≥ 0.95: 17 out of 50+ (34%)
- Datasets with accuracy ≤ 0.50: 10 out of 50+ (20%)
4.2. Performance of OIKAN Regressor Across Symbolic and Noisy Tasks
- 344_mv: R2 = 0.992, RMSE = 0.950
- 523_analcatdata_neavote: R2 = 0.938, RMSE = 0.885
- 215_2dplanes: R2 = 0.937, RMSE = 1.098
- 210_cloud: R2 = 0.920, RMSE = 0.353
- 624_fri_c0_100_5: R2 = 0.875, RMSE = 0.327
- 227_cpu_small: RMSE = 189.2, R2 undefined (possibly due to numerical instability)
- 218_house_8L: RMSE = 46,944.5, R2 = 0.215
- 573_cpu_act: RMSE = 164.1, R2 undefined
- Median training time: ~3.5 s
- Median training memory: ~14 MB
- Median prediction time: <0.01 s
- Median prediction memory: <0.05 MB
4.3. Comparative Analysis with Baseline Models
4.4. Interpretability Assessment
5. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Category | Description | Link |
|---|---|---|
| Source Code | Official GitHub repository with full implementation and examples | https://github.com/silvermete0r/oikan (accessed on 1 September 2025) |
| Documentation | Official library documentation page with usage and tutorials | https://silvermete0r.github.io/oikan/ (accessed on 1 September 2025) |
| Baseline Model (Interpretable) | DecisionTree model from scikit-learn | https://scikit-learn.org/stable/modules/tree.html (accessed on 1 September 2025) |
| Baseline Model (Black-box) | XGBoost documentation | https://xgboost.readthedocs.io (accessed on 1 September 2025) |
| Benchmark Results | Updated PMLB benchmarking notebook comparing OIKAN, XGBoost, and DecisionTree | https://www.kaggle.com/code/armanzhalgasbayev/oikan-v0-0-3-auto-benchmarking-sr/ (accessed on 1 September 2025) |
| Credit Score Classification | Credit score classification using a public credit-score dataset. | https://www.kaggle.com/code/armanzhalgasbayev/oikan-ml-credit-score-classification (accessed on 1 October 2025) |
References
- Kautz, H. The third AI summer: AAAI Robert S. Engelmore memorial lecture. AI Mag. 2022, 43, 105–125. [Google Scholar]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef]
- Cranmer, M.; Sanchez Gonzalez, A.; Battaglia, P.; Xu, R.; Cranmer, K.; Spergel, D.; Ho, S. Discovering symbolic models from deep learning with inductive biases. In Proceedings of the 34th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 6–12 December 2020; Curran Associates Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 17429–17442. [Google Scholar]
- Iten, R.; Metger, T.; Wilming, H.; Del Rio, L.; Renner, R. Discovering physical concepts with neural networks. Phys. Rev. Lett. 2020, 124, 010508. [Google Scholar] [CrossRef]
- Makke, N.; Chawla, S. Interpretable scientific discovery with symbolic regression: A review. Artif. Intell. Rev. 2024, 57, 2. [Google Scholar] [CrossRef]
- La Cava, W.; Singh, T.R.; Taggart, J.; Suri, S.; Moore, J.H. Learning concise representations for regression by evolving networks of trees. arXiv 2018, arXiv:1807.00981. [Google Scholar]
- Udrescu, S.M.; Tan, A.; Feng, J.; Neto, O.; Wu, T.; Tegmark, M. AI Feynman 2.0: Pareto-optimal symbolic regression exploiting graph modularity. In Proceedings of the 34th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 6–12 December 2020; Curran Associates Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 4860–4871. [Google Scholar]
- Johnston, W.J.; Fusi, S. Abstract representations emerge naturally in neural networks trained to perform multiple tasks. Nat. Commun. 2023, 14, 1040. [Google Scholar] [CrossRef]
- Elmoznino, E.; Bonner, M.F. High-performing neural network models of visual cortex benefit from high latent dimensionality. PLoS Comput. Biol. 2024, 20, e1011792. [Google Scholar] [CrossRef] [PubMed]
- Hornik, K. Approximation capabilities of multilayer feedforward networks. Neural Netw. 1991, 4, 251–257. [Google Scholar] [CrossRef]
- Kolmogorov, A.N. On the representations of continuous functions of many variables by superposition of continuous functions of one variable and addition. Dokl. Akad. Nauk USSR 1957, 114, 953–956. [Google Scholar]
- Liu, Z.; Wang, Y.; Vaidya, S.; Ruehle, F.; Halverson, J.; Soljačić, M.; Tegmark, M. Kan: Kolmogorov-Arnold networks. arXiv 2024, arXiv:2404.19756. [Google Scholar]
- Cherednichenko, O.; Poptsova, M. Kolmogorov–Arnold networks for genomic tasks. Brief. Bioinform. 2025, 26, bbaf129. [Google Scholar] [CrossRef]
- La Cava, W.G.; Lee, P.C.; Ajmal, I.; Ding, X.; Solanki, P.; Cohen, J.B.; Herman, D.S. A flexible symbolic regression method for constructing interpretable clinical prediction models. NPJ Digit. Med. 2023, 6, 107. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.; Fan, W.; Liu, X.; Li, Y.; Jain, S.; Liu, Y.; Jain, A.; Tang, J. Trustworthy AI: A computational perspective. ACM Trans. Intell. Syst. Technol. 2022, 14, 1–59. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, Y.; Ton, J.-F.; Zhang, X.; Guo, R.; Cheng, H.; Klochkov, Y.; Taufiq, M.F.; Li, H. Trustworthy LLMs: A survey and guideline for evaluating large language models’ alignment. arXiv 2023, arXiv:2308.05374. [Google Scholar]
- Yu, R.; Yu, W.; Wang, X. Kan or MLP: A fairer comparison. arXiv 2024, arXiv:2407.16674. [Google Scholar] [CrossRef]
- Shukla, K.; Toscano, J.D.; Wang, Z.; Zou, Z.; Karniadakis, G.E. A comprehensive and fair comparison between MLP and KAN representations for differential equations and operator networks. Comput. Methods Appl. Mech. Eng. 2024, 431, 117290. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, X. LSS-SKAN: Efficient Kolmogorov-Arnold Networks based on single-parameterized function. arXiv 2024, arXiv:2410.14951. [Google Scholar]
- Mintisan. Awesome-KAN: A Curated List of Resources Related to Kolmogorov–Arnold Networks (KANs). Available online: https://github.com/mintisan/awesome-kan (accessed on 19 June 2025).
- Ji, T.; Hou, Y.; Zhang, D. A comprehensive survey on Kolmogorov Arnold Networks (KAN). arXiv 2024, arXiv:2407.11075. [Google Scholar] [CrossRef]
- Liu, J. Kolmogorov-Arnold networks for symbolic regression and time series prediction. J. Mach. Learn. Res. 2024, 25, 95–110. [Google Scholar]
- Xu, L. Time-Kolmogorov-Arnold Networks and multi-task Kolmogorov-Arnold Networks for time series prediction. J. Time Ser. Anal. 2024, 45, 200–220. [Google Scholar]
- Wang, Y.; Sun, J.; Bai, J.; Anitescu, C.; Eshaghi, M.S.; Zhuang, X.; Liu, Y. Kolmogorov Arnold Informed Neural Network: A physics-informed deep learning framework for solving forward and inverse problems based on Kolmogorov Arnold Networks. arXiv 2024, arXiv:2406.11045. [Google Scholar] [CrossRef]
- Zhalgasbayev, A.; Khaimuldin, N. Optimized Interpretable Kolmogorov-Arnold Networks (OIKAN) for Neuro-Symbolic Machine Learning. In Proceedings of the 2nd International Students Conference “Digital Generation-2025”; Astana IT University: Astana, Kazakhstan, 2025; pp. 638–646. ISBN 978-601-7911-72-0. [Google Scholar]
- Harmon, I.; Weinstein, B.; Bohlman, S.; White, E.; Wang, D.Z. A neuro-symbolic framework for tree crown delineation and tree species classification. Remote Sens. 2024, 16, 4365. [Google Scholar] [CrossRef]
- Hoehndorf, R.; Pesquita, C.; Zhapa-Camacho, F. Neuro-symbolic AI in life sciences. In Handbook on Neurosymbolic AI and Knowledge Graphs; IOS Press: Amsterdam, The Netherlands, 2025; pp. 924–951. [Google Scholar]
- Schmidt, M.; Lipson, H. Distilling free-form natural laws from experimental data. Science 2009, 324, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Fu, W.; Kang, J.; Guo, J.; Guo, J. Bayesian symbolic regression. arXiv 2019, arXiv:1910.08892. [Google Scholar]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Sette, S.; Boullart, L. Genetic programming: Principles and applications. Eng. Appl. Artif. Intell. 2001, 14, 727–736. [Google Scholar] [CrossRef]
- Björck, Å. Least squares methods. In Handbook of Numerical Analysis; Elsevier: Amsterdam, The Netherlands, 1990; Volume 1, pp. 465–652. [Google Scholar]
- Bertsimas, D.; Pauphilet, J.; Van Parys, B. Sparse regression. Stat. Sci. 2020, 35, 555–578. [Google Scholar]
- Udrescu, S.M.; Tegmark, M. AI Feynman: A physics-inspired method for symbolic regression. Sci. Adv. 2020, 6, eaay2631. [Google Scholar] [CrossRef]
- Yi, K.; Wu, J.; Gan, C.; Torralba, A.; Kohli, P.; Tenenbaum, J. Neural-symbolic VQA: Disentangling reasoning from vision and language understanding. In Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montréal, QC, Canada, 3–8 December 2018; Curran Associates Inc.: Red Hook, NY, USA, 2018; Volume 31, pp. 1039–1050. [Google Scholar]
- Vsevolodovna, R.I.M.; Monti, M. Enhancing Large Language Models through Neuro-Symbolic Integration and Ontological Reasoning. arXiv 2025, arXiv:2504.07640. [Google Scholar] [CrossRef]
- He, Y.; Xie, Y.; Yuan, Z.; Sun, L. MLP-KAN: Unifying Deep Representation and Function Learning. arXiv 2024, arXiv:2410.03027. [Google Scholar] [CrossRef]
- Garcez, A.D.A.; Gori, M.; Lamb, L.C.; Serafini, L.; Spranger, M.; Tran, S.N. Neural-symbolic computing: An effective methodology for principled integration of machine learning and reasoning. arXiv 2019, arXiv:1905.06088. [Google Scholar] [CrossRef]
- Ellis, K.; Wong, L.; Nye, M.; Sable-Meyer, M.; Cary, L.; Anaya Pozo, L.; Tenenbaum, J.B. DreamCoder: Growing generalizable, interpretable knowledge with wake–sleep Bayesian program learning. Philos. Trans. R. Soc. A 2023, 381, 20220050. [Google Scholar] [CrossRef] [PubMed]
- Acharya, K.; Raza, W.; Dourado, C.; Velasquez, A.; Song, H.H. Neurosymbolic reinforcement learning and planning: A survey. IEEE Trans. Artif. Intell. 2023, 5, 1939–1953. [Google Scholar] [CrossRef]
- Colelough, B.C.; Regli, W. Neuro-symbolic AI in 2024: A systematic review. arXiv 2025, arXiv:2501.05435. [Google Scholar] [CrossRef]
- Nawaz, U.; Anees-ur-Rahaman, M.; Saeed, Z. A review of neuro-symbolic AI integrating reasoning and learning for advanced cognitive systems. Intell. Syst. Appl. 2025, 26, 200541. [Google Scholar] [CrossRef]
- Ennab, M.; Mcheick, H. Designing an interpretability-based model to explain the artificial intelligence algorithms in healthcare. Diagnostics 2022, 12, 1557. [Google Scholar] [CrossRef]
- de Franca, F.O.; Virgolin, M.; Kommenda, M.; Majumder, M.S.; Cranmer, M.; Espada, G.; La Cava, W.G. Interpretable Symbolic Regression for Data Science: Analysis of the 2022 Competition. arXiv 2023, arXiv:2304.01117. [Google Scholar] [CrossRef]
- Cranmer, M. Interpretable machine learning for science with PySR and SymbolicRegression. arXiv 2023, arXiv:2305.01582. [Google Scholar]
- La Cava, W.; Burlacu, B.; Virgolin, M.; Kommenda, M.; Orzechowski, P.; de França, F.O.; Moore, J.H. Contemporary symbolic regression methods and their relative performance. Adv. Neural Inf. Process. Syst. 2021, 2021, 1. [Google Scholar] [PubMed]
- gplearn: Genetic Programming in Python. Available online: https://github.com/trevorstephens/gplearn (accessed on 19 June 2025).
- Kim, S.; Lu, P.Y.; Mukherjee, S.; Gilbert, M.; Jing, L.; Čeperić, V.; Soljačić, M. Integration of neural network-based symbolic regression in deep learning for scientific discovery. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4166–4177. [Google Scholar] [CrossRef] [PubMed]
- pykan: Kolmogorov–Arnold Networks in Python. Available online: https://github.com/KindXiaoming/pykan (accessed on 19 June 2025).
- Kulkarni, T.D.; Saeedi, A.; Gautam, S.; Gershman, S.J. Deep successor reinforcement learning. arXiv 2016, arXiv:1606.02396. [Google Scholar] [CrossRef]
- Broløs, K.R.; Machado, M.V.; Cave, C.; Kasak, J.; Stentoft-Hansen, V.; Batanero, V.G.; Wilstrup, C. An approach to symbolic regression using Feyn. arXiv 2021, arXiv:2104.05417. [Google Scholar] [CrossRef]
- SRTransformer: Symbolic Regression with Transformers. Available online: https://github.com/yinghanlong/SRtransformer (accessed on 19 June 2025).
- Eureqa (DataRobot Documentation). Available online: https://docs.datarobot.com/en/docs/modeling/analyze-models/describe/eureqa.html (accessed on 19 June 2025).
- Schmidt-Hieber, J. The Kolmogorov–Arnold representation theorem revisited. Neural Netw. 2021, 137, 119–126. [Google Scholar] [CrossRef]
- Bebis, G.; Georgiopoulos, M. Feed-forward neural networks. IEEE Potentials 1994, 13, 27–31. [Google Scholar] [CrossRef]
- McHutchon, A.; Rasmussen, C. Gaussian process training with input noise. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Granada, Spain, 12–15 December 2011; Curran Associates Inc.: Red Hook, NY, USA, 2011; Volume 24. [Google Scholar]
- Tay, J.K.; Narasimhan, B.; Hastie, T. Elastic net regularization paths for all generalized linear models. J. Stat. Softw. 2023, 106, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.S.; La Cava, W.; Orzechowski, P.; Urbanowicz, R.J.; Moore, J.H. PMLB: A large benchmark suite for machine learning evaluation and comparison. BioData Min. 2017, 10, 1–13. [Google Scholar] [CrossRef]
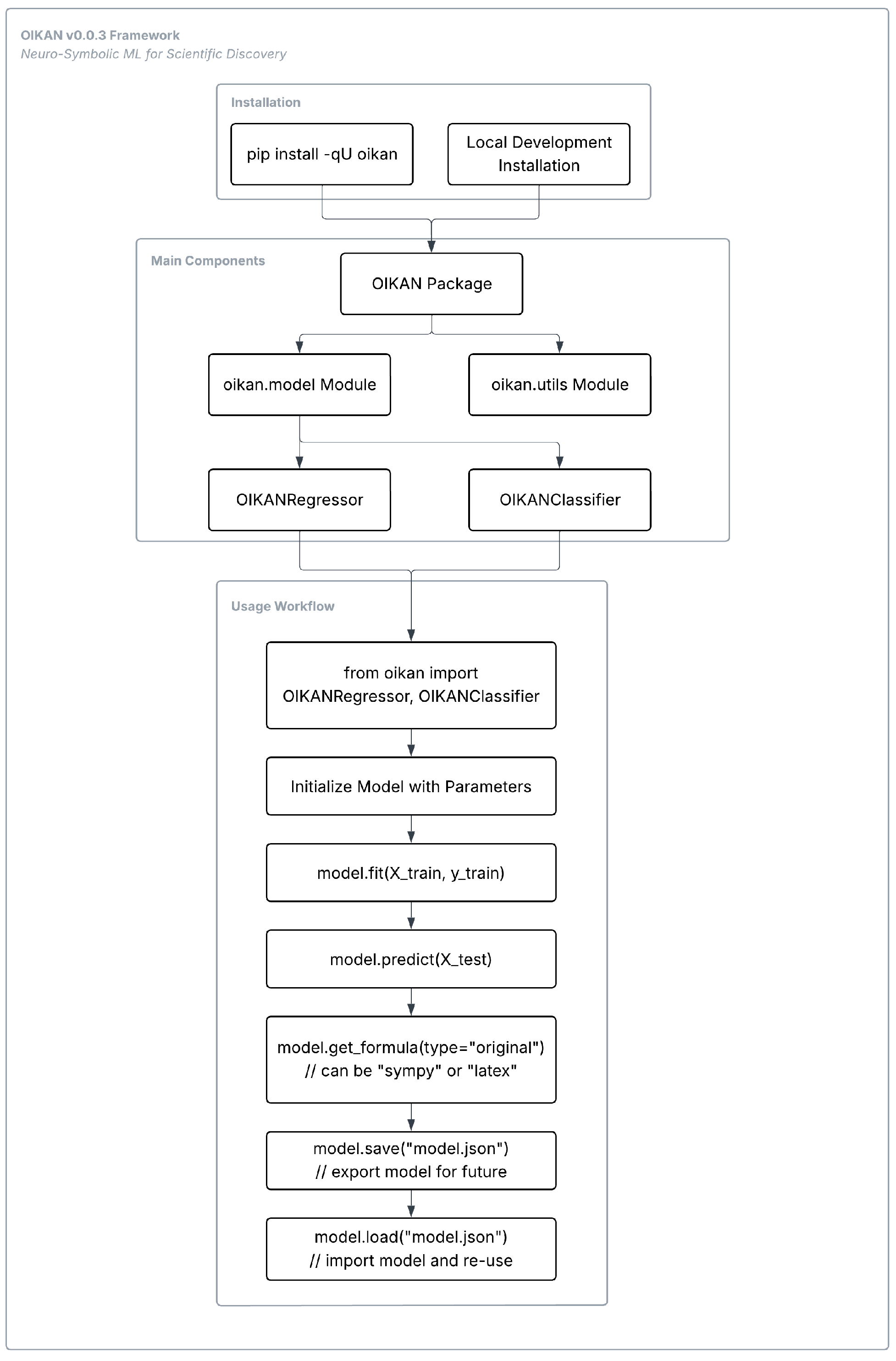

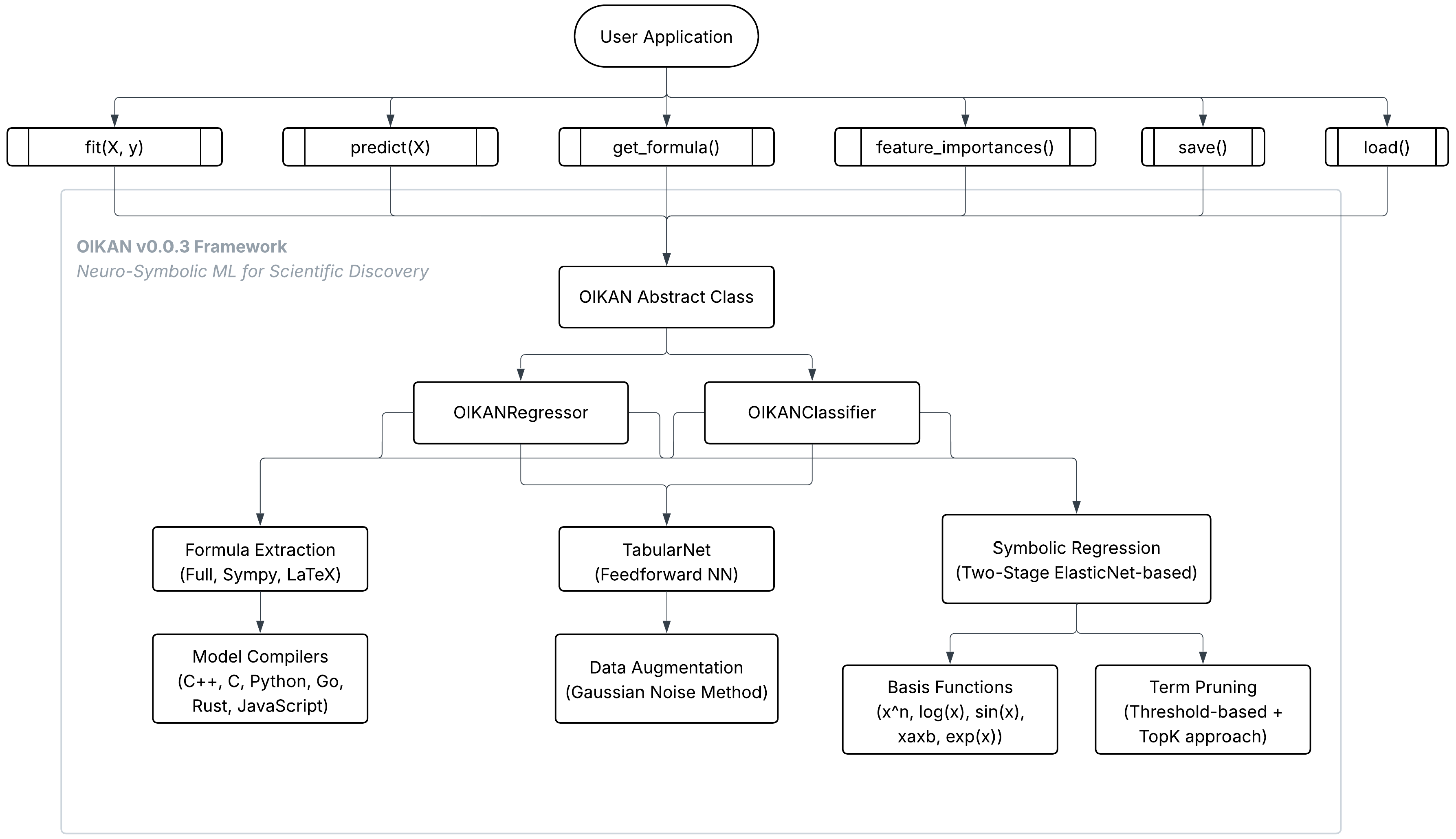


| Step | Description | Purpose |
|---|---|---|
| Data Preprocessing and Augmentation | A feedforward NN (TabularNet) analyzes input data to capture initial patterns. Gaussian noise is added to the data, creating multiple perturbed versions to augment the dataset. The NN then generates predictions or logits for the augmented data. | Enhances model robustness by introducing controlled variability, allowing the model to learn generalized patterns. The NN provides a preliminary mapping of data relationships, which is refined in later steps. |
| Two-Stage Symbolic Regression | Stage 1 (Coarse Model): Polynomial features (degree 2) are generated from the augmented data. An ElasticNet model, combining L1 (LASSO) and L2 (Ridge) regularization, is fitted to compute feature importances based on coefficients. The top-k features are selected. Stage 2 (Refined Model): Non-linear basis functions are added for the top-k features. A second ElasticNet model is fitted to the combined polynomial and non-linear features. | Constructs an interpretable model by first identifying the most relevant features (Stage 1) and then enhancing the model with non-linear transformations (Stage 2). ElasticNet ensures sparsity and robustness, balancing model complexity and accuracy. |
| Model Compilation | The coefficients and basic functions from the refined model are used to construct a symbolic mathematical expression, representing the relationship between features and the target. The expression is simplified using SymPy and supports formats like original, SymPy, or LATEX. | Produces a human-readable, interpretable formula that encapsulates the learned relationships, enabling efficient predictions and easy inspection of the model’s logic. |
| Model Export and Use | The symbolic model, including basis functions, coefficients, and feature metadata, is saved in JSON format. The model can be loaded and used in multiple languages (C, C++, JavaScript, Rust, Go, Python). | Facilitates model portability and deployment across diverse platforms, ensuring accessibility and usability for different applications while maintaining interpretability and efficiency. |
| Parameter | Stage | Description | Values |
|---|---|---|---|
| hidden_sizes | NN Training | List of hidden layer sizes for the MLP. | List of integers, default = [64, 64] |
| activation | NN Training | Activation function for the neural network layers. | Options: ‘relu’, ‘tanh’, ‘leaky_relu’, ‘elu’, ‘swish’, ‘gelu’, default=‘relu’ |
| epochs | NN Training | Number of epochs for training the neural network. | Integer, default = 100 |
| lr | NN Training | Learning rate for the optimizer. | Float, default = 0.001 |
| batch_size | NN Training | Batch size for mini-batch training. | Integer, default = 32 |
| evaluate_nn | NN Training | Whether to evaluate the NN before full training. | Boolean, default = False |
| augmentation_factor | Data Augmentation | Number of augmented samples generated per original sample. | Integer, default = 10 |
| sigma | Data Augmentation | Standard deviation of Gaussian noise added during data augmentation. | Float, default = 0.1 |
| alpha | Symbolic Regression | L1 regularization strength for Lasso in symbolic regression. | Float, default = 0.1 |
| top_k | Symbolic Regression | Number of top features to select in hierarchical symbolic regression. | Integer, default = 5 |
| verbose | General | Whether to display training progress for neural network and symbolic regression. | Boolean, default = False |
| random_state | General | Random seed for reproducibility. | Integer, default = None |
| Requirement | Details |
|---|---|
| Python | Version 3.7 or higher |
| Operating System | Platform independent (Windows/macOS/Linux) |
| Memory | Recommended minimum 4 GB RAM |
| Disk Space | ~100 MB for installation (including dependencies) |
| GPU | Optional (for faster training) |
| Dependencies | torch, numpy, scikit-learn, sympy, tqdm |
| Metrics | Formula | Type | Description |
|---|---|---|---|
| R2 Score (Coefficient of Determination) | Regression | Proportion of variance explained by the model. Higher (0 to 1) is better. | |
| Root Mean Squared Error (RMSE) | Regression | Measures average prediction error magnitude in target units. Lower is better. | |
| Mean Absolute Percentage Error (MAPE) | Regression | Average percentage error of predictions. Lower is better. | |
| Accuracy | 7 | Classification | Proportion of correct predictions. Higher is better, but less reliable for imbalanced data. |
| Precision (Weighted) | Classification | Weighted proportion of positive predictions that are correct. Higher is better. | |
| F1-Score (Weighted) | Classification | Weighted harmonic mean of precision and recall. Higher is better for balanced performance. |
| Dataset | #Feat. * | Rows | Accuracy | Precision | F1 |
|---|---|---|---|---|---|
| iris | 4 | 1200 | 1.000 | 1.000 | 1.000 |
| analcatdata_creditscore | 6 | 1040 | 1.000 | 1.000 | 1.000 |
| corral | 6 | 1280 | 1.000 | 1.000 | 1.000 |
| prnn_crabs | 7 | 1600 | 1.000 | 1.000 | 1.000 |
| mushroom | 22 | 64,990 | 1.000 | 1.000 | 1.000 |
| analcatdata_authorship | 70 | 6720 | 1.000 | 1.000 | 1.000 |
| clean2 | 168 | 52,780 | 1.000 | 1.000 | 1.000 |
| dermatology | 34 | 2920 | 0.986 | 0.988 | 0.986 |
| optdigits | 64 | 44,960 | 0.985 | 0.985 | 0.985 |
| new_thyroid | 5 | 1720 | 0.977 | 0.978 | 0.976 |
| dis | 29 | 30,170 | 0.975 | 0.961 | 0.968 |
| mfeat_karhunen | 64 | 16,000 | 0.973 | 0.973 | 0.972 |
| mfeat_pixel | 240 | 16,000 | 0.968 | 0.968 | 0.967 |
| hypothyroid | 25 | 25,300 | 0.961 | 0.957 | 0.959 |
| monk3 | 6 | 4430 | 0.955 | 0.956 | 0.955 |
| balance_scale | 4 | 5000 | 0.952 | 0.955 | 0.944 |
| clean1 | 168 | 3800 | 0.948 | 0.948 | 0.948 |
| kr_vs_kp | 36 | 25,560 | 0.947 | 0.947 | 0.947 |
| page_blocks | 10 | 43,780 | 0.944 | 0.945 | 0.937 |
| ionosphere | 34 | 2800 | 0.930 | 0.932 | 0.929 |
| analcatdata_lawsuit | 4 | 2110 | 0.925 | 0.914 | 0.916 |
| coil2000 | 85 | 78,570 | 0.909 | 0.892 | 0.900 |
| backache | 32 | 1440 | 0.889 | 0.902 | 0.865 |
| car_evaluation | 6 | 13,820 | 0.887 | 0.891 | 0.884 |
| prnn_synth | 2 | 2000 | 0.880 | 0.880 | 0.880 |
| mfeat_zernike | 47 | 16,000 | 0.860 | 0.862 | 0.851 |
| ecoli | 7 | 2610 | 0.833 | 0.811 | 0.791 |
| lymphography | 18 | 1180 | 0.833 | 0.851 | 0.831 |
| tic_tac_toe | 9 | 7660 | 0.823 | 0.822 | 0.821 |
| analcatdata_boxing2 | 3 | 1050 | 0.815 | 0.813 | 0.813 |
| confidence | 3 | 1026 | 0.800 | 0.778 | 0.775 |
| biomed | 8 | 1670 | 0.786 | 0.835 | 0.735 |
| lupus | 3 | 1035 | 0.778 | 0.781 | 0.769 |
| phoneme | 5 | 43,230 | 0.765 | 0.754 | 0.734 |
| adult | 14 | 78,146 | 0.760 | 0.738 | 0.745 |
| labor | 16 | 1035 | 0.750 | 0.771 | 0.756 |
| hepatitis | 19 | 1240 | 0.742 | 0.742 | 0.742 |
| led7 | 7 | 25,600 | 0.734 | 0.743 | 0.730 |
| appendicitis | 7 | 1008 | 0.727 | 0.529 | 0.612 |
| hayes_roth | 4 | 1280 | 0.719 | 0.753 | 0.710 |
| sonar | 60 | 1660 | 0.714 | 0.716 | 0.714 |
| mfeat_fourier | 76 | 16,000 | 0.670 | 0.731 | 0.633 |
| saheart | 9 | 3690 | 0.656 | 0.688 | 0.555 |
| monk1 | 6 | 4440 | 0.652 | 0.650 | 0.649 |
| haberman | 3 | 2440 | 0.645 | 0.592 | 0.609 |
| monk2 | 6 | 4800 | 0.620 | 0.393 | 0.481 |
| collins | 23 | 3880 | 0.598 | 0.605 | 0.557 |
| ring | 20 | 59,200 | 0.585 | 0.778 | 0.511 |
| analcatdata_fraud | 11 | 1023 | 0.556 | 0.309 | 0.397 |
| movement_libras | 90 | 2880 | 0.556 | 0.687 | 0.588 |
| penguins | 7 | 2660 | 0.537 | 0.327 | 0.395 |
| analcatdata_boxing1 | 3 | 1056 | 0.500 | 0.500 | 0.500 |
| flags | 43 | 1420 | 0.472 | 0.487 | 0.462 |
| mfeat_morphological | 6 | 16,000 | 0.445 | 0.331 | 0.357 |
| bupa | 5 | 2760 | 0.435 | 0.189 | 0.264 |
| parity5+5 | 10 | 8990 | 0.418 | 0.421 | 0.405 |
| mfeat_factors | 216 | 16,000 | 0.385 | 0.521 | 0.346 |
| parity5 | 5 | 1000 | 0.286 | 0.082 | 0.127 |
| schizo | 14 | 2720 | 0.221 | 0.338 | 0.190 |
| analcatdata_dmft | 4 | 6370 | 0.181 | 0.195 | 0.169 |
| Dataset | #Feat. | Rows | Train (s) | Train (MB) | Pred (s) | Pred (MB) |
|---|---|---|---|---|---|---|
| iris | 4 | 1200 | 1.147 | 1.756 | 0.001 | 0.007 |
| analcatdata_creditscore | 6 | 1040 | 0.859 | 2.276 | 0.001 | 0.006 |
| corral | 6 | 1280 | 1.113 | 2.788 | 0.002 | 0.012 |
| prnn_crabs | 7 | 1600 | 1.387 | 4.004 | 0.002 | 0.014 |
| mushroom | 22 | 64,990 | 100.766 | 646.607 | 0.029 | 2.147 |
| analcatdata_authorship | 70 | 6720 | 131.617 | 567.542 | 0.062 | 2.041 |
| clean2 | 168 | 52,780 | 2944.120 | 24,386.306 | 1.165 | 149.450 |
| dermatology | 34 | 2920 | 12.370 | 70.326 | 0.026 | 0.182 |
| optdigits | 64 | 44,960 | 1042.342 | 3157.350 | 0.078 | 13.036 |
| new_thyroid | 5 | 1720 | 1.650 | 3.265 | 0.001 | 0.012 |
| dis | 29 | 30,170 | 55.173 | 488.690 | 0.021 | 1.070 |
| mfeat_karhunen | 64 | 16,000 | 426.694 | 1129.190 | 0.071 | 5.215 |
| mfeat_pixel | 240 | 16,000 | 4278.023 | 14,991.724 | 0.591 | 40.449 |
| hypothyroid | 25 | 25,300 | 41.107 | 317.996 | 0.018 | 0.837 |
| monk3 | 6 | 4430 | 4.173 | 9.545 | 0.001 | 0.026 |
| balance_scale | 4 | 5000 | 4.892 | 7.276 | 0.002 | 0.029 |
| clean1 | 168 | 3800 | 277.464 | 1765.348 | 0.420 | 7.985 |
| kr_vs_kp | 36 | 25,560 | 48.079 | 611.387 | 0.023 | 1.051 |
| page_blocks | 10 | 43,780 | 54.460 | 140.837 | 0.016 | 0.684 |
| ionosphere | 34 | 2800 | 4.917 | 67.644 | 0.012 | 0.092 |
| analcatdata_lawsuit | 4 | 2110 | 2.003 | 3.036 | 0.001 | 0.012 |
| coil2000 | 85 | 78570 | 595.198 | 9532.018 | 0.094 | 12.436 |
| backache | 32 | 1440 | 2.655 | 34.219 | 0.010 | 0.029 |
| car_evaluation | 6 | 13820 | 14.807 | 29.910 | 0.008 | 0.145 |
| prnn_synth | 2 | 2000 | 1.758 | 1.352 | 0.001 | 0.008 |
| mfeat_zernike | 47 | 16,000 | 293.332 | 630.603 | 0.049 | 3.516 |
| ecoli | 7 | 2610 | 2.563 | 6.565 | 0.002 | 0.028 |
| lymphography | 18 | 1180 | 1.779 | 10.394 | 0.004 | 0.023 |
| tic_tac_toe | 9 | 7660 | 7.662 | 24.942 | 0.006 | 0.072 |
| analcatdata_boxing2 | 3 | 1050 | 1.179 | 1.110 | 0.001 | 0.004 |
| confidence | 3 | 1026 | 0.649 | 1.124 | 0.001 | 0.004 |
| biomed | 8 | 1670 | 1.835 | 4.796 | 0.002 | 0.023 |
| lupus | 3 | 1035 | 0.844 | 1.097 | 0.001 | 0.003 |
| phoneme | 5 | 43,230 | 38.545 | 74.135 | 0.004 | 0.217 |
| adult | 14 | 78,146 | 410.054 | 409.513 | 0.049 | 9.926 |
| labor | 16 | 1035 | 0.827 | 7.542 | 0.004 | 0.008 |
| hepatitis | 19 | 1240 | 1.592 | 11.894 | 0.005 | 0.029 |
| led7 | 7 | 25,600 | 27.383 | 61.526 | 0.006 | 0.318 |
| appendicitis | 7 | 1008 | 0.899 | 2.640 | 0.003 | 0.007 |
| hayes_roth | 4 | 1280 | 1.163 | 1.866 | 0.001 | 0.008 |
| sonar | 60 | 1660 | 3.661 | 110.948 | 0.008 | 0.042 |
| mfeat_fourier | 76 | 16,000 | 141.373 | 1570.264 | 0.035 | 1.673 |
| saheart | 9 | 3690 | 3.806 | 12.037 | 0.002 | 0.043 |
| monk1 | 6 | 4440 | 4.012 | 9.573 | 0.001 | 0.031 |
| haberman | 3 | 2440 | 2.169 | 2.594 | 0.001 | 0.010 |
| monk2 | 6 | 4800 | 4.396 | 10.346 | 0.001 | 0.027 |
| collins | 23 | 3880 | 21.469 | 49.628 | 0.017 | 0.153 |
| ring | 20 | 59,200 | 77.061 | 503.507 | 0.008 | 0.451 |
| analcatdata_fraud | 11 | 1023 | 0.584 | 4.340 | 0.001 | 0.002 |
| movement_libras | 90 | 2880 | 256.894 | 400.099 | 0.101 | 1.753 |
| penguins | 7 | 2660 | 2.932 | 6.651 | 0.002 | 0.026 |
| analcatdata_boxing1 | 3 | 1056 | 0.860 | 1.116 | 0.001 | 0.004 |
| flags | 43 | 1420 | 6.841 | 54.917 | 0.014 | 0.055 |
| mfeat_morphological | 6 | 16000 | 18.853 | 35.406 | 0.004 | 0.113 |
| bupa | 5 | 2760 | 2.669 | 5.274 | 0.001 | 0.018 |
| parity5+5 | 10 | 8990 | 7.889 | 33.289 | 0.004 | 0.032 |
| mfeat_factors | 216 | 16,000 | 5980.859 | 12,166.962 | 0.899 | 59.125 |
| parity5 | 5 | 1000 | 0.287 | 1.900 | 0.001 | 0.002 |
| schizo | 14 | 2720 | 4.063 | 16.103 | 0.004 | 0.071 |
| analcatdata_dmft | 4 | 6370 | 6.122 | 9.273 | 0.002 | 0.039 |
| Dataset | #Feat. | Rows | RMSE | R2 | MAPE |
|---|---|---|---|---|---|
| 344_mv | 10 | 97,842 | 0.950 | 0.992 | 2.772 |
| 523_analcatdata_neavote | 2 | 1040 | 0.885 | 0.938 | 0.446 |
| 215_2dplanes | 10 | 97,842 | 1.098 | 0.937 | 1.509 |
| 210_cloud | 5 | 1032 | 0.353 | 0.920 | 0.407 |
| 624_fri_c0_100_5 | 5 | 1040 | 0.327 | 0.875 | 0.453 |
| 649_fri_c0_500_5 | 5 | 4000 | 0.382 | 0.862 | 2.156 |
| 229_pwLinear | 10 | 1600 | 1.386 | 0.862 | 0.418 |
| 595_fri_c0_1000_10 | 10 | 8000 | 0.386 | 0.855 | 0.958 |
| 230_machine_cpu | 6 | 1670 | 87.062 | 0.851 | 0.732 |
| 1027_ESL | 4 | 3900 | 0.507 | 0.836 | 0.082 |
| 590_fri_c0_1000_50 | 50 | 8000 | 0.399 | 0.825 | 1.034 |
| 609_fri_c0_1000_5 | 5 | 8000 | 0.438 | 0.823 | 1.495 |
| 1096_FacultySalaries | 4 | 1000 | 1.936 | 0.797 | 0.036 |
| 561_cpu | 7 | 1670 | 104.397 | 0.797 | 0.278 |
| 529_pollen | 4 | 30,780 | 1.477 | 0.786 | 3.445 |
| 294_satellite_image | 36 | 51,480 | 1.073 | 0.763 | 0.360 |
| 579_fri_c0_250_5 | 5 | 2000 | 0.439 | 0.757 | 2.838 |
| 635_fri_c0_250_10 | 10 | 2000 | 0.478 | 0.757 | 2.842 |
| 201_pol | 48 | 96,000 | 22.270 | 0.708 | 4.76 × 1016 |
| 564_fried | 10 | 97,842 | 2.693 | 0.705 | 0.229 |
| 621_fri_c0_100_10 | 10 | 1040 | 0.589 | 0.687 | 0.810 |
| 599_fri_c2_1000_5 | 5 | 8000 | 0.605 | 0.670 | 1.108 |
| 192_vineyard | 2 | 1025 | 2.503 | 0.665 | 0.138 |
| 1089_USCrime | 13 | 1036 | 20.204 | 0.663 | 0.090 |
| 589_fri_c2_1000_25 | 25 | 8000 | 0.635 | 0.623 | 0.952 |
| 612_fri_c1_1000_5 | 5 | 8000 | 0.600 | 0.615 | 1.584 |
| 623_fri_c4_1000_10 | 10 | 8000 | 0.618 | 0.609 | 1.610 |
| 4544_GeographicalOriginalofMusic | 117 | 8470 | 0.580 | 0.597 | 4.051 |
| 620_fri_c1_1000_25 | 25 | 8000 | 0.642 | 0.592 | 1.125 |
| 592_fri_c4_1000_25 | 25 | 8000 | 0.613 | 0.570 | 1.322 |
| 586_fri_c3_1000_25 | 25 | 8000 | 0.691 | 0.569 | 1.448 |
| 588_fri_c4_1000_100 | 100 | 8000 | 0.678 | 0.555 | 1.224 |
| 583_fri_c1_1000_50 | 50 | 8000 | 0.657 | 0.530 | 2.462 |
| 616_fri_c4_500_50 | 50 | 4000 | 0.691 | 0.528 | 1.074 |
| 605_fri_c2_250_25 | 25 | 2000 | 0.686 | 0.498 | 1.127 |
| 581_fri_c3_500_25 | 25 | 4000 | 0.768 | 0.493 | 6.904 |
| 617_fri_c3_500_5 | 5 | 4000 | 0.808 | 0.417 | 1.925 |
| 225_puma8NH | 8 | 65,530 | 4.265 | 0.413 | 2.799 |
| 1029_LEV | 4 | 8000 | 0.718 | 0.396 | 2.96 × 1014 |
| 1030_ERA | 4 | 8000 | 1.607 | 0.376 | 0.425 |
| 527_analcatdata_election2000 | 14 | 1007 | 96,238.090 | 0.344 | 0.708 |
| 1028_SWD | 10 | 8000 | 0.672 | 0.307 | 0.145 |
| 547_no2 | 7 | 4000 | 0.627 | 0.301 | 0.142 |
| 218_house_8L | 8 | 91,135 | 46,944.468 | 0.215 | 2.71 × 1017 |
| 591_fri_c1_100_10 | 10 | 1040 | 0.687 | 0.124 | 1.454 |
| 228_elusage | 2 | 1012 | 26.929 | - | 0.589 |
| 557_analcatdata_apnea1 | 3 | 3800 | 3925.899 | - | 1.89 × 1018 |
| 556_analcatdata_apnea2 | 3 | 3800 | 4036.276 | - | 1.85 × 1018 |
| 485_analcatdata_vehicle | 4 | 1026 | 373.732 | - | 0.821 |
| 522_pm10 | 7 | 4000 | 0.915 | - | 0.278 |
| 227_cpu_small | 12 | 65,530 | 189.186 | - | 4.24 × 1015 |
| 562_cpu_small | 12 | 65,530 | 48.787 | - | 2.24 × 1015 |
| 542_pollution | 15 | 1008 | 267.912 | - | 0.262 |
| 574_house_16H | 16 | 91,135 | 53,044.147 | - | 4.88 × 1017 |
| 197_cpu_act | 21 | 65,530 | 22.030 | - | 1.19 × 1015 |
| 573_cpu_act | 21 | 65,530 | 164.100 | - | 2.62 × 1015 |
| Dataset | #Feat. | Rows | Train (s) | Train (MB) | Pred (s) | Pred (MB) |
|---|---|---|---|---|---|---|
| 344_mv | 10 | 97,842 | 282.833 | 301.584 | 0.013 | 3.197 |
| 523_analcatdata_neavote | 2 | 1040 | 0.806 | 0.715 | 0.000 | 0.002 |
| 215_2dplanes | 10 | 97,842 | 266.519 | 301.576 | 0.008 | 1.827 |
| 210_cloud | 5 | 1032 | 0.766 | 1.950 | 0.000 | 0.004 |
| 624_fri_c0_100_5 | 5 | 1040 | 0.750 | 1.963 | 0.001 | 0.004 |
| 649_fri_c0_500_5 | 5 | 4000 | 3.216 | 7.437 | 0.001 | 0.015 |
| 229_pwLinear | 10 | 1600 | 1.341 | 5.944 | 0.002 | 0.018 |
| 595_fri_c0_1000_10 | 10 | 8000 | 6.397 | 29.555 | 0.001 | 0.028 |
| 230_machine_cpu | 6 | 1670 | 1.547 | 3.613 | 0.001 | 0.015 |
| 1027_ESL | 4 | 3900 | 3.340 | 5.539 | 0.001 | 0.018 |
| 590_fri_c0_1000_50 | 50 | 8000 | 7.390 | 357.336 | 0.001 | 0.035 |
| 609_fri_c0_1000_5 | 5 | 8000 | 6.389 | 14.828 | 0.001 | 0.031 |
| 1096_FacultySalaries | 4 | 1000 | 0.539 | 1.446 | 0.001 | 0.003 |
| 561_cpu | 7 | 1670 | 1.575 | 4.159 | 0.001 | 0.018 |
| 529_pollen | 4 | 30,780 | 26.735 | 43.379 | 0.004 | 0.143 |
| 294_satellite_image | 36 | 51,480 | 103.953 | 1222.647 | 0.063 | 6.982 |
| 579_fri_c0_250_5 | 5 | 2000 | 1.681 | 3.741 | 0.001 | 0.008 |
| 635_fri_c0_250_10 | 10 | 2000 | 1.749 | 7.422 | 0.001 | 0.010 |
| 201_pol | 48 | 96,000 | 235.943 | 3885.826 | 0.084 | 16.249 |
| 564_fried | 10 | 97,842 | 267.517 | 301.579 | 0.010 | 2.806 |
| 621_fri_c0_100_10 | 10 | 1040 | 0.748 | 3.885 | 0.001 | 0.006 |
| 599_fri_c2_1000_5 | 5 | 8000 | 6.481 | 14.826 | 0.001 | 0.030 |
| 192_vineyard | 2 | 1025 | 0.504 | 0.699 | 0.001 | 0.002 |
| 1089_USCrime | 13 | 1036 | 0.608 | 5.518 | 0.004 | 0.011 |
| 589_fri_c2_1000_25 | 25 | 8000 | 6.805 | 106.004 | 0.002 | 0.047 |
| 612_fri_c1_1000_5 | 5 | 8000 | 6.678 | 14.826 | 0.001 | 0.032 |
| 623_fri_c4_1000_10 | 10 | 8000 | 6.391 | 29.553 | 0.001 | 0.030 |
| 4544_GeographicalOriginalofMusic | 117 | 8470 | 121.030 | 1926.686 | 0.022 | 0.279 |
| 620_fri_c1_1000_25 | 25 | 8000 | 6.669 | 106.002 | 0.001 | 0.035 |
| 592_fri_c4_1000_25 | 25 | 8000 | 7.189 | 106.000 | 0.002 | 0.043 |
| 586_fri_c3_1000_25 | 25 | 8000 | 6.689 | 106.006 | 0.001 | 0.033 |
| 588_fri_c4_1000_100 | 100 | 8000 | 11.997 | 1340.217 | 0.008 | 0.123 |
| 583_fri_c1_1000_50 | 50 | 8000 | 7.678 | 357.328 | 0.008 | 0.088 |
| 616_fri_c4_500_50 | 50 | 4000 | 4.011 | 182.768 | 0.009 | 0.081 |
| 605_fri_c2_250_25 | 25 | 2000 | 1.791 | 30.500 | 0.005 | 0.033 |
| 581_fri_c3_500_25 | 25 | 4000 | 3.504 | 57.044 | 0.003 | 0.031 |
| 617_fri_c3_500_5 | 5 | 4000 | 3.323 | 7.436 | 0.001 | 0.013 |
| 225_puma8NH | 8 | 65,530 | 55.508 | 159.550 | 0.006 | 0.394 |
| 1029_LEV | 4 | 8000 | 6.511 | 11.306 | 0.001 | 0.029 |
| 1030_ERA | 4 | 8000 | 6.586 | 11.302 | 0.001 | 0.040 |
| 527_analcatdata_election2000 | 14 | 1007 | 0.711 | 5.975 | 0.004 | 0.015 |
| 1028_SWD | 10 | 8000 | 7.321 | 29.554 | 0.002 | 0.049 |
| 547_no2 | 7 | 4000 | 3.689 | 9.905 | 0.001 | 0.032 |
| 218_house_8L | 8 | 91,135 | 164.753 | 218.753 | 0.009 | 2.444 |
| 591_fri_c1_100_10 | 10 | 1040 | 0.741 | 3.883 | 0.001 | 0.006 |
| 228_elusage | 2 | 1012 | 0.498 | 0.692 | 0.000 | 0.002 |
| 557_analcatdata_apnea1 | 3 | 3800 | 3.182 | 3.872 | 0.001 | 0.021 |
| 556_analcatdata_apnea2 | 3 | 3800 | 3.160 | 3.868 | 0.001 | 0.021 |
| 485_analcatdata_vehicle | 4 | 1026 | 0.510 | 1.489 | 0.001 | 0.004 |
| 522_pm10 | 7 | 4000 | 3.650 | 9.903 | 0.001 | 0.028 |
| 227_cpu_small | 12 | 65,530 | 63.710 | 258.117 | 0.018 | 0.998 |
| 562_cpu_small | 12 | 65,530 | 62.503 | 258.115 | 0.005 | 0.867 |
| 542_pollution | 15 | 1008 | 0.675 | 6.639 | 0.005 | 0.016 |
| 574_house_16H | 16 | 91,135 | 176.405 | 539.557 | 0.021 | 6.381 |
| 197_cpu_act | 21 | 65,530 | 77.496 | 602.564 | 0.029 | 2.597 |
| 573_cpu_act | 21 | 65,530 | 78.804 | 602.562 | 0.020 | 2.781 |
| Performance Tier | OIKAN Count | Notable Datasets |
|---|---|---|
| Perfect (1.000) | 6 | mushroom, iris, analcatdata_authorship, analcatdata_creditscore, corral, prnn_crabs |
| Excellent (0.95+) | 17 | monk3 (0.955), balance_scale (0.952), dermatology (0.986) |
| Good (0.80–0.94) | 18 | tic_tac_toe (0.823), car_evaluation (0.887), ionosphere (0.930) |
| Fair (0.60–0.79) | 12 | adult (0.760), penguins (0.537), phoneme (0.765) |
| Poor (<0.60) | 8 | analcatdata_dmft (0.181), parity5 (0.286) |
| Model | Dataset | Accuracy | Precision | F1 |
|---|---|---|---|---|
| OIKAN | iris (150 × 4) | 1 | 1 | 1 |
| ElasticNet | 1 | 1 | 1 | |
| XGBoost | 0.966667 | 0.969444 | 0.966514 | |
| DecisionTree | 1 | 1 | 1 | |
| OIKAN | monk3 (432 × 6) | 0.955 | 0.956 | 0.995 |
| ElasticNet | 0.702703 | 0.716642 | 0.702703 | |
| XGBoost | 0.981982 | 0.981982 | 0.981982 | |
| DecisionTree | 0.972973 | 0.97308 | 0.972946 | |
| OIKAN | mushroom (8124 × 22) | 1 | 1 | 1 |
| ElasticNet | 0.953231 | 0.953235 | 0.95322 | |
| XGBoost | 1 | 1 | 1 | |
| DecisionTree | 1 | 1 | 1 | |
| OIKAN | car_evaluation (1728 × 6) | 0.887 | 0.891 | 0.884 |
| ElasticNet | 0.812139 | 0.801617 | 0.803303 | |
| XGBoost | 0.979769 | 0.985781 | 0.981074 | |
| DecisionTree | 0.965318 | 0.973908 | 0.967508 | |
| OIKAN | kr_vs_kp (3196 × 36) | 0.947 | 0.947 | 0.947 |
| ElasticNet | 0.95 | 0.951953 | 0.950049 | |
| XGBoost | 0.989063 | 0.989099 | 0.989059 | |
| DecisionTree | 0.985938 | 0.986152 | 0.985926 | |
| OIKAN | coil2000 (9822 × 85) | 0.909 | 0.892 | 0.900 |
| ElasticNet | 0.937405 | 0.892946 | 0.910041 | |
| XGBoost | 0.926209 | 0.896749 | 0.909317 | |
| DecisionTree | 0.887023 | 0.89294 | 0.889946 |
| Model | Dataset | RMSE | R2 | MAPE |
|---|---|---|---|---|
| OIKAN | 1027_ESL (488 × 4) | 1.141872 | 0.170539 | 0.192851 |
| ElasticNet | 0.781992 | 0.610985 | 0.140495 | |
| XGBoost | 0.639834 | 0.739567 | 0.092441 | |
| DecisionTree | 0.707947 | 0.681167 | 0.092292 | |
| OIKAN | 192_vineyard (632 × 4) | 2.694328 | 0.611247 | 0.125973 |
| ElasticNet | 2.454929 | 0.677262 | 0.148458 | |
| XGBoost | 3.013396 | 0.513721 | 0.169473 | |
| DecisionTree | 4.012056 | 0.138001 | 0.217817 | |
| OIKAN | 225_puma8NH (8192 × 8) | 5.878939 | -0.11466 | 1.53969 |
| ElasticNet | 4.66571 | 0.29793 | 2.446583 | |
| XGBoost | 3.503805 | 0.604064 | 3.433912 | |
| DecisionTree | 4.642225 | 0.30498 | 3.676655 | |
| OIKAN | 197_cpu_act (8192 × 21) | 1.19 × 1011 | −4.8 × 1019 | 1.67 × 1024 |
| ElasticNet | 8.917987 | 0.734598 | 4.3 × 1015 | |
| XGBoost | 2.482231 | 0.979438 | 8.13 × 1013 | |
| DecisionTree | 3.518798 | 0.95868 | 1.37 × 1013 | |
| OIKAN | 215_2dplanes (40k × 10) | 2.16976 | 0.753329 | 2.666862 |
| ElasticNet | 3.211894 | 0.459474 | 2.692004 | |
| XGBoost | 1.018963 | 0.945598 | 1.315541 | |
| DecisionTree | 1.370601 | 0.901573 | 1.936285 | |
| OIKAN | 229_pwLinear (10k × 10) | 1.657709 | 0.8018 | 0.639679 |
| ElasticNet | 2.434435 | 0.572552 | 0.766036 | |
| XGBoost | 1.846041 | 0.754207 | 0.644108 | |
| DecisionTree | 2.413889 | 0.579736 | 0.829767 |
| Model & Parameters | Training Time (Total) | Weighted F1-Score | Accuracy | Formula |
|---|---|---|---|---|
| OIKANClassifier (with 5× Data Augmentation) | 30m 47s | 0.47 | 0.56 | Yes |
| OIKANClassifier (without Data Augmentation) | 2.8 s | 0.63 | 0.64 | Yes |
| XGBClassifier | 4.5 s | 0.75 | 0.75 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yedilkhan, D.; Zhalgasbayev, A.; Saleshova, S.; Khaimuldin, N. OIKAN: A Hybrid AI Framework Combining Symbolic Inference and Deep Learning for Interpretable Information Retrieval Models. Algorithms 2025, 18, 639. https://doi.org/10.3390/a18100639
Yedilkhan D, Zhalgasbayev A, Saleshova S, Khaimuldin N. OIKAN: A Hybrid AI Framework Combining Symbolic Inference and Deep Learning for Interpretable Information Retrieval Models. Algorithms. 2025; 18(10):639. https://doi.org/10.3390/a18100639
Chicago/Turabian StyleYedilkhan, Didar, Arman Zhalgasbayev, Sabina Saleshova, and Nursultan Khaimuldin. 2025. "OIKAN: A Hybrid AI Framework Combining Symbolic Inference and Deep Learning for Interpretable Information Retrieval Models" Algorithms 18, no. 10: 639. https://doi.org/10.3390/a18100639
APA StyleYedilkhan, D., Zhalgasbayev, A., Saleshova, S., & Khaimuldin, N. (2025). OIKAN: A Hybrid AI Framework Combining Symbolic Inference and Deep Learning for Interpretable Information Retrieval Models. Algorithms, 18(10), 639. https://doi.org/10.3390/a18100639







