Qualitative Perturbation Analysis and Machine Learning: Elucidating Bacterial Optimization of Tryptophan Production
Abstract
1. Introduction
1.1. Related Work
1.2. Research Overview
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Conditions
2.2. Growth Kinetics
2.3. pIAAMHs Induction for IAA Production
2.4. IAA/Tryptophan Determination
2.5. Genome-Scale Metabolic Network Model of E. coli
2.6. Primary Software Utilized
2.7. Compilation of Reference Reactions from the Literature
2.8. Generation of Optima Reactions for Tryptophan Biosynthesis with pFBA
- : Reaction representing biomass formation.
- : Stochiometric matrix.
- : A vector containing the flux values of individual reactions.
- : Flux of the target reaction to be maximized.
- : Flux of when it is maximized.
- : Minimized flux of reaction i.
- : Lower bound of reaction i.
- : The flux of reaction that was evaluated as pFBA optima.
- : The flux of reaction j when is maximized.
- : The upper bound of reaction as evaluated to be pFBA optima.
- : The upper bound of reaction j.
- : The maximized value of reaction k.
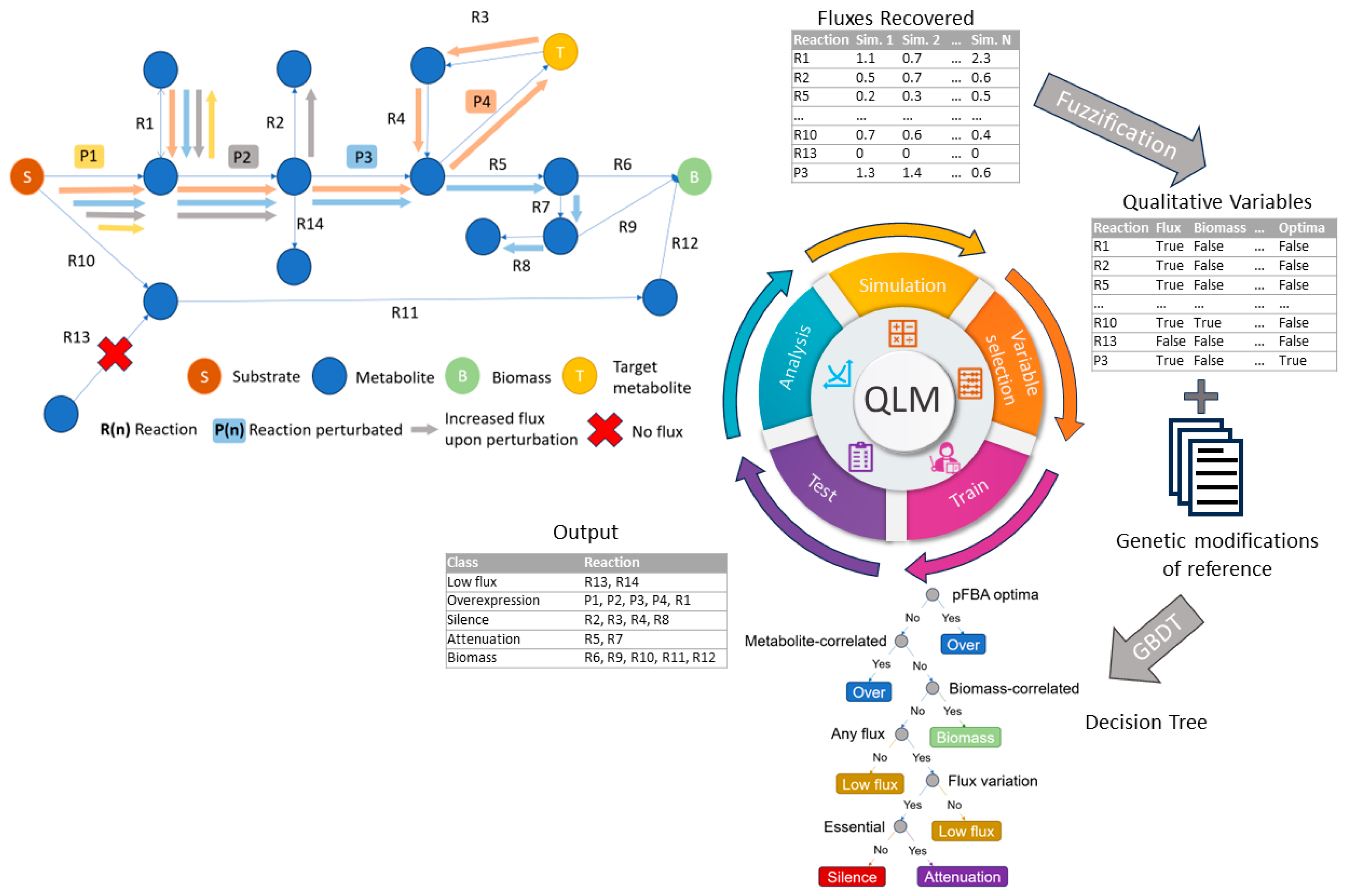
2.9. Perturbating Reactions with FSEOF
- : Flux of when it is minimized.
- : Flux of a pFBA optima reaction subjected to perturbation.
- : Maximum flux of in each simulation.
- : Flux of when the is maximized.
- : Perturbation factor, this was set to 1.5 in simulations.
- : Number of enforced flux levels for FSEOF (default = 10).
2.10. Use of QPA to Transform the Perturbation Fluxes to Reaction–Metabolite Relationships
2.11. Use of QLM and GBDT to Train the QPAML Model with Experimentally Validated Tryptophan Reactions
| Algorithm 1: QPAML model construction algorithm. |
| Input: |
|
Truth label vector y |
|
Qualitative variables X |
| Output |
|
QPAML model F |
|
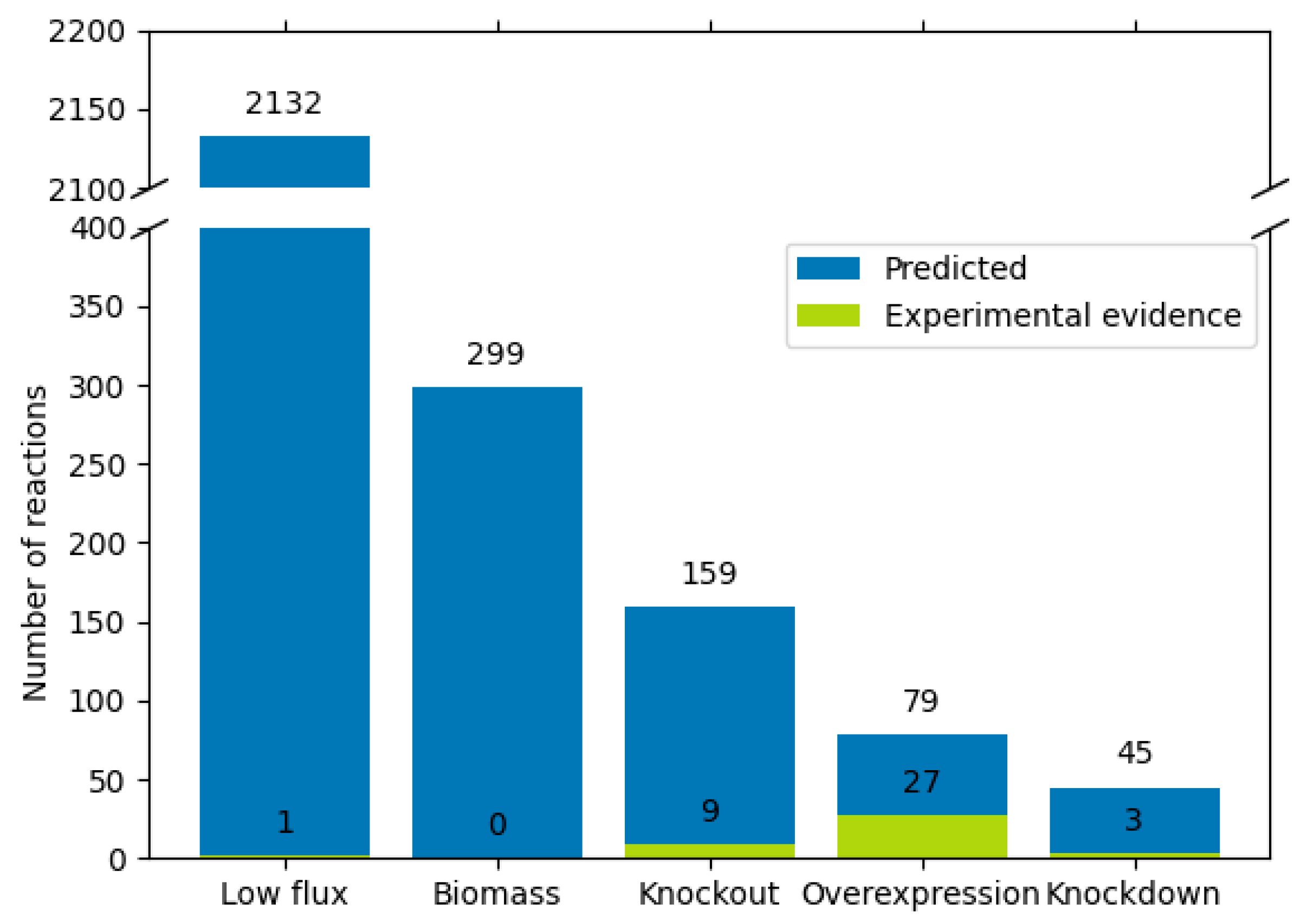
2.12. Classification of 322 Reactions to Produce Tryptophan and Other 30 Endogenous Metabolites
| Algorithm 2: Algorithm for classification of reactions. |
| Input: |
| Target reaction vtarget |
| Output |
| Predicted class vector Y |
|
2.13. Final Qualitative Variables
- : Flux of reaction.
- m: Slope of line, most be greater than 0.
- b: Intersection at origin, must be 0.
- : Flux of biomass.
2.14. Distance Measurement of Relevant Reactions Respect the 76 pFBA Optima Reactions for Tryptophan Production
3. Results
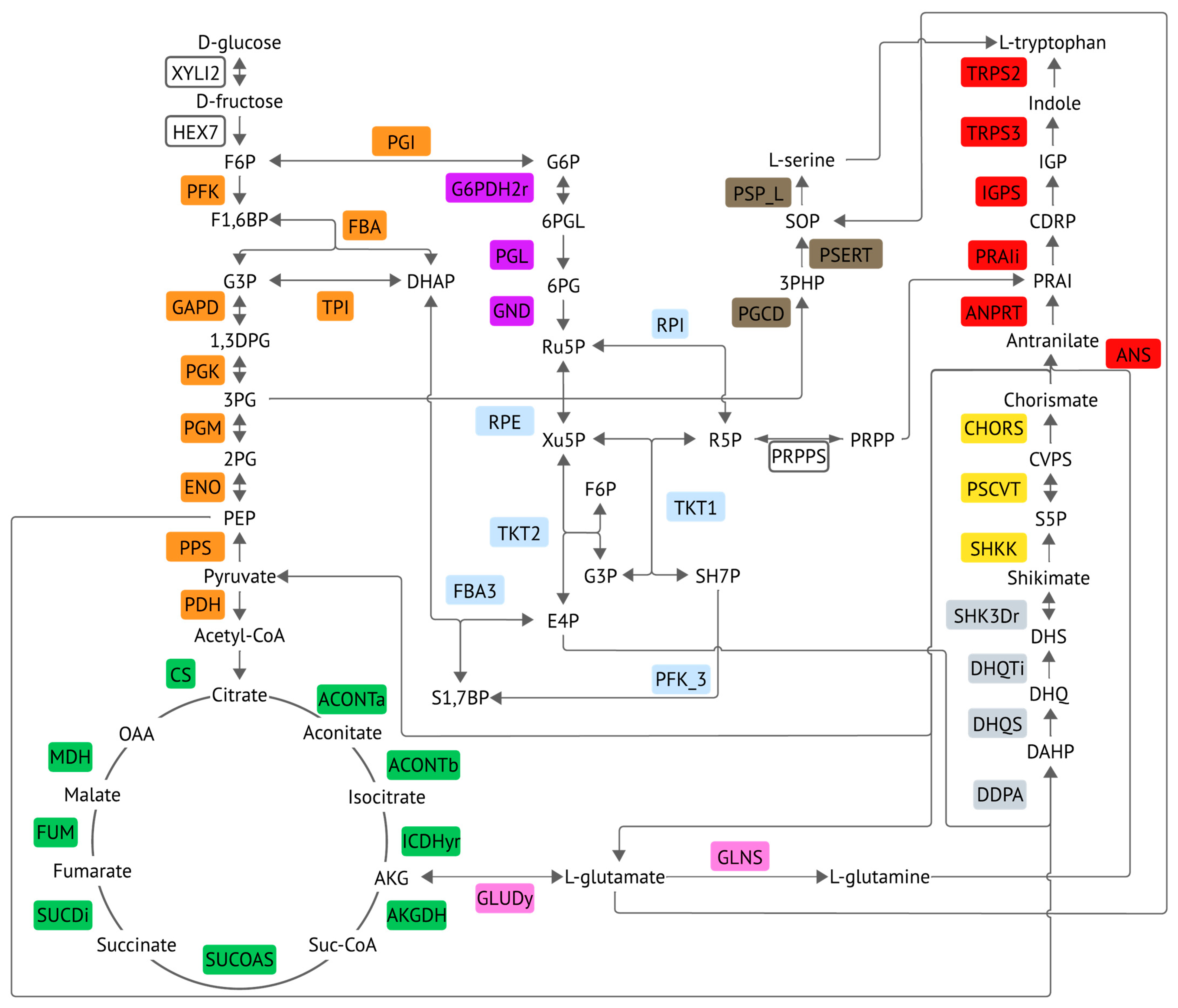
3.1. pFBA Implementation to Define the Core Pathway from Glucose to Tryptophan
3.2. Analysis of Perturbations in Fluxes Propagated by FSEOF
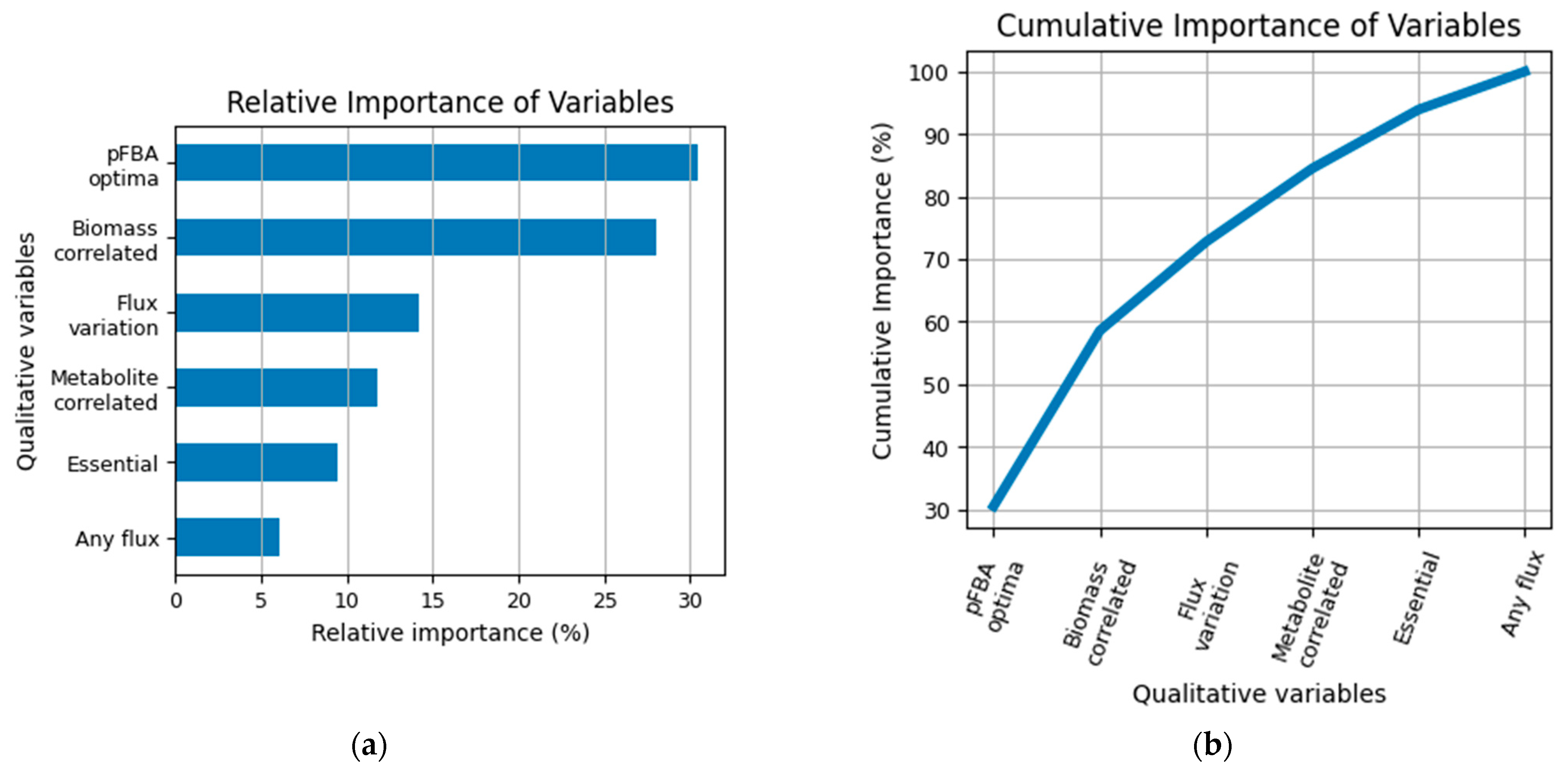
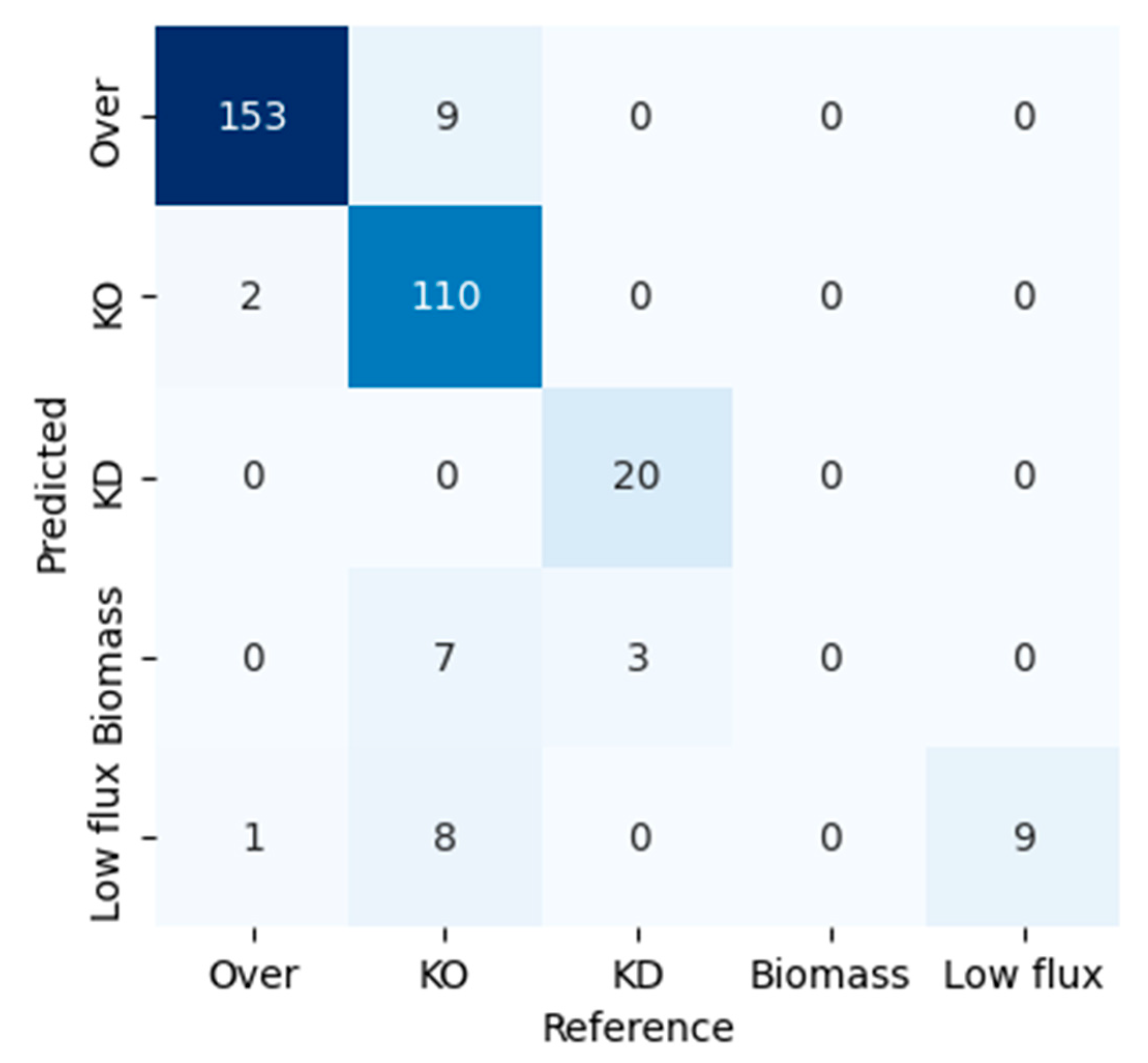
3.3. Conversion of the Flux Variables to a Classification Model Based on QPAML
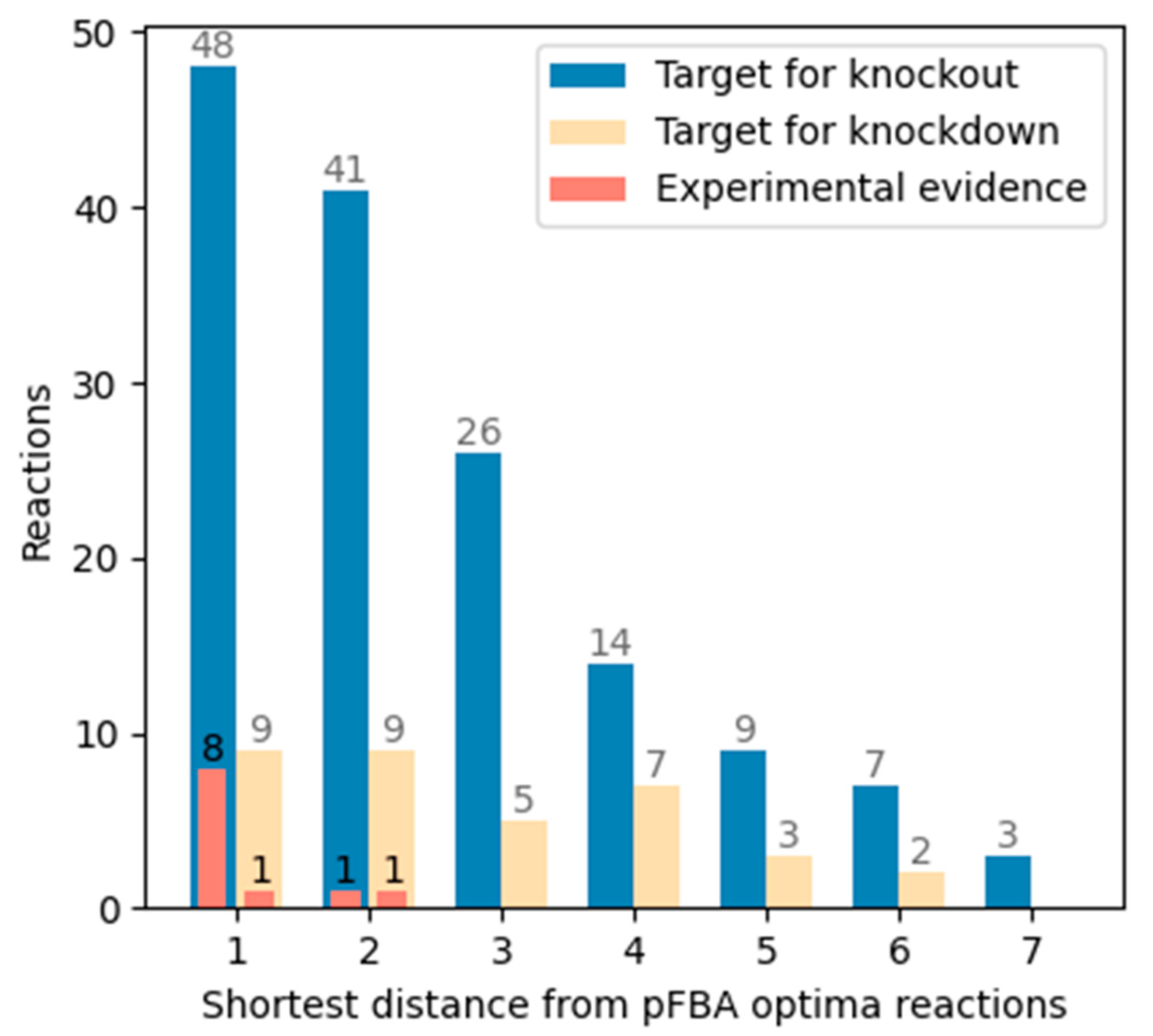
3.4. Shortest Distance from pFBA Optima Reactions
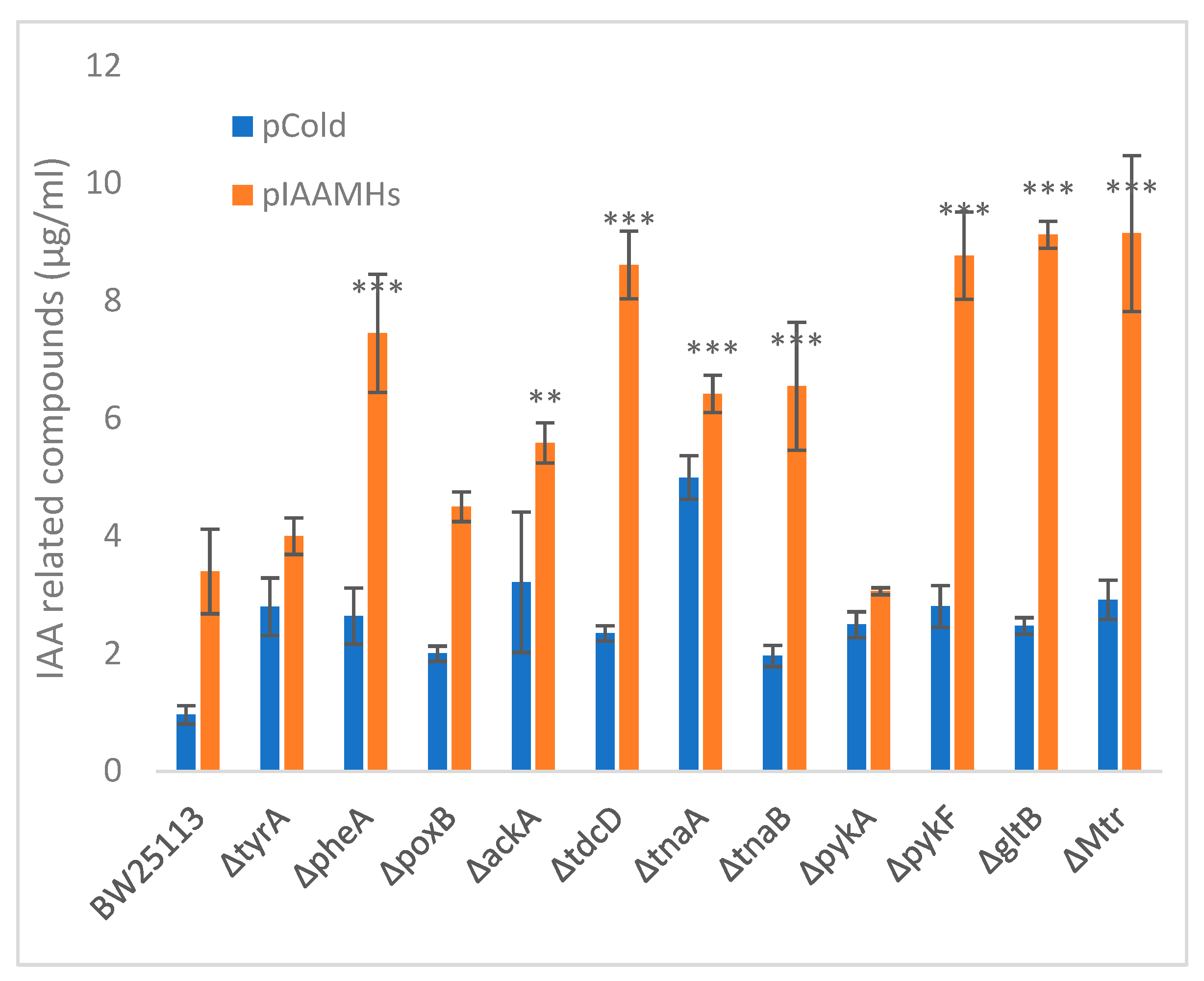
3.5. Production of Indole-3-Acetic Acid by the Keio Mutants
4. Discussion
4.1. Interpretation of the Bacterial Response to Enzymatic Perturbations
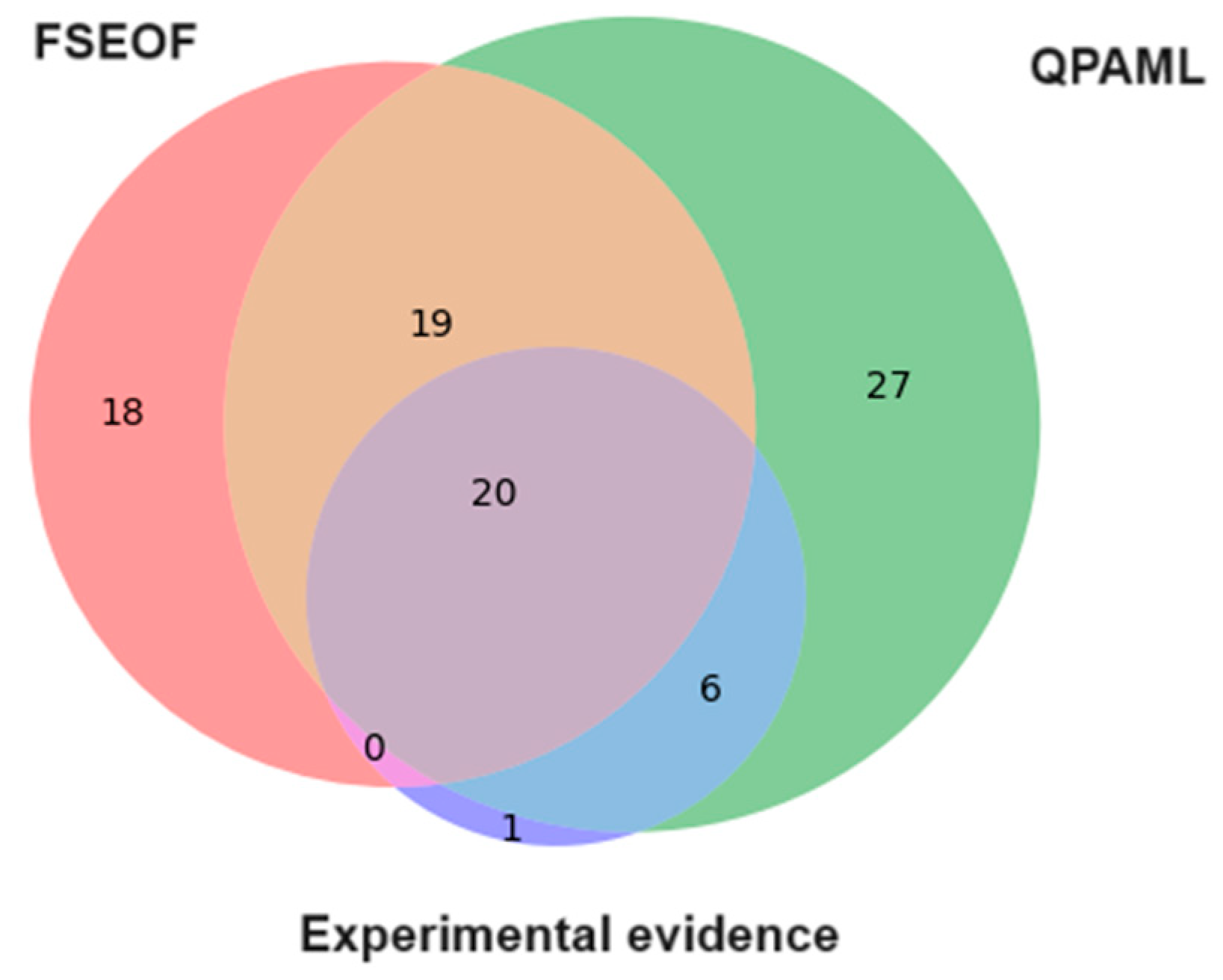
4.2. Comparison of FSEOF and QPAML
4.3. Limitations of the QPAML Classification Model
4.4. Identification of Branching Reactions
4.5. Application of the QPAML Model in Other Metabolic Pathways
4.6. Prioritization in Inactivating Reactions and Objective Function
4.7. Model Interpretation and Strategies Used to Produce Tryptophan
4.8. Advantages of the QPAML Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gang, S.; Sharma, S.; Saraf, M.; Buck, M.; Schumacher, J. Analysis of Indole-3-Acetic Acid (IAA) Production in Klebsiella by LC-MS/MS and the Salkowski Method. Bio-Protocol 2019, 9, e3230. [Google Scholar] [CrossRef] [PubMed]
- Pharkya, P.; Maranas, C.D. An Optimization Framework for Identifying Reaction Activation/Inhibition or Elimination Candidates for Overproduction in Microbial Systems. Metab. Eng. 2006, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, M.; Ma, C.; Zeng, A.-P. Discovery of Feed-Forward Regulation in L-Tryptophan Biosynthesis and Its Use in Metabolic Engineering of E. coli for Efficient Tryptophan Bioproduction. Metab. Eng. 2018, 47, 434–444. [Google Scholar] [CrossRef]
- Castro-López, D.A.; González de la Vara, L.E.; Santillán, M.; Martínez-Antonio, A. A Molecular Dynamic Model of Tryptophan Overproduction in Escherichia coli. Fermentation 2022, 8, 560. [Google Scholar] [CrossRef]
- Panichkin, V.B.; Livshits, V.A.; Biryukova, I.V.; Mashko, S.V. Metabolic Engineering of Escherichia coli for L-Tryptophan Production. Appl. Biochem. Microbiol. 2016, 52, 783–809. [Google Scholar] [CrossRef]
- Du, L.; Zhang, Z.; Xu, Q.; Chen, N. Central Metabolic Pathway Modification to Improve L-Tryptophan Production in Escherichia coli. Bioengineered 2019, 10, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.-J.; Zou, C.; Zhu, Y.-X.; Dai, J.; Chen, S.; Wu, D.; Wu, J.; Chen, J. Development of L-Tryptophan Production Strains by Defined Genetic Modification in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2011, 38, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, J.-Z.; Zhang, W.-G. Advances and Prospects in Metabolic Engineering of Escherichia coli for L-Tryptophan Production. World J. Microbiol. Biotechnol. 2022, 38, 22. [Google Scholar] [CrossRef]
- Li, Z.; Ding, D.; Wang, H.; Liu, L.; Fang, H.; Chen, T.; Zhang, D. Engineering Escherichia coli to Improve Tryptophan Production via Genetic Manipulation of Precursor and Cofactor Pathways. Synth. Syst. Biotechnol. 2020, 5, 200–205. [Google Scholar] [CrossRef]
- Zhao, C.; Cheng, L.; Xu, Q.; Wang, J.; Shen, Z.; Chen, N. Improvement of the Production of L-Tryptophan in Escherichia coli by Application of a Dissolved Oxygen Stage Control Strategy. Ann. Microbiol. 2016, 66, 843–854. [Google Scholar] [CrossRef]
- Xu, Q.; Bai, F.; Chen, N.; Bai, G. Gene Modification of the Acetate Biosynthesis Pathway in Escherichia coli and Implementation of the Cell Recycling Technology to Increase L-Tryptophan Production. PLoS ONE 2017, 12, e0179240. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cheng, L.-K.; Wang, J.; Liu, Q.; Shen, T.; Chen, N. Genetic Engineering of Escherichia coli to Enhance Production of L-Tryptophan. Appl. Microbiol. Biotechnol. 2013, 97, 7587–7596. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Liu, Q.; Xie, X.; Xu, Q.; Chen, N. Improved Production of Tryptophan in Genetically Engineered Escherichia coli with TktA and PpsA Overexpression. J. Biomed. Biotechnol. 2012, 2012, 605219. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Duan, X.; Wu, J. L-Tryptophan Production in Escherichia coli Improved by Weakening the Pta-AckA Pathway. PLoS ONE 2016, 11, e0158200. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, B.-B.; Xu, J.-Z.; Zhang, W.-G. Engineering of Shikimate Pathway and Terminal Branch for Efficient Production of L-Tryptophan in Escherichia coli. Int. J. Mol. Sci. 2023, 24, 11866. [Google Scholar] [CrossRef] [PubMed]
- Schoppel, K.; Trachtmann, N.; Korzin, E.J.; Tzanavari, A.; Sprenger, G.A.; Weuster-Botz, D. Metabolic Control Analysis Enables Rational Improvement of E. coli L-Tryptophan Producers but Methylglyoxal Formation Limits Glycerol-Based Production. Microb. Cell Fact. 2022, 21, 201. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, R.; Zhang, Y.; Yang, Y.; Lin, Y.; Yan, Y. Developing a Pyruvate-Driven Metabolic Scenario for Growth-Coupled Microbial Production. Metab. Eng. 2019, 55, 191–200. [Google Scholar] [CrossRef]
- Alter, T.B.; Ebert, B.E. Determination of Growth-Coupling Strategies and Their Underlying Principles. BMC Bioinform. 2019, 20, 447. [Google Scholar] [CrossRef] [PubMed]
- von Kamp, A.; Klamt, S. Growth-Coupled Overproduction Is Feasible for Almost All Metabolites in Five Major Production Organisms. Nat. Commun. 2017, 8, 15956. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 In-frame, Single-gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Tepper, N.; Shlomi, T. Predicting Metabolic Engineering Knockout Strategies for Chemical Production: Accounting for Competing Pathways. Bioinformatics 2010, 26, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Reed, J.L. OptORF: Optimal Metabolic and Regulatory Perturbations for Metabolic Engineering of Microbial Strains. BMC Syst. Biol. 2010, 4, 53. [Google Scholar] [CrossRef]
- Pharkya, P.; Burgard, A.P.; Maranas, C.D. OptStrain: A Computational Framework for Redesign of Microbial Production Systems. Genome Res. 2004, 14, 2367–2376. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Reed, J.L.; Maravelias, C.T. Large-Scale Bi-Level Strain Design Approaches and Mixed-Integer Programming Solution Techniques. PLoS ONE 2011, 6, e24162. [Google Scholar] [CrossRef]
- Ranganathan, S.; Suthers, P.F.; Maranas, C.D. OptForce: An Optimization Procedure for Identifying All Genetic Manipulations Leading to Targeted Overproductions. PLoS Comput. Biol. 2010, 6, e1000744. [Google Scholar] [CrossRef]
- Burgard, A.P.; Pharkya, P.; Maranas, C.D. Optknock: A Bilevel Programming Framework for Identifying Gene Knockout Strategies for Microbial Strain Optimization. Biotechnol. Bioeng. 2003, 84, 647–657. [Google Scholar] [CrossRef]
- Patil, K.R.; Rocha, I.; Förster, J.; Nielsen, J. Evolutionary Programming as a Platform for in Silico Metabolic Engineering. BMC Bioinform. 2005, 6, 308. [Google Scholar] [CrossRef]
- Ohno, S.; Shimizu, H.; Furusawa, C. FastPros: Screening of Reaction Knockout Strategies for Metabolic Engineering. Bioinformatics 2014, 30, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.E.; Hixson, K.K.; Conrad, T.M.; Lerman, J.A.; Charusanti, P.; Polpitiya, A.D.; Adkins, J.N.; Schramm, G.; Purvine, S.O.; Lopez-Ferrer, D.; et al. Omic Data from Evolved E. coli Are Consistent with Computed Optimal Growth from Genome-scale Models. Mol. Syst. Biol. 2010, 6, 390. [Google Scholar] [CrossRef]
- Choi, H.S.; Lee, S.Y.; Kim, T.Y.; Woo, H.M. In Silico Identification of Gene Amplification Targets for Improvement of Lycopene Production. Appl. Environ. Microbiol. 2010, 76, 3097–3105. [Google Scholar] [CrossRef]
- Doroshenko, V.; Airich, L.; Vitushkina, M.; Kolokolova, A.; Livshits, V.; Mashko, S. YddG from Escherichia coli Promotes Export of Aromatic Amino Acids. FEMS Microbiol. Lett. 2007, 275, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Diaz, H.; Arrasate, S.; Gomez-SanJuan, A.; Sotomayor, N.; Lete, E.; Besada-Porto, L.; Ruso, J. General Theory for Multiple Input-Output Perturbations in Complex Molecular Systems. 1. Linear QSPR Electronegativity Models in Physical, Organic, and Medicinal Chemistry. Curr. Top. Med. Chem. 2013, 13, 1713–1741. [Google Scholar] [CrossRef] [PubMed]
- Diéguez-Santana, K.; Casañola-Martin, G.M.; Green, J.R.; Rasulev, B.; González-Díaz, H. Predicting Metabolic Reaction Networks with Perturbation-Theory Machine Learning (PTML) Models. Curr. Top. Med. Chem. 2021, 21, 819–827. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosio, B. Qualitative Perturbation Analysis. In Qualitative Process Theory Using Linguistic Variables; Springer: New York, NY, USA, 1989; pp. 120–141. [Google Scholar]
- De Mori, R.; Prager, R. Perturbation Analysis with Qualitative Models. Int. Jt. Conf. Artif. Intell. Organ. 1989, 2, 1180–1186. [Google Scholar]
- Zhang, C.; Zhang, Y.; Shi, X.; Almpanidis, G.; Fan, G.; Shen, X. On Incremental Learning for Gradient Boosting Decision Trees. Neural Process Lett. 2019, 50, 957–987. [Google Scholar] [CrossRef]
- Sahu, A.; Blätke, M.-A.; Szymański, J.J.; Töpfer, N. Advances in Flux Balance Analysis by Integrating Machine Learning and Mechanism-Based Models. Comput. Struct. Biotechnol. J. 2021, 19, 4626–4640. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Preparation and Transformation of Competent E. coli Using Calcium Chloride. Cold Spring Harb. Protoc. 2006, 2006, pdb.prot3932. [Google Scholar] [CrossRef]
- Salkowski, E. Ueber Das Verhalten Der Skatolcarbonsäure Im Organismus. Biol. Chem. 1885, 9, 23–33. [Google Scholar] [CrossRef]
- Glickmann, E.; Dessaux, Y. A Critical Examination of the Specificity of the Salkowski Reagent for Indolic Compounds Produced by Phytopathogenic Bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef]
- Love, J.; Selker, R.; Marsman, M.; Jamil, T.; Dropmann, D.; Verhagen, J.; Ly, A.; Gronau, Q.F.; Smíra, M.; Epskamp, S.; et al. JASP: Graphical Statistical Software for Common Statistical Designs. J. Stat. Softw. 2019, 88, 1–17. [Google Scholar] [CrossRef]
- Simensen, V.; Schulz, C.; Karlsen, E.; Bråtelund, S.; Burgos, I.; Thorfinnsdottir, L.B.; García-Calvo, L.; Bruheim, P.; Almaas, E. Experimental Determination of Escherichia coli Biomass Composition for Constraint-Based Metabolic Modeling. PLoS ONE 2022, 17, e0262450. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Binkley, R.; Kim, H.U.; Lee, S.Y. Metabolic Engineering of Escherichia coli for the Enhanced Production of L-tyrosine. Biotechnol. Bioeng. 2018, 115, 2554–2564. [Google Scholar] [CrossRef] [PubMed]
- Decottignies-Le Maréchal, P.; Calderón-Seguin, R.; Vandecasteele, J.P.; Azerad, R. Synthesis of L-Tryptophan by Immobilized Escherichia coli Cells. Eur. J. Appl. Microbiol. Biotechnol. 1979, 7, 33–44. [Google Scholar] [CrossRef]
- Long, M.; Xu, M.; Ma, Z.; Pan, X.; You, J.; Hu, M.; Shao, Y.; Yang, T.; Zhang, X.; Rao, Z. Significantly Enhancing Production of Trans-4-Hydroxy-L-Proline by Integrated System Engineering in Escherichia coli. Sci. Adv. 2020, 6, eaba2383. [Google Scholar] [CrossRef]
- Qian, Z.; Xia, X.; Lee, S.Y. Metabolic Engineering of Escherichia coli for the Production of Cadaverine: A Five Carbon Diamine. Biotechnol. Bioeng. 2011, 108, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Li, Y.; Ma, Q.; Li, Q.; Jia, Z.; Yang, B.; Xu, Q.; Fan, X.; Zhang, C.; Chen, N.; et al. Metabolic Engineering of Escherichia coli for High-Yield Uridine Production. Metab. Eng. 2018, 49, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, W.; Chen, H.; Liu, B.; Lin, B.; Tao, Y. Metabolic Engineering for Efficient Supply of Acetyl-CoA from Different Carbon Sources in Escherichia coli. Microb. Cell Fact. 2019, 18, 130. [Google Scholar] [CrossRef] [PubMed]
- Song, C.W.; Kim, D.I.; Choi, S.; Jang, J.W.; Lee, S.Y. Metabolic Engineering of Escherichia coli for the Production of Fumaric Acid. Biotechnol. Bioeng. 2013, 110, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Ma, N.; Zhu, K.; Mao, Y.; Wei, X.; Zhao, Y. Balancing the Carbon Flux Distributions between the TCA Cycle and Glyoxylate Shunt to Produce Glycolate at High Yield and Titer in Escherichia coli. Metab. Eng. 2018, 46, 28–34. [Google Scholar] [CrossRef]
- Jantama, K.; Haupt, M.J.; Svoronos, S.A.; Zhang, X.; Moore, J.C.; Shanmugam, K.T.; Ingram, L.O. Combining Metabolic Engineering and Metabolic Evolution to Develop Nonrecombinant Strains of Escherichia coli C That Produce Succinate and Malate. Biotechnol. Bioeng. 2008, 99, 1140–1153. [Google Scholar] [CrossRef]
- Moon, S.Y.; Hong, S.H.; Kim, T.Y.; Lee, S.Y. Metabolic Engineering of Escherichia coli for the Production of Malic Acid. Biochem. Eng. J. 2008, 40, 312–320. [Google Scholar] [CrossRef]
- Dong, X.; Chen, X.; Qian, Y.; Wang, Y.; Wang, L.; Qiao, W.; Liu, L. Metabolic Engineering of Escherichia coli W3110 to Produce L-malate. Biotechnol. Bioeng. 2017, 114, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Zha, W.; Rubin-Pitel, S.B.; Shao, Z.; Zhao, H. Improving Cellular Malonyl-CoA Level in Escherichia coli via Metabolic Engineering. Metab. Eng. 2009, 11, 192–198. [Google Scholar] [CrossRef]
- Chen, X.; Li, M.; Zhou, L.; Shen, W.; Algasan, G.; Fan, Y.; Wang, Z. Metabolic Engineering of Escherichia coli for Improving Shikimate Synthesis from Glucose. Bioresour. Technol. 2014, 166, 64–71. [Google Scholar] [CrossRef]
- Stols, L.; Donnelly, M.I. Production of Succinic Acid through Overexpression of NAD(+)-Dependent Malic Enzyme in an Escherichia coli Mutant. Appl. Environ. Microbiol. 1997, 63, 2695–2701. [Google Scholar] [CrossRef]
- Ebrahim, A.; Lerman, J.A.; Palsson, B.O.; Hyduke, D.R. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst. Biol. 2013, 7, 74. [Google Scholar] [CrossRef]
- Cardoso, J.G.R.; Jensen, K.; Lieven, C.; Lærke Hansen, A.S.; Galkina, S.; Beber, M.; Özdemir, E.; Herrgård, M.J.; Redestig, H.; Sonnenschein, N. Cameo: A Python Library for Computer Aided Metabolic Engineering and Optimization of Cell Factories. ACS Synth. Biol. 2018, 7, 1163–1166. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-Learn: Machine Learning in Python Pedregosa, Varoquaux, Gramfort et Al. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hagberg, A.A.; Schult, D.A.; Swart, P.J. Exploring Network Structure, Dynamics, and Function Using NetworkX. In Proceedings of the 7th Python in Science conference (SciPy 2008), Pasadena, CA, USA, 19–24 August 2008; Varoquaux, G., Vaught, T., Millman, J., Eds.; Los Alamos National Laboratory (LANL): Los Alamos, NM, USA, 2008; pp. 11–15. [Google Scholar]
- Deng, M.-D.; Severson, D.K.; Grund, A.D.; Wassink, S.L.; Burlingame, R.P.; Berry, A.; Running, J.A.; Kunesh, C.A.; Song, L.; Jerrell, T.A.; et al. Metabolic Engineering of Escherichia coli for Industrial Production of Glucosamine and N-Acetylglucosamine. Metab. Eng. 2005, 7, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhu, Y.; Yuan, Z.; Liu, G.; Sun, Z.; Du, S.; Liu, H.; Li, Y.; Liu, H.; Zhou, Z. Evaluation of Metabolic Engineering Strategies on 2-Ketoisovalerate Production by Escherichia coli. Appl. Environ. Microbiol. 2022, 88, e00976-22. [Google Scholar] [CrossRef]
- Farmer, W.R.; Liao, J.C. Reduction of Aerobic Acetate Production by Escherichia coli. Appl. Environ. Microbiol. 1997, 63, 3205–3210. [Google Scholar] [CrossRef] [PubMed]
- Balderas-Hernández, V.E.; Sabido-Ramos, A.; Silva, P.; Cabrera-Valladares, N.; Hernández-Chávez, G.; Báez-Viveros, J.L.; Martínez, A.; Bolívar, F.; Gosset, G. Metabolic Engineering for Improving Anthranilate Synthesis from Glucose in Escherichia coli. Microb. Cell Fact. 2009, 8, 19. [Google Scholar] [CrossRef]
- Piao, X.; Wang, L.; Lin, B.; Chen, H.; Liu, W.; Tao, Y. Metabolic Engineering of Escherichia coli for Production of L-Aspartate and Its Derivative β-Alanine with High Stoichiometric Yield. Metab. Eng. 2019, 54, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jantama, K.; Moore, J.C.; Shanmugam, K.T.; Ingram, L.O. Production of L-Alanine by Metabolically Engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2007, 77, 355–366. [Google Scholar] [CrossRef]
- Chang, D.-E.; Jung, H.-C.; Rhee, J.-S.; Pan, J.-G. Homofermentative Production of D- or L-Lactate in Metabolically Engineered Escherichia coli RR1. Appl. Environ. Microbiol. 1999, 65, 1384–1389. [Google Scholar] [CrossRef]
- Deng, Y.; Mao, Y.; Zhang, X. Metabolic Engineering of E. coli for Efficient Production of Glycolic Acid from Glucose. Biochem. Eng. J. 2015, 103, 256–262. [Google Scholar] [CrossRef]
- Ginesy, M.; Belotserkovsky, J.; Enman, J.; Isaksson, L.; Rova, U. Metabolic Engineering of Escherichia coli for Enhanced Arginine Biosynthesis. Microb. Cell Fact. 2015, 14, 29. [Google Scholar] [CrossRef]
- Liu, H.; Fang, G.; Wu, H.; Li, Z.; Ye, Q. L-Cysteine Production in Escherichia coli Based on Rational Metabolic Engineering and Modular Strategy. Biotechnol. J. 2018, 13, 1700695. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, G.; Takumi, K. Cysteine Degradation Gene YhaM, Encoding Cysteine Desulfidase, Serves as a Genetic Engineering Target to Improve Cysteine Production in Escherichia coli. AMB Express 2017, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hou, Y.; Wang, Y.; Li, Z. Enhancement of Sulfur Conversion Rate in the Production of L-Cysteine by Engineered Escherichia coli. J. Agric. Food Chem. 2020, 68, 250–257. [Google Scholar] [CrossRef]
- Wu, H.; Tian, D.; Fan, X.; Fan, W.; Zhang, Y.; Jiang, S.; Wen, C.; Ma, Q.; Chen, N.; Xie, X. Highly Efficient Production of L-Histidine from Glucose by Metabolically Engineered Escherichia coli. ACS Synth. Biol. 2020, 9, 1813–1822. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Oh, J.E.; Lee, K.H.; Kim, J.Y.; Lee, S.Y. Rational Design of Escherichia coli for L-Isoleucine Production. ACS Synth. Biol. 2012, 1, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-F.; Liu, Z.-Q.; Jin, L.-Q.; Tang, X.-L.; Shen, Z.-Y.; Yin, H.-H.; Zheng, Y.-G. Metabolic Engineering of Escherichia coli for Microbial Production of L-Methionine. Biotechnol. Bioeng. 2017, 114, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Yakandawala, N.; Romeo, T.; Friesen, A.D.; Madhyastha, S. Metabolic Engineering of Escherichia coli to Enhance Phenylalanine Production. Appl. Microbiol. Biotechnol. 2008, 78, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Báez-Viveros, J.L.; Osuna, J.; Hernández-Chávez, G.; Soberón, X.; Bolívar, F.; Gosset, G. Metabolic Engineering and Protein Directed Evolution Increase the Yield of L-phenylalanine Synthesized from Glucose in Escherichia coli. Biotechnol. Bioeng. 2004, 87, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kang, P.; Liu, S.; Zhao, Y.; Wang, Z.; Chen, T. GlyA Gene Knock-out in Escherichia coli Enhances L-Serine Production without Glycine Addition. Biotechnol. Bioprocess Eng. 2017, 22, 390–396. [Google Scholar] [CrossRef]
- Wang, C.; Wu, J.; Shi, B.; Shi, J.; Zhao, Z. Improving L-Serine Formation by Escherichia coli by Reduced Uptake of Produced l-Serine. Microb. Cell Fact. 2020, 19, 66. [Google Scholar] [CrossRef] [PubMed]
- Mundhada, H.; Seoane, J.M.; Schneider, K.; Koza, A.; Christensen, H.B.; Klein, T.; Phaneuf, P.V.; Herrgard, M.; Feist, A.M.; Nielsen, A.T. Increased Production of L-Serine in Escherichia coli through Adaptive Laboratory Evolution. Metab. Eng. 2017, 39, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.-N.T.; Eom, G.T.; Hong, S.H. Improving L-Serine Production in Escherichia coli via Synthetic Protein Scaffold of SerB, SerC, and EamA. Biochem. Eng. J. 2019, 148, 138–142. [Google Scholar] [CrossRef]
- Lee, K.H.; Park, J.H.; Kim, T.Y.; Kim, H.U.; Lee, S.Y. Systems Metabolic Engineering of Escherichia coli for L-threonine Production. Mol. Syst. Biol. 2007, 3, 149. [Google Scholar] [CrossRef]
- Dong, X.; Quinn, P.J.; Wang, X. Metabolic Engineering of Escherichia coli and Corynebacterium Glutamicum for the Production of L-Threonine. Biotechnol. Adv. 2011, 29, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Sung, B.H.; Kim, M.S.; Blattner, F.R.; Yoon, B.H.; Kim, J.H.; Kim, S.C. Metabolic Engineering of a Reduced-Genome Strain of Escherichia coli for L-Threonine Production. Microb. Cell Fact. 2009, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, K.H.; Kim, T.Y.; Lee, S.Y. Metabolic Engineering of Escherichia coli for the Production of L-Valine Based on Transcriptome Analysis and in Silico Gene Knockout Simulation. Proc. Natl. Acad. Sci. USA 2007, 104, 7797–7802. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Xia, X.; Lee, S.Y. Metabolic Engineering of Escherichia coli for the Production of Putrescine: A Four Carbon Diamine. Biotechnol. Bioeng. 2009, 104, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Moxley, W.C.; Eiteman, M.A. Pyruvate Production by Escherichia coli by Use of Pyruvate Dehydrogenase Variants. Appl. Environ. Microbiol. 2021, 87, e00487-21. [Google Scholar] [CrossRef]
- Thakker, C.; Martínez, I.; San, K.; Bennett, G.N. Succinate Production in Escherichia coli. Biotechnol. J. 2012, 7, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Bennett, G.N.; San, K.-Y. Metabolic Engineering of Aerobic Succinate Production Systems in Escherichia coli to Improve Process Productivity and Achieve the Maximum Theoretical Succinate Yield. Metab. Eng. 2005, 7, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lee, D.-Y.; Kim, T.Y.; Kim, B.H.; Lee, J.; Lee, S.Y. Metabolic Engineering of Escherichia coli for Enhanced Production of Succinic Acid, Based on Genome Comparison and in Silico Gene Knockout Simulation. Appl. Environ. Microbiol. 2005, 71, 7880–7887. [Google Scholar] [CrossRef]
- Heirendt, L.; Arreckx, S.; Pfau, T.; Mendoza, S.N.; Richelle, A.; Heinken, A.; Haraldsdóttir, H.S.; Wachowiak, J.; Keating, S.M.; Vlasov, V.; et al. Creation and Analysis of Biochemical Constraint-Based Models Using the COBRA Toolbox v.3.0. Nat. Protoc. 2019, 14, 639–702. [Google Scholar] [CrossRef]
- Hartline, C.J.; Schmitz, A.C.; Han, Y.; Zhang, F. Dynamic Control in Metabolic Engineering: Theories, Tools, and Applications. Metab. Eng. 2021, 63, 126–140. [Google Scholar] [CrossRef]
- D’Ambrosio, B. Fuzzy Logic Control. In Qualitative Process Theory Using Linguistic Variables; Springer: New York, NY, USA, 1989; pp. 5–15. [Google Scholar]
- Burgard, A.P.; Nikolaev, E.V.; Schilling, C.H.; Maranas, C.D. Flux Coupling Analysis of Genome-Scale Metabolic Network Reconstructions. Genome Res. 2004, 14, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Bundy, A.; Wallen, L. Breadth-First Search. In Catalogue of Artificial Intelligence Tools; Springer: Berlin/Heidelberg, Germany, 1984; p. 13. [Google Scholar]
- Westers, H. Genome Engineering Reveals Large Dispensable Regions in Bacillus Subtilis. Mol. Biol. Evol. 2003, 20, 2076–2090. [Google Scholar] [CrossRef]
- Kakuda, H.; Hosono, K.; Shiroishi, K.; Ichihara, S. Identification and Characterization of the AckA (Acetate Kinase A)-Pta (Phosphotransacetylase) Operon and Complementation Analysis of Acetate Utilization by an AckA-Pta Deletion Mutant of Escherichia coli. J. Biochem. 1994, 116, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Ball, E. Heteroauxin and the Growth of Escherichia coli. J. Bacteriol. 1938, 36, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.S.; Broadbelt, L.J.; Hatzimanikatis, V. Thermodynamics-Based Metabolic Flux Analysis. Biophys. J. 2007, 92, 1792–1805. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Su, X.; Zhao, B.; Li, G.; Hu, P.; Zhang, J.; Hu, L. Fuzzy-Based Deep Attributed Graph Clustering. IEEE Trans. Fuzzy Syst. 2024, 32, 1951–1964. [Google Scholar] [CrossRef]
- Bologna, F.P.; Campos-Bermudez, V.A.; Saavedra, D.D.; Andreo, C.S.; Drincovich, M.F. Characterization of Escherichia coli EutD: A Phosphotransacetylase of the Ethanolamine Operon. J. Microbiol. 2010, 48, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Flood, J.J.; Kristofich, M.R.; Gidfar, C.; Morgenthaler, A.B.; Fuhrer, T.; Sauer, U.; Snyder, D.; Cooper, V.S.; Ebmeier, C.C.; et al. Hidden Resources in the Escherichia coli Genome Restore PLP Synthesis and Robust Growth after Deletion of the Essential Gene PdxB. Proc. Natl. Acad. Sci. USA 2019, 116, 24164–24173. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yang, W.; Wang, B.; Liu, Q.; Zhong, X.; Gao, Q.; Liu, J.; Huang, J.; Lin, B.; Tao, Y. Metabolic Engineering of Escherichia coli for Efficient Production of L-Alanyl-l-Glutamine. Microb. Cell Fact. 2020, 19, 129. [Google Scholar] [CrossRef]
- Causey, T.B.; Shanmugam, K.T.; Yomano, L.P.; Ingram, L.O. Engineering Escherichia coli for Efficient Conversion of Glucose to Pyruvate. Proc. Natl. Acad. Sci. USA 2004, 101, 2235–2240. [Google Scholar] [CrossRef]
- Sarkar, D.; Siddiquee, K.A.Z.; Araúzo-Bravo, M.J.; Oba, T.; Shimizu, K. Effect of Cra Gene Knockout Together with Edd and IclR Genes Knockout on the Metabolism in Escherichia coli. Arch. Microbiol. 2008, 190, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Ba, F.; Ji, X.; Huang, S.; Zhang, Y.; Liu, W.; Liu, Y.; Ling, S.; Li, J. Engineering Escherichia coli to Utilize Erythritol as Sole Carbon Source. Adv. Sci. 2023, 10, 2207008. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, P.; Delépine, B.; Faulon, J.-L. Extended Metabolic Space Modeling. In Synthetic Metabolic Pathways: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2018; pp. 83–96. [Google Scholar]
- Faust, K.; Croes, D.; van Helden, J. Metabolic Pathfinding Using RPAIR Annotation. J. Mol. Biol. 2009, 388, 390–414. [Google Scholar] [CrossRef] [PubMed]
- Tarasava, K.; Lee, S.H.; Chen, J.; Köpke, M.; Jewett, M.C.; Gonzalez, R. Reverse β-Oxidation Pathways for Efficient Chemical Production. J. Ind. Microbiol. Biotechnol. 2022, 49, kuac003. [Google Scholar] [CrossRef] [PubMed]
| Name | Description | Reference |
|---|---|---|
| Strain | ||
| BW25113 * | Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW0855-1 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔpoxB772::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW3179-3 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔgltB740::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW1843-2 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔpykA779::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW1666-3 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔpykF751::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW5806-1 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔtdcD731::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW2293-1 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔackA778::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW2581-1 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔtyrA763::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW2580-1 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔpheA762::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW3686-7 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, ΔtnaA739::kan, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW5619-1 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, ΔtnaB740::kan, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| JW3130-1 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, Δmtr-773::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [20] |
| Plasmids | ||
| pCold IV | Ampr Cold Shock Expression System | Takara Bio Inc., San Jose, CA, USA |
| pIAAMHs | pCold IV derivative; iaaMH | Biological engineering laboratory |
| Reaction | Number of Relationships | Qualitative Variable | |||
|---|---|---|---|---|---|
| Positive | Negative | No Relation | No Clear | ||
| R1 | 74 | 0 | 2 | 0 | Metabolite-correlated |
| R2 | 0 | 0 | 76 | 0 | No flux |
| R3 | 0 | 76 | 0 | 0 | Possible Biomass-correlated |
| R4 | 23 | 49 | 4 | 0 | Negatively metabolite-correlated |
| R5 | 0 | 0 | 74 | 2 | Flux variation |
| Reaction | Enzyme | L-Tryptophan Precursor | Reference |
|---|---|---|---|
| GHMT2r | Glycine hydroxymethyltransferase | L-serine | [78,79,80] |
| SERD_L | L-serine deaminase | L-serine | [5,78,79,80] |
| HPYRP | 3-phosphohydroxypyruvate phosphatase | L-serine | [102] |
| GLUN | Glutaminase | L-glutamine | [103] |
| ICL | Isocitrate lyase | L-glutamate (L-proline) | [45] |
| FRD2 | Fumarate reductase | L-glutamate (L-alanine) | [66] |
| FRD3 | Fumarate reductase | L-glutamate (L-alanine) | [66] |
| MGSA | Methylglyoxal synthase | L-glutamate (L-alanine) | [66] |
| ACALD | Acetaldehyde dehydrogenase | L-glutamate (L-alanine) | [66] |
| LDH_D | D-lactate dehydrogenase | L-glutamate (L-alanine) | [66] |
| PFL | Pyruvate formate lyase | Pyruvate | [104] |
| EDD | 6-phosphogluconate dehydratase | Phosphoenolpyruvate | [105] |
| F6PA | Fructose 6-phosphate aldolase | Phosphoenolpyruvate | [105] |
| TALA | Transaldolase | D-erythrose-4-phosphate | [106] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos-Valdovinos, M.A.; Salas-Navarrete, P.C.; Amores, G.R.; Hernández-Orihuela, A.L.; Martínez-Antonio, A. Qualitative Perturbation Analysis and Machine Learning: Elucidating Bacterial Optimization of Tryptophan Production. Algorithms 2024, 17, 282. https://doi.org/10.3390/a17070282
Ramos-Valdovinos MA, Salas-Navarrete PC, Amores GR, Hernández-Orihuela AL, Martínez-Antonio A. Qualitative Perturbation Analysis and Machine Learning: Elucidating Bacterial Optimization of Tryptophan Production. Algorithms. 2024; 17(7):282. https://doi.org/10.3390/a17070282
Chicago/Turabian StyleRamos-Valdovinos, Miguel Angel, Prisciluis Caheri Salas-Navarrete, Gerardo R. Amores, Ana Lilia Hernández-Orihuela, and Agustino Martínez-Antonio. 2024. "Qualitative Perturbation Analysis and Machine Learning: Elucidating Bacterial Optimization of Tryptophan Production" Algorithms 17, no. 7: 282. https://doi.org/10.3390/a17070282
APA StyleRamos-Valdovinos, M. A., Salas-Navarrete, P. C., Amores, G. R., Hernández-Orihuela, A. L., & Martínez-Antonio, A. (2024). Qualitative Perturbation Analysis and Machine Learning: Elucidating Bacterial Optimization of Tryptophan Production. Algorithms, 17(7), 282. https://doi.org/10.3390/a17070282






