Challenges in Reducing Bias Using Post-Processing Fairness for Breast Cancer Stage Classification with Deep Learning
Abstract
1. Introduction
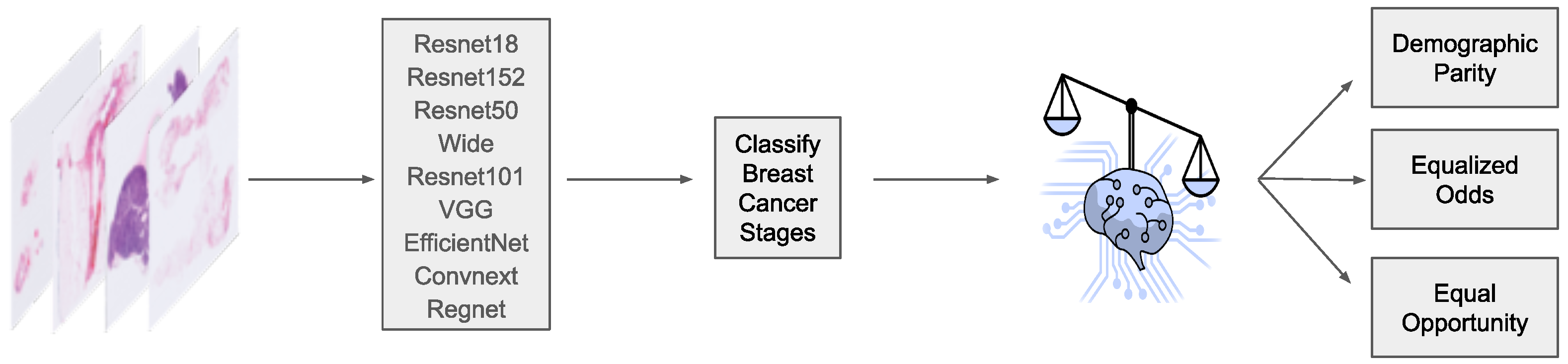
2. Materials and Methods
2.1. Dataset
2.2. Machine Learning Models
2.3. Fairness Definitions
2.3.1. Equalized Odds
2.3.2. Equal Opportunity
2.4. Fairness Adjustments
2.5. Evaluation Procedure
3. Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Golatkar, A.; Anand, D.; Sethi, A. Classification of breast cancer histology using deep learning. In Proceedings of the Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, 27–29 June 2018; Proceedings 15. Springer: Berlin/Heidelberg, Germany, 2018; pp. 837–844. [Google Scholar]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast cancer histopathological image classification using Convolutional Neural Networks. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 2560–2567. [Google Scholar] [CrossRef]
- Boag, W.; Suresh, H.; Celi, L.A.; Szolovits, P.; Ghassemi, M. Racial Disparities and Mistrust in End-of-Life Care. Proc. Mach. Learn. Res. 2018, 85, 587–602. [Google Scholar]
- Adamson, A.S.; Smith, A. Machine Learning and Health Care Disparities in Dermatology. JAMA Dermatol. 2018, 154, 1247–1248. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Hardt, M.; Howell, M.; Corrado, G.; Chin, M. Ensuring Fairness in Machine Learning to Advance Health Equity. Ann. Intern. Med. 2018, 169, 866–887. [Google Scholar] [CrossRef] [PubMed]
- Velagapudi, L.; Mouchtouris, N.; Baldassari, M.; Nauheim, D.; Khanna, O.; Al Saiegh, F.; Herial, N.; Gooch, M.; Tjoumakaris, S.; Rosenwasser, R.; et al. Discrepancies in Stroke Distribution and Dataset Origin in Machine Learning for Stroke. J. Stroke Cerebrovasc. Dis. Off. J. Natl. Stroke Assoc. 2021, 30, 105832. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.R.; Wen, P.Y.; Lang-Orsini, M.; Chukwueke, U.N. World Health Organization 2021 classification of central nervous system tumors and implications for therapy for adult-type gliomas: A review. JAMA Oncol. 2022, 8, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Bardhan, I.R.; Chen, H.; Karahanna, E. Connecting systems, data, and people: A multidisciplinary research roadmap for chronic disease management. Manag. Inf. Syst. Q. 2020, 44, 185–200. [Google Scholar]
- Bostrom, N.; Yudkowsky, E. The ethics of artificial intelligence. In The Cambridge Handbook of Artificial Intelligence; Frankish, K., Ramsey, W.M., Eds.; Cambridge University Press: Cambridge, UK, 2014; pp. 316–334. [Google Scholar] [CrossRef]
- Futoma, J.; Simons, M.; Panch, T.; Doshi velez, F.; Celi, L. The myth of generalisability in clinical research and machine learning in health care. Lancet Digit. Health 2020, 2, e489–e492. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, B.; O’Neil, C.; LaGatta, T. Conscientious Classification: A Data Scientist’s Guide to Discrimination-Aware Classification. Big Data 2017, 5, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Bellamy, R.K.E.; Dey, K.; Hind, M.; Hoffman, S.C.; Houde, S.; Kannan, K.; Lohia, P.; Martino, J.; Mehta, S.; Mojsilovic, A.; et al. AI Fairness 360: An Extensible Toolkit for Detecting, Understanding, and Mitigating Unwanted Algorithmic Bias. arXiv 2018, arXiv:1810.01943. [Google Scholar]
- Berk, R.; Heidari, H.; Jabbari, S.; Joseph, M.; Kearns, M.; Morgenstern, J.; Neel, S.; Roth, A. A Convex Framework for Fair Regression. arXiv 2017, arXiv:1706.02409. [Google Scholar]
- Bifulco, C.; Piening, B.; Bower, T.; Robicsek, A.; Weerasinghe, R.; Lee, S.; Foster, N.; Juergens, N.; Risley, J.; Nachimuthu, S.; et al. Identifying High-risk Breast Cancer Using Digital Pathology Images: A Nightingale Open Science Dataset. Nightingale Open Science. 2021. Available online: https://docs.ngsci.org/datasets/brca-psj-path/ (accessed on 26 March 2024).
- Zhu, H.; Chen, B.; Yang, C. Understanding Why ViT Trains Badly on Small Datasets: An Intuitive Perspective. arXiv 2023, arXiv:2302.03751. [Google Scholar]
- Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data Efficient and Weakly Supervised Computational Pathology on Whole Slide Images. arXiv 2020, arXiv:2004.09666. [Google Scholar] [CrossRef] [PubMed]
- Hardt, M.; Price, E.; Srebro, N. Equality of Opportunity in Supervised Learning. arXiv 2016, arXiv:1610.02413. [Google Scholar]
- Putzel, P.; Lee, S. Blackbox Post-Processing for Multiclass Fairness. arXiv 2022, arXiv:2201.04461. [Google Scholar]
- Lee, S. Scotthlee/Fairness: Now with Support for Multiclass Outcomes. 2022. Available online: https://zenodo.org/records/6127503 (accessed on 26 March 2024).
- Zhang, H.; Dullerud, N.; Roth, K.; Oakden-Rayner, L.; Pfohl, S.; Ghassemi, M. Improving the Fairness of Chest X-ray Classifiers. PMLR 2022, 174, 204–233. [Google Scholar]
- Ghassemi, M.; Mohamed, S. Machine learning and health need better values. Npj Digit. Med. 2022, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.; Johansson, F.D.; Sontag, D. Why Is My Classifier Discriminatory? arXiv 2018, arXiv:1805.12002. [Google Scholar]



| Train | Validation | Test | |
|---|---|---|---|
| Biopsies | 328 | 41 | 41 |
| Images | 3273 | 342 | 367 |
| White | 234 | 28 | 32 |
| non-White | 94 | 13 | 9 |
| Biopsies with stage | |||
| 0 | 41% | 49% | 61% |
| III, IV | 59% | 51% | 39% |
| Train | Validation | Test | |
|---|---|---|---|
| Biopsies | 800 | 100 | 100 |
| Images | 8847 | 967 | 1032 |
| White | 483 | 66 | 61 |
| non-White | 317 | 34 | 39 |
| Biopsies with stage | |||
| 0 | 18% | 16% | 18% |
| I, II | 58% | 60% | 66% |
| III, IV | 23% | 24% | 16% |
| Models | Groups | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| ResNet18 | White | ||||
| non-White | |||||
| ResNet50 | White | ||||
| non-White | |||||
| Wide ResNet101 | White | ||||
| non-White | |||||
| ResNet152 | White | ||||
| non-White | |||||
| VGG | White | ||||
| non-White | |||||
| EfficientNet | White | ||||
| non-White | |||||
| ConvNeXt | White | ||||
| non-White | |||||
| RegNet | White | ||||
| non-White | |||||
| Ensemble | White | ||||
| non-White | |||||
| Slide Level | White | ||||
| non-White |
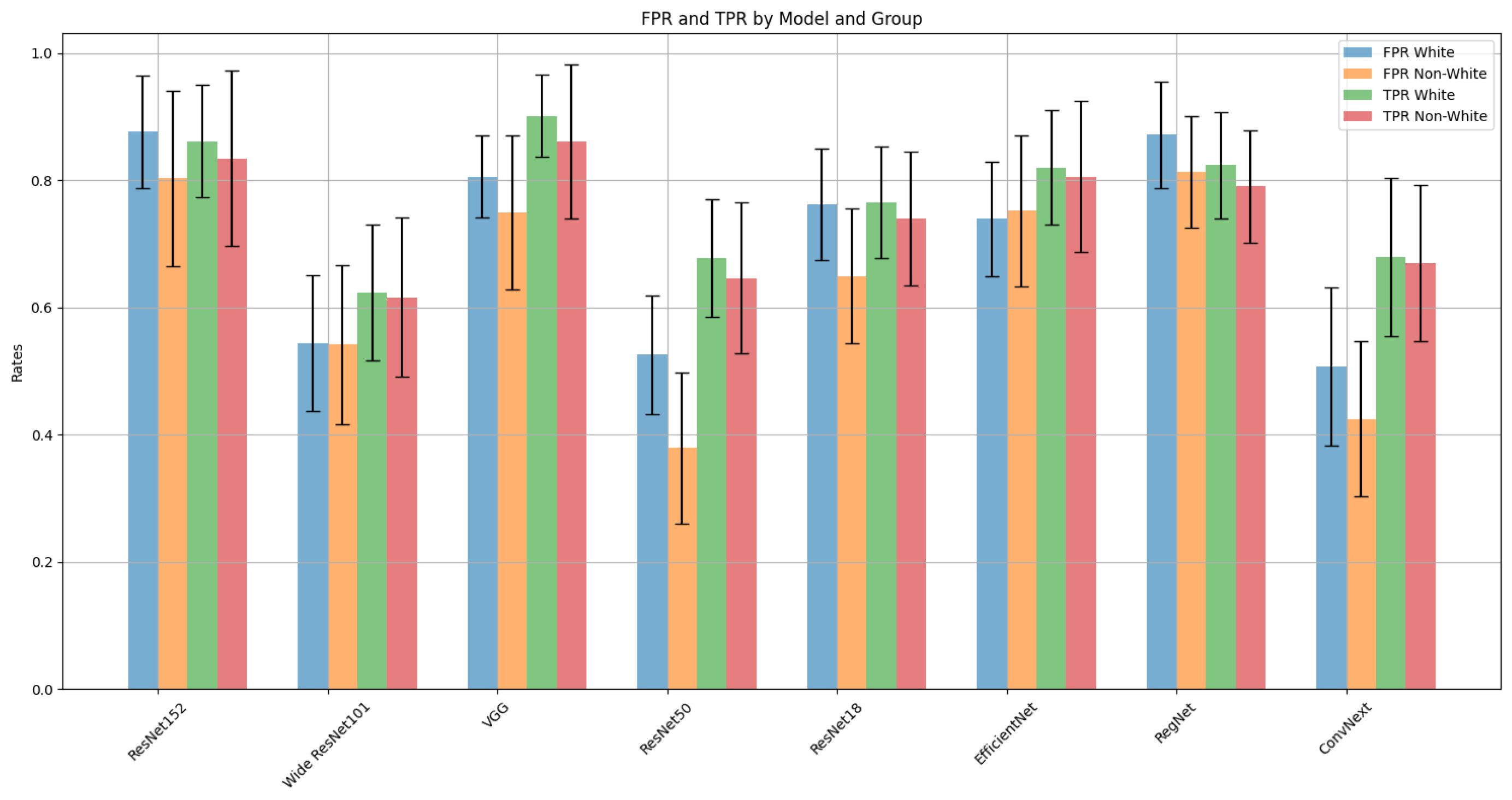
| Model | FPR | TPR | ||
|---|---|---|---|---|
| t-statistic | p-value | t-statistic | p-value | |
| ResNet152 | 1.93 | 0.0606 | 1.93 | 0.0607 |
| Wide ResNet101 | 0.06 | 0.9507 | 0.32 | 0.7474 |
| VGG | 1.87 | 0.0693 | 3.04 | 0.0043 |
| ResNet50 | 3.93 | 0.0003 | 1.46 | 0.1514 |
| ResNet18 | 3.24 | 0.0024 | 1.33 | 0.1923 |
| EfficientNet | −0.35 | 0.7272 | 0.84 | 0.4034 |
| RegNet | 1.83 | 0.0746 | 1.87 | 0.0696 |
| ConvNeXt | 1.76 | 0.0860 | 0.47 | 0.6399 |
| Model | Group | FPR | TPR | Loss | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pre-Adjust | Post-Adjust | Post-Adjust | Pre-Adjust | Post-Adjust | Post-Adjust | Pre-Adjust | Post-Adjust | Post-Adjust | ||
| Odds | Opportunity | Odds | Opportunity | Odds | Opportunity | |||||
| ResNet152 | White | 0.8889 | 0.8889 | 0.8652 | 0.8667 | 0.8165 | 0.8652 | 0.2732 | 0.3161 | 0.2752 |
| non-White | 0.6786 | 0.7500 | 0.7778 | 0.8214 | 0.8148 | 0.8889 | 0.2732 | 0.3161 | 0.2752 | |
| Wide_ResNet101 | White | 0.4444 | 0.5556 | 0.3745 | 0.5556 | 0.6367 | 0.6255 | 0.5683 | 0.4033 | 0.4142 |
| non-White | 0.1071 | 0.5714 | 0.0000 | 0.5714 | 0.6296 | 0.5926 | 0.5683 | 0.4033 | 0.4142 | |
| VGG | White | 0.8000 | 0.8000 | 0.8989 | 0.8000 | 0.8989 | 0.8989 | 0.2077 | 0.2371 | 0.2289 |
| non-White | 0.8214 | 0.6786 | 1.0000 | 0.7500 | 0.8889 | 0.9259 | 0.2077 | 0.2371 | 0.2289 | |
| ResNet50 | White | 0.5333 | 0.5333 | 0.6742 | 0.5333 | 0.6742 | 0.6742 | 0.2842 | 0.3706 | 0.3651 |
| non-White | 0.5357 | 0.4286 | 0.9630 | 0.5357 | 0.6296 | 0.5926 | 0.2842 | 0.3706 | 0.3651 | |
| ResNet18 | White | 0.7556 | 0.7556 | 0.7790 | 0.7556 | 0.7790 | 0.7790 | 0.2896 | 0.3188 | 0.3324 |
| non-White | 0.8571 | 0.7500 | 0.9630 | 0.6786 | 0.8148 | 0.7037 | 0.2896 | 0.3188 | 0.3324 | |
| EfficientNet | White | 0.7556 | 0.7556 | 0.8165 | 0.7556 | 0.7865 | 0.8165 | 0.2896 | 0.3188 | 0.2970 |
| non-White | 0.7143 | 0.7500 | 0.7778 | 0.7143 | 0.7778 | 0.8148 | 0.2896 | 0.3188 | 0.2970 | |
| RegNet | White | 0.8667 | 0.8667 | 0.824 | 0.9778 | 1.0000 | 0.8240 | 0.3333 | 0.3661 | 0.3106 |
| non-White | 1.0000 | 0.8214 | 1.000 | 1.0000 | 0.9630 | 0.8148 | 0.3333 | 0.3661 | 0.3106 | |
| ConvNeXt | White | 0.5111 | 0.5111 | 0.6704 | 0.5111 | 0.6704 | 0.6704 | 0.3497 | 0.3678 | 0.3488 |
| non-White | 0.6786 | 0.3571 | 1.0000 | 0.5000 | 0.6296 | 0.7407 | 0.3497 | 0.3678 | 0.3488 | |
| Models | Groups | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| ResNet18 | White | ||||
| non-White | |||||
| ResNet50 | White | ||||
| non-White | |||||
| Wide ResNet101 | White | ||||
| non-White | |||||
| ResNet152 | White | ||||
| non-White | |||||
| VGG | White | ||||
| non-White | |||||
| EfficientNet | White | ||||
| non-White | |||||
| ConvNeXt | White | ||||
| non-White | |||||
| RegNet | White | ||||
| non-White | |||||
| Ensemble | White | ||||
| non-White |
| Model | Race | Class | FPR Pre-Adjust | FPR Post-Adjust | TPR Pre-Adjust | TPR Post-Adjust |
|---|---|---|---|---|---|---|
| RegNet | non-White | 0 | 0.3577 | 0.0759 | 0.9375 | 0.1989 |
| 1 | 0.1034 | 0.6321 | 0.0459 | 0.7160 | ||
| 2 | 0.5598 | 0.2060 | 0.5000 | 0.2891 | ||
| White | 0 | 0.4375 | 0.0759 | 0.5000 | 0.1989 | |
| 1 | 0.0738 | 0.6321 | 0.0549 | 0.7160 | ||
| 2 | 0.4019 | 0.2060 | 0.5639 | 0.2981 | ||
| ResNet50 | non-White | 0 | 0.3462 | 0.3304 | 0.5625 | 0.4375 |
| 1 | 0.5172 | 0.5402 | 0.5872 | 0.5489 | ||
| 2 | 0.0769 | 0.1154 | 0.0238 | 0.1219 | ||
| White | 0 | 0.3304 | 0.3304 | 0.4375 | 0.4375 | |
| 1 | 0.5772 | 0.5402 | 0.5824 | 0.5489 | ||
| 2 | 0.0748 | 0.1154 | 0.0902 | 0.1219 | ||
| ConvNeXt | non-White | 0 | 0.2692 | 0.5265 | 0.8125 | 0.8034 |
| 1 | 0.3966 | 0.4441 | 0.3899 | 0.4262 | ||
| 2 | 0.2735 | 0.0312 | 0.5000 | 0.0068 | ||
| White | 0 | 0.2098 | 0.5265 | 0.5625 | 0.8034 | |
| 1 | 0.4228 | 0.4441 | 0.4066 | 0.4262 | ||
| 2 | 0.2243 | 0.0312 | 0.4511 | 0.0068 | ||
| ResNet152 | non-White | 0 | 0.0500 | 0.4895 | 0.0000 | 0.7500 |
| 1 | 0.5000 | 0.4986 | 0.5046 | 0.4946 | ||
| 2 | 0.4402 | 0.0000 | 0.5000 | 0.0000 | ||
| White | 0 | 0.0357 | 0.4895 | 0.0000 | 0.7500 | |
| 1 | 0.5101 | 0.4986 | 0.5275 | 0.4946 | ||
| 2 | 0.3832 | 0.0000 | 0.5038 | 0.0000 | ||
| ResNet101 | non-White | 0 | 0.2231 | 0.4420 | 0.1250 | 0.5005 |
| 1 | 0.6552 | 0.2012 | 0.5596 | 0.2422 | ||
| 2 | 0.2137 | 0.3196 | 0.1429 | 0.3292 | ||
| White | 0 | 0.2545 | 0.4420 | 0.1250 | 0.5005 | |
| 1 | 0.4832 | 0.2012 | 0.4176 | 0.2422 | ||
| 2 | 0.2897 | 0.3196 | 0.3008 | 0.3292 | ||
| EfficientNet | non-White | 0 | 0.2462 | 0.3849 | 0.6875 | 0.6581 |
| 1 | 0.5172 | 0.2245 | 0.4404 | 0.3421 | ||
| 2 | 0.2821 | 0.2646 | 0.2143 | 0.3787 | ||
| White | 0 | 0.2009 | 0.3849 | 0.5000 | 0.6581 | |
| 1 | 0.5705 | 0.2245 | 0.3956 | 0.3421 | ||
| 2 | 0.3084 | 0.2646 | 0.2481 | 0.3787 | ||
| ResNet18 | non-White | 0 | 0.0885 | 0.3341 | 0.0000 | 0.4074 |
| 1 | 0.1034 | 0.5663 | 0.1835 | 0.5816 | ||
| 2 | 0.7436 | 0.0812 | 0.7857 | 0.0952 | ||
| White | 0 | 0.0670 | 0.3341 | 0.1250 | 0.4074 | |
| 1 | 0.1812 | 0.5663 | 0.1429 | 0.5816 | ||
| 2 | 0.8131 | 0.0812 | 0.7218 | 0.0952 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soltan, A.; Washington, P. Challenges in Reducing Bias Using Post-Processing Fairness for Breast Cancer Stage Classification with Deep Learning. Algorithms 2024, 17, 141. https://doi.org/10.3390/a17040141
Soltan A, Washington P. Challenges in Reducing Bias Using Post-Processing Fairness for Breast Cancer Stage Classification with Deep Learning. Algorithms. 2024; 17(4):141. https://doi.org/10.3390/a17040141
Chicago/Turabian StyleSoltan, Armin, and Peter Washington. 2024. "Challenges in Reducing Bias Using Post-Processing Fairness for Breast Cancer Stage Classification with Deep Learning" Algorithms 17, no. 4: 141. https://doi.org/10.3390/a17040141
APA StyleSoltan, A., & Washington, P. (2024). Challenges in Reducing Bias Using Post-Processing Fairness for Breast Cancer Stage Classification with Deep Learning. Algorithms, 17(4), 141. https://doi.org/10.3390/a17040141







