Abstract
The importance of online recommender systems for drugs, medical professionals, and hospitals is growing. Today, the majority of people use online consultations for drug recommendations for all types of health issues. Emergencies such as pandemics, floods, or cyclones can be helped by the medical recommender system. In the era of machine learning (ML), recommender systems produce more accurate, quick, and reliable clinical predictions with minimal costs. As a result, these systems maintain better performance, integrity, and privacy of patient data in the decision-making process and provide precise information at any time. Therefore, we present drug recommender systems with a stacked artificial neural network (ANN) model to improve the fairness and safety of treatment for infectious diseases. To reduce side effects, drugs are recommended based on a patient’s previous health profile, lifestyle, and habits. The proposed system produced results with 97.5% accuracy. A system such as this could be useful in recommending safe medicines to patients, especially during health emergencies.
1. Introduction
Online consultations require the patient to describe their symptoms to the doctor. A spike in virtual medical services has been reported in the wake of the novel coronavirus disease (COVID-19) [1]. Diabetes, hypertension, and heart disease are all associated with an increased risk of virus infections. The availability of health care professionals 24/7, no need for travel, security, privacy, and drug recommendations are all advantages of virtual medical services. The recommender system allows for improvements in medical services in disparate areas [2]. Often, finding a physician in remote areas can be tricky, so recommender systems have been created to help.
Health-related recommender systems can make an early diagnosis, predict disease progression, and make appropriate recommendations according to the health status of patients [3,4]. Machine learning (ML) greatly improves the quality of medical recommender systems by providing suggestions that are based on patient needs and feedback [5,6]. By using sentiment analysis and feature engineering, the drug recommender system can dispense medicine according to a specific condition. Emotions, such as attitudes and opinions, are separated and extracted from language through sentiment analysis [7].
By using the recommender system, information overload can be solved, and e-government and e-learning can be improved [8]. Depending on an individual’s health status, these recommender systems prescribe medications, diagnose diseases, and refer them to the relevant health care. An ML-driven recommendation system generates appropriate recommendations using parameters such as blood pressure, gender, cholesterol levels, and blood sugar for diseases such as colds, fevers, and cardiac deaths [9]. The healthcare system built on the Internet of Things (IoT) coupled with an oncology interface has provided nutrition information to individuals [10].
Depending on the patient’s medical history, a decision support system can assist a doctor in prescribing a drug. In contrast, the recommendation system suggests the same based on an analysis of previous usage patterns [11]. Four types of recommender systems exist, including content-driven filtering, collaborative filtering, knowledge-driven recommender systems, and hybrid recommender systems [12,13]. Since the drug recommendation framework includes medical terminology, such as infection names, side effects, and synthetic names, only a limited number of papers are available.
In this work, we proposed the development of a drug recommender system (DRS) for different diseases to maintain good patient health and longevity. We addressed the unfairness in drug usage by DRS for severe chronic diseases by improving the recommendation accuracy by the integration of ML knowledge. A further discussion was carried out on how the proposed DRS integrates the person’s health profile and automation of meditation, and drug dosages. The system performance was calculated based on different metrics such as accuracy, sensitivity, and specificity. The performance was further compared with other existing ML models to validate its efficiency.
2. Related Work
From the existing literature, different health recommender systems (HRS) are available. Collaborative filtering utilizes past user behavior to examine similar profiles and determine preferences to make clear recommendations. A hospital recommendation system was proposed by Fedelucio et al. [14] based on the treatments, consulted physicians, hospitals, and patient health indicators of a patient. An alternative hybrid recommender system based on available information on family doctors and available patients was suggested [15].
Various HRS help to support medical treatment and prognosis [16]. Recommendations made on content-based filtering are dependent on specific features only. Different features selected using rough set feature reduction can predict diabetes [17]. Content-driven models are used to evaluate radial doses and weights for elements in cancer treatments [18]. It is reported that content-based models achieve a better performance than traditional models in predicting the risk of heart attack [19].
A model called iCARE uses collaborative filtering and hybrid learning to predict disease risk based on a patient’s previous illnesses [20]. The risks of delivery for pregnant women can be predicted using a collaborative filtering algorithm that includes Mahalanobis distance and fuzzy membership [21]. Ontologies and methods of problem-solving are fundamental components of knowledge-based systems [22]. Based on the knowledge of users and products, knowledge-based filtering selects products that are suitable for users [23]. Meanwhile, hybrid systems combine different filtering approaches [24].
Demographic filtering offers recommendations based on demographic data such as age, gender, nationality, and residency [25]. In medical emergencies such as the COVID-19 pandemic, older people have a higher risk of complications and contracting serious illnesses if they are untreated. Through information filtering, the HRS can handle such emergencies by collecting patient messages and recommending treatment [26]. With the help of the patient’s demographic information, these messages for smoking cessation users used hybrid filtering to assess similarity.
A semantic web is a fast-evolving technology that utilizes a content-based recommendation system with machine-readable annotations [27]. The social-based filtering algorithm considers information about an individual’s neighborhood, along with similar tastes [28]. To prescribe the most appropriate treatment to patients, semantic clustering assesses the similarities between records, taking into account the patient’s demographics, location, and medical complications [29].
The DRS offers medicine based on patient reviews using sentiment analysis and feature engineering. The risk level classification identifies a patient’s immune system and recommends medicines if the patient has a low immune system [30]. Doulaverakis et al. [31] proposed GalenOWL, a semantic-driven online framework with the help of a specialist to manage drug recommendations based on the past profile of the patient. By considering worldwide standards such as ICD-10 and UNII, this framework converts clinical data and drug interactions to ontological terms. Cloud-assisted drug recommendation (CADRE) also considers the patient’s side effects and shifts to the cloud to advance the quality of the patient’s experience [32]. No particular DRS system was developed for the COVID-19 emergency. Therefore, we aimed to develop a DRS modeling framework by incorporating a stacked artificial neural network (ANN) for the fair and safe usage of drugs in pandemics.
3. Methods and Materials
3.1. Dataset
We prepared the drug selection dataset for the COVID-19 treatment from different sources including drug banks, news reports, and the existing literature [33,34,35,36,37]. Primarily, potential drugs recommended for COVID-19 by the World Health Organization (WHO) including Remdesivir, Umifenovir, acetaminophen, and Favipiravir were considered. Later, the data from studies [34,35,36,37] helped us to create interactions between the drugs and the collected clinical data. Other demographic patient information including gender, age, height, weight, exercise habits, country, food habits, COVID-19 infected data, and the co-morbidities were collected and are presented in Table 1. The incomplete or missing data columns were excluded from a given dataset. Patient symptoms based on individual questionnaires and lab reports were collected and drug categorization was completed for the drug using the stacked ANN. The recommendation of antibacterial drugs was completed by mapping the patient’s health history.

Table 1.
Features of the lifestyle and health status of a person.
3.1.1. Data Pre-Processing
The symmetric uncertainty feature selection measure was applied to find the association between different features in the feature space. Under the candidate feature (fi) we evaluated the information shared between the selected feature (fs) and fi. The relevance of the class of the independent features was measured by:
Symmetric uncertainty = ; here, I is the mutual information and E is the entropy of the features.
The entropy of fi is calculated by =
= ; Here , are the possible values of I and S, respectively; P(i) and P(s) signify the distribution of I and S, and P(i,s) signifies the joint distribution.
3.1.2. Symptom Extraction and Severity Rating
We divided the survey data into parts of speech to extract the symptoms, and the extracted words were mapped to the disease dictionary with the symptoms and drugs. The dictionary of medical terms was adopted from the Systematized Nomenclature of Medicine and Clinical Terms (SNOWMED-CT) [38,39]. The mapping of the corresponding symptom entities was completed using the infection severity. The infection severity was estimated under the guidance of the pharmacist.
To standardize the feature = ( …, ), we used the min–max scaling to normalize the features using = ; where is the rating specific to the infection symptom in are the maximum and the minimum ratings in , = .
3.1.3. Drug Target
To create the drug labels, we used the categorized drug information and described them based on the patient attributes. To measure the frequency (FREQ) of the occurrence of the drug based on the symptom feature for all the cases:
= ; here is the frequency of the drug (dp) to the symptom feature (sp); is the number of cases containing the dp and sp; and is the number of cases containing the drug .
The drug can be represented as = {}. The k-means clusters were applied in drug categorization [40,41,42]. We assembled the information about diseases and drug interactions for a better representation by:
; here represents the association of drug (t) to drug set (b).
The drug prediction was performed based on the drug correlation presented by ; here is the activation function, represents the element-wise product, represents the correlation between drug d and drug q.
The drug target aggregation operation associates a drug with a specific disease by ; here represents drug d after the g aggregation operation.
To aggregate the drug target information by aggregating the k layers by ; here t represents the target of the disease symptom in drug symptom, represents the initialized symptom t.
The Artificial Neural Network (ANN) model was incorporated to perform the drug selection based on the patient’s symptoms. When a new patient (pnew) appears, the model collects patient symptomatic data for a similarity check of the existing patient records. Then, the training phase is performed and the symptom drug classifier is classified according to the patients’ symptoms. Medications are displayed based on side effects and co-morbidities, and a drug is chosen based on the drug attributes. This is analyzed by:
; here are the features for the new cases and represent the old cases.
3.2. Recommendation Algorithm
To suggest drugs for the specific disease, the DRS recommends antibacterial drugs based on the individual’s past health status and present risk level. The matching of the drug with the active individual features is completed using the following equation:
here is the feature set of the individual and is the mean over the set of features of I.
If the diseased individual has allergies, high blood pressure, and poor health, adverse side effects of the drug may lead to death or morbidity [43]. The probability of the drug side effect is calculated by:
; here Sx and Sy are the side effects of the drug Dx, prediction score = , and is the average risk of all the diseased individuals with a risk factor of k.
The fair drug recommendation system takes into account health conditions, preferences, race, and gender. Based on the weighted binary singular value decomposition, a stacked ANN is proposed. Figure 1 illustrates the framework for the proposed drug recommendation system. A normal health condition or a worsened state is indicated by the input parameter values. For the recommendation algorithm, the current state of a parameter is crucial. Each parameter value can belong to a different class according to the proposed technique. Every user has a unique health profile, which is compared to a user who had a similar health condition in the past. Based on the user’s immune system and preferences, the stacked ANN model identifies the appropriate set of medications. Our recommendations were based on a comparison of the individuals’ health profiles and individual preferences with those of similar individuals. In Algorithm 1, all the individuals with the same features are grouped into a recommendation cluster.
| Algorithm 1. Drug recommendation algorithm |
| Input: Patient data |
| Output: Recommended drug |
|
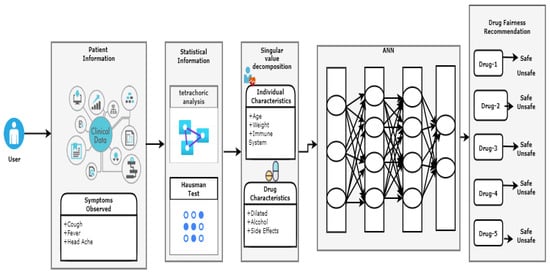
Figure 1.
Block diagram for the proposed recommender system.
3.3. Software and Hardware Specifications
The experiments were conducted on a PC with an Intel i9, 7890XE processor with 18 physical and 36 logical cores, 128 GB of memory, and Windows 10 Enterprise as the operating system. Anaconda IDE was used to write the Python code.
4. Results
4.1. Fairness Drug Recommendation for SARS-CoV-2 Infectious Diseases Using ML and Regression
Calculating high prediction values involves three steps including obtaining, filtering, and choosing the highest prediction values. The correlation between the recommendation errors and the threshold should be calculated. We used a statistical test to identify the possible predictors to improve the fairness of drug recommendations by considering the characteristics of the patient and the severity of the disease. A threshold of one standard deviation was applied to each diagnostic feature based on symmetric uncertainty. Tetrachoric analysis was used to estimate the correlation between the binary covariant features. The Conway Maxwell Poisson regression was employed to identify the drug characteristics and the clinical evidence. The Hausman test was also applied to assess the change in the effectiveness prediction score for the individual and the drug, as shown in Figure 2.
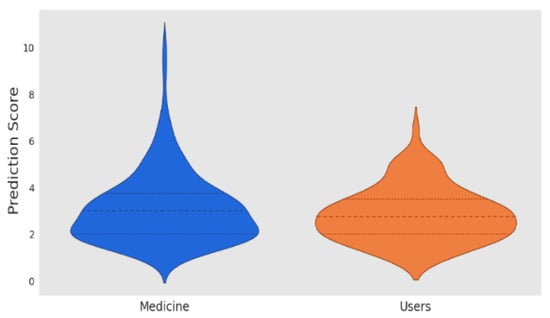
Figure 2.
Prediction score.
4.2. Fairness of Drug Side-Effects Predictions Using Deep Learning and Regression
Drug features were extracted to predict drug side effects associated with drug names and clinical characteristics. A bilingual evaluation understudy metric was used to identify the associated drug side effects. The following drugs are tabulated along with their side effects in Table 2.

Table 2.
Predicted drugs for specific diseases with the side effects.
The selection of an adequate threshold is crucial to fine-tune the medical recommendation process. It is recommended that the parameter range is defined as (0,1), where 0 indicates complete fairness and 1 indicates no fairness. An assessment of the average error for the recommendations is necessary for a better scale of accuracy. We chose 0.4 as the threshold value for the optimal selection, and 0.5 as the default parameter value. Table 3 summarizes the proposed drug recommender system with different hidden layers.

Table 3.
Performance of the stacked ANN with different hidden layers for the drug recommendation system.
The stacked ANN was implemented for a drug recommendation system with varying hidden layers, and the error rate was compared as the number of hidden layers was increased. A minimum error rate was observed when three hidden layers were used. To evaluate the proposed models, the Relu activation function and Adam optimizer with 150 epochs and 32 batch sizes were used. Table 4 presents a comparison of the performance estimates for the different models. The performance metrics were applied to evaluate the drug recommendation performance. We observed that the proposed model performed better than other traditional ML algorithms in terms of accuracy, precision, sensitivity, and specificity.

Table 4.
Comparison of the performance metrics of machine learning-based recommender systems.
5. Discussion
For example, a DRS for migraine patients can help doctors write the appropriate and accurate drugs for the patients based on their severity and importance [44]. The proposed DRS consists of individuals who are 80% similar to the present diseased individual. An additional system for diabetic patients using collaborative filtering can recognize patients that closely match the active patient by considering features such as insulin, glucose, body mass index, and blood pressure [45]. By using information about a patient’s profile, combined with ontologies and rule-based decision making, these systems can recommend anti-diabetes medicines with dose restrictions [46].
A patient with COVID-19 and susceptible virus-related fatigue was treated with chloroquine, as reported in [47]. Lopinavir significantly decreased the incidence of ARDS in patients with SARS-CoV-2 infection [48]. Nafamostat has been reported as a treatment for pancreatitis and abnormal coagulation that occur frequently in COVID-19 patients [49]. In the early phases of the COVID-19 treatment and for the treatment of influenza, camostat is used [50]. Gastric ulcers are treated with farotine, which has few side effects and an adequate efficacy and reduces iNOS activity [51]. With its immune system boosting effects, Nitazoxanide improves the respiratory distress associated with SARS-CoV-2 [52]. Combined with other COVID-19 treatments, ivermectin lowers mortality rates and hospital stays for moderate COVID-19 patients [53].
Researchers used resilient distributed dataset programming to implement the density peak-based clustering algorithm in their study [54]. A system for identifying sickness and treatment association rules was proposed to identify disease diagnosis recommendations. Unfortunately, the outbreak of coronavirus has limited the availability of legitimate clinical resources, such as doctors, nurses, and equipment. Due to the distress of the medical profession, a lot of people are dying. Shen et al. [55] outlined a system for performing infectious disease diagnoses and clinical decisions. In the proposed system, antibiotic usage is recommended using the naive Bayes classifier, and the ontological relations and rules are accurately stored in Neo4j.
Fairness is the major bias arising from recommender systems. Certain characteristics, such as race, gender, age, qualification, or property, are not represented equally in the dataset. In the case of unbalanced data, it is possible to highlight overrepresented groups in the rankings while reducing visibility for underrepresented groups. According to previous studies, the feedback loop causes the high usage medicines to become more popular and the low usage medicines to become less popular. Using the Type 2 fuzzy ontology and the wearable sensors for diabetes treatments, another study has reported that drugs and diet plans are available to patients [56]. Based on the findings of the authors, the proposed system is effective in extracting diabetic patients’ risk factors and recommending drug therapy.
In this paper, the accuracy of the proposed system framework gradually increased to 98.5%, which indicates that the ANN is the accepted model for drug recommender systems. After pre-processing, the patient feature spaces and the drug-drug interaction are obtained from the drug-target association. Health care data provide drug information for infectious diseases to the medical recommender system. This system compares the patient’s choices based on their similarity and has a superior level of accuracy to other state-of-the-art technologies. Since the proposed methodology builds both the interactions between a disease and the drug and the interactions between a drug and a drug, as well as between a drug and target, the hybrid restricted Boltzmann machine outperformed the logistic regression by approximately 3.9%.
6. Conclusions
This work involved deep learning techniques to make unbiased and fair drug recommendations. The loss function is used with the input data to improve fairness and accuracy. When a patient with comorbidities comes for a recommendation, we obtain the patient’s lab test results. A diagnosis is made by the DRS based on the features of the patient, and we rely on this diagnosis to determine the drug category in the system. The architecture uses statistical analysis to improve accuracy by adjusting the threshold value, which also balances fairness.
Author Contributions
Conceptualization, U.B. and G.B.; methodology, U.B.; software, G.B.; validation, G.B. and U.B.; formal analysis, U.B. and N.C.; investigation, U.B.; resources, G.B.; data curation, U.B.; writing—original draft preparation, U.B.; writing—review and editing, G.B.; visualization, U.B.; supervision, G.B.; project administration, G.B.; funding acquisition, G.B. and N.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
No author has any potential conflict of interest during the submission and publication of the manuscript.
References
- Battineni, G.; Sagaro, G.G.; Chintalapudi, N.; Di Canio, M.; Amenta, F. Assessment of Awareness and Knowledge on Novel Coronavirus (COVID-19) Pandemic among Seafarers. Healthcare 2021, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Goh, J.M.; Gao, G.; Agarwal, R. The creation of social value: Can an online health community reduce rural–urban health disparities? MIS Q. 2016, 40, 247–263. [Google Scholar] [CrossRef] [Green Version]
- Cook, S.F.; Bies, R.R. Disease Progression Modeling: Key Concepts and Recent Developments. Curr. Pharmacol. Rep. 2016, 2, 221–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koren, Y. Factorization Meets the Neighborhood: A Multifaceted Collaborative Filtering Model. In Proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Las Vegas, NV, USA, 24–27 August 2008; pp. 426–434. [Google Scholar]
- Ye, Q.; Hsieh, C.Y.; Yang, Z.; Kang, Y.; Chen, J.; Cao, D.; He, S.; Hou, T. A unified drug-target interaction prediction framework based on knowledge graph and recommendation system. Nat. Commun. 2021, 12, 6775. [Google Scholar] [CrossRef]
- Fox, S.; Duggan, M. Health Online 2013; Pew Research Internet Project Report: Washington, DC, USA, 2013. [Google Scholar]
- Chintalapudi, N.; Angeloni, U.; Battineni, G.; di Canio, M.; Marotta, C.; Rezza, G.; Sagaro, G.G.; Silenzi, A.; Amenta, F. LASSO Regression Modeling on Prediction of Medical Terms among Seafarers’ Health Documents Using Tidy Text Mining. Bioengineering 2022, 9, 124. [Google Scholar] [CrossRef]
- Lu, J.; Wu, D.; Mao, M.; Wang, W.; Zhang, G. Recommender system application developments: A survey. Decis. Support Syst. 2015, 74, 12–32. [Google Scholar] [CrossRef]
- Huang, F.; Wang, S.; Chan, C.-C. Predicting disease by using data mining based on healthcare information system. In Proceedings of the 2012 IEEE International Conference on granular computing, Washington, DC, USA, 11–13 August 2012; pp. 191–194. [Google Scholar]
- Subramaniyaswamy, V.; Manogaran, G.; Logesh, R.; Vijayakumar, V.; Chilamkurti, N.; Malathi, D.; Senthilselvan, N. An ontology driven personalized food recommendation in IoT-based healthcare system. J. Supercomput. 2019, 75, 3184–3216. [Google Scholar] [CrossRef]
- Liang, T.P. Recommender systems for decision support. Expert Syst. Appl. 2008, 45, 385–386. [Google Scholar]
- Adomavicius, G.; Tuzhilin, A. Toward the Next Generation of Recommender Systems: A Survey of the State of the Art and Possible Extensions. IEEE Trans. Knowl. Data Eng. 2005, 17, 734–749. [Google Scholar] [CrossRef]
- Esfandiari, N.; Babavalian, R.; Moghadam, E.; Tabar, V. Knowledge discovery in medicine: Current issue and future trend. Expert Syst. Appl. 2014, 41, 4434–4463. [Google Scholar] [CrossRef]
- Narducci, F.; Musto, C.; Polignano, M.; de Gemmis, M.; Lops, P.; Semeraro, G. A Recommender System for Connecting Patients to the Right Doctors in the Healthnet Social Network. In Proceedings of the 24th International Conference on World Wide Web, Florence, Italy, 18–22 May 2015; pp. 81–82. [Google Scholar]
- Han, Q.; Ji, M.; de Troya, I.M.d.R.; Gaur, M.; Zejnilovic, L. A Hybrid Recommender System for Patient-Doctor matchmaking in Primary Care. In Proceedings of the 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy, 1–4 October 2018; pp. 481–490. [Google Scholar]
- Hassan, S.; Syed, Z. From Netflix to heart attacks: Collaborative filtering in medical datasets. In Proceedings of the HI’10: ACM International Health Informatics Symposium, IHI’10: ACM International Health Informatics Symposium, Arlington, VA, USA, 11–12 November 2010; pp. 128–134. [Google Scholar]
- Teodorovic, D.; Selmic, M.; Mijatovic, L. Combining case-based reasoning with Bee Colony Optimization for dose planning in well differentiated thyroid cancer treatment. Expert Syst. Appl. 2013, 40, 2147–2155. [Google Scholar] [CrossRef]
- Savova, G.K.; Masanz, J.J.; Ogren, P.V.; Zheng, J.; Sohn, S.; Kipper, K.C.; Chute, C.G. Mayo clinical text analysis and knowledge extraction system(cTAKES): Architecture component evaluation and applications. J. Am. Med. Inform. Assoc. 2010, 17, 507–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, D.A.; Chawla, N.V.; Christakis, A.; Barabasi, A.L. Time to CARE: A collaborative engine for practical disease prediction. Data Min. Knowl. Discov. 2010, 20, 388–415. [Google Scholar] [CrossRef]
- Komkhao, M.; Lu, J.; Zhang, L. Determine Pattern Similarity in a Medical Recommender System. In International Conference on Data and Knowledge Engineering; Springer: Berlin/Heidelberg, Germany, 2012; pp. 103–114. [Google Scholar]
- Lu, X.; Huang, Z.; Duan, H. Supporting adaptive clinical treatment processes through recommendations. Comput. Methods Programs Biomed. 2012, 107, 413–424. [Google Scholar] [CrossRef]
- Caorsar, D.; Sleeman, D.H. Developing Knowledge Based System Using the Semantic web. In Proceedings of the International BCS Conference, London, UK, 22–24 September 2008; pp. 29–40. [Google Scholar]
- Burke, R. Knowledge recommender system. Encycl. Libr. Inf. Syst. 2000, 69 (Suppl. 32), 175–186. [Google Scholar]
- Wiesner, M.; Pfeifer, D. Health recommender systems: Concepts requirements technical basics and challenges. Int. J. Environ. Res. Public Health 2014, 11, 2580–2607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodadilla, J.; Ortega, F.; Hernando, A.; Gutierrez, A. Recommender system survey. Knowl. Based Syst. 2013, 46, 109–132. [Google Scholar] [CrossRef]
- Masaba, B.B.; Moturi, J.K.; Taiswa, J.; Mmusi-Phetoe, R.M. Devolution of healthcare system in Kenya: Progress and challenges. Public Health 2020, 189, 135–140. [Google Scholar] [CrossRef]
- Berners, L.; Hendler, J.; Lassila, O. The semantic web. Sci. Am. 2001, 284, 28–37. [Google Scholar] [CrossRef]
- Shardanad, U.; Maes, P. Social information filtering: Algorithms for automating word of mouth. Experts Syst. Appl. 1995, 95, 210–217. [Google Scholar]
- Leilei, S.; Chuanren, L.; Chonghui, G.; Hui, X.; Yanming, X. Data-Driven Automatic Treatment Regimen Development and Recommendation. In Proceedings of the International Conference on Knowledge Discovery and Data Mining(SIGKDD2016), San Francisco, CA, USA, 13–17 August 2016; pp. 1865–1874. [Google Scholar]
- Shimada, K.; Takada, H.; Mitsuyama, S.; Matsuo, H.; Otake, H.; Kunishima, H.; Kanemitsu, K.; Kaku, M. Drug recommendation system for patients with infectious diseases. AMIA Annu. Symp. Proc. 2005, 2005, 1112. [Google Scholar]
- Doulaverakis, C.; Nikolaidis, G.; Kleontas, A.; Kompatsiaris, I. GalenOWL: Ontology based drug recommendations discovery. J. Biomed. Semat. 2012, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.Z.; Dafang, H.; Mohammad, A.; Atif, P.; Limei, P. CADRE: Cloud assisted drug recommendation service for online pharmacies. Mob. Netw. Appl. 2014, 20, 348–355. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, Y.; Shen, Y.; Huang, W.; Martin, F.; Cheng, F. Network based drug repurposing for novel coronavirus2019-ncov/SARS-CoV-2. Nat. Cell Discov. 2020, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zheng, B.; Chen, B.; Butte, A.J.; Swamidass, S.J.; Lu, Z. A survey of current trends in computational drug repositioning. Brief. Bioinform. 2016, 17, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.; Zhu, S.; Liu, Y.; Zhou, R.; Nussinov, C.F. DeerDR: A network based deep learning approach to in silico drug repositionoing. Bioinformatics 2019, 35, 5191–5198. [Google Scholar] [CrossRef]
- Aliper, A.; Plis, S.; Artemov, A.; Ulloa, P.; Mamoshina, A.; Zhavoronkov, A. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol. Pharmcy 2016, 13, 2524–2530. [Google Scholar] [CrossRef] [Green Version]
- Haifeng, L.; Hongfei, L.; Chen, S.; Liang, Y.; Yuan, L.; Bo, X.; Zhihao, Y.; Jian, W.; Yuanyuan, S. A network representation approach for COVID-19 drug recommendation. Methods 2022, 198, 3–10. [Google Scholar]
- SNOMED CT Standard Ontology Based on the Ontology for General Medical Science. Available online: https://bioportal.bioontology.org/ontologies/SCTO (accessed on 16 May 2022).
- Available online: https://go.drugbank.com/drugs (accessed on 8 January 2022).
- Wang, X.; Sontag, D.; Wang, F. Unsupervised Learning of Disease Progression Models. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 85–94. [Google Scholar]
- Kumar, N.K.; Vigneswari, D. A Drug Recommendation System for Multi-Disease in Health Care using Machine Learning. In Advances in Communication and Computational Technology; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–12. [Google Scholar]
- Stark, B.; Knahl, C.; Aydin, M.; Samarah, M.; Elish, K.O. Better choice: A migraine drug recommendation system based on Neo4J. In Proceedings of the 2017 2nd IEEE International Conference on Computational Intelligence and Applications (ICCIA), Beijing, China, 8–11 September 2017; pp. 382–386. [Google Scholar]
- Qian, Z.; Guangquan, Z.; Jie, L.; Wu, D. A framework of hybrid recommender system for personalized clinical prescription. In Proceedings of the 10th International Conference on Intelligent Systems and Knowledge Engineering (ISKE), Taipei, Taiwan, 24–27 November 2015; pp. 1–7. [Google Scholar]
- Schäfer, H.; Hors-Fraile, S.; Karumur, R.P.; Valdez, A.C.; Said, A.; Torkamaan, H.; Ulmer, T.; Trattner, C. Towards health (aware) recommender systems. In Proceedings of the DH’17: International Conference on Digital Health, London, UK, 2–5 July 2017; pp. 157–161. [Google Scholar]
- Bankhele, S.; Mhaske, A.; Bhat, S.; Shinde, S.V. A diabetic healthcare recommendation system. Int. J. Comput. Appl. 2017, 167, 14–18. [Google Scholar] [CrossRef]
- Mahmoud, N.; Elbeh, H. Irs-t2d: Individualize recommendation system for type2 diabetes medication based on ontology and swrl. In Proceedings of the 10th International Conference on Informatics and Systems, Giza, Egypt, 9–11 May 2016; pp. 203–209. [Google Scholar]
- Toutet, F.; de Lamnallerie, X. Of chloroquine and COVID-19. Antivir. Res. 2020, 177, 104762. [Google Scholar]
- Meini, S.; Pagotto, A.; Longo, B.; Vendramin, I.; Pecori, D.; Tascini, C. Role of Lopinavir/Ritonavir in the Treatment of COVID-19: A Review of Current Evidence, Guideline Recommendations and Perceptives. J. Clin. Med. 2020, 9, 2050. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, W.; Yoneda, T.; Koba, H.; Ueda, T.; Tsuji, N.; Ogawa, H.; Asakura, H. Potential mechanisms of nafamostat therapy for severe COVID-19 pneumonia with disseminated intravascular coagulation. Int. J. Infect. Dis. 2021, 102, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Breining, P.; Frolund, A.L.; Hojen, J.F.; Gunst, J.D.; Staerke, N.B.; Saedder, E.; Thomas, M.C.; Little, P.; Nielsen, L.P.; Sogaard, O.S.; et al. Camostat mesylate against SARS-CoV-2 and COVID-19 Rationale, dosing and safety. Basic Clin. Pharmacol. Toxicol. 2021, 128, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Sehitoglu, M.H.; Oztopuz, O.; Karaboga, I.; Ovali, M.A.; Uzun, M. Human Acid has a protective effect on gastric ulcer by alleviating inflammation in rats. Cytol. Genet. 2022, 56, 84–97. [Google Scholar] [CrossRef]
- Amit, S.; Lokhande, P.; Devarajan, V. A review on possible mechanistic insights of Nitazoxanide for repurposing in COVID-19. Eur. J. Pharmacol. 2021, 891, 173748. [Google Scholar]
- Sherief, A.E.; Noor, R.A.; Badawi, R.; Eslam, M.K.; Soliman, S.E.S.; Mohamed, S.A.E.G.; Elbahnasawy, M.; Moustafa, E.F.; Hassany, S.M.; Medhat, M.A.; et al. Clinical study evaluating the efficacy of ivermectin in COVID-19 treatment: A randomized controlled study. J. Med. Virol. 2021, 93, 5833–5838. [Google Scholar]
- Chen, J.; Li, K.; Rong, H.; Bilal, K.; Yang, N.; Li, K. A disease diagnosis and treatment recommendation system based on big data mining and cloud computing. Inf. Sci. 2018, 435, 124–149. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Yuan, K.; Chen, D.; Colloc, J.; Yang, M.; Li, Y.; Lei, K. An ontology-driven clinical decision support system (IDDAP) for infectious disease diagnosis and antibiotic prescription. Artif. Intell. Med. 2018, 86, 20–32. [Google Scholar] [CrossRef]
- Ali, F.; Islam, S.M.R.; Kwak, D.; Khan, P.; Ullah, N.; Yoo, S.-J.; Kwak, K.S. Type-2 fuzzy ontology-aided recommender systems for IoT–based healthcare. Comput. Commun. 2018, 119, 138–155. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).