PUB-SalNet: A Pre-Trained Unsupervised Self-Aware Backpropagation Network for Biomedical Salient Segmentation
Abstract
1. Introduction
- We propose the novel PUB-SalNet model for biomedical salient segmentation, which is a completely unsupervised method utilizing weights and knowledge from pre-training and attention-guided refinement during back propagation.
- We aggregate a new biomedical data set called SalSeg-CECT, featuring rich salient objects, different SNR settings, and various resolutions, which also serves for pre-training and fine-tuning for other complex biomedical tasks.
- Extensive experiments show that the proposed PUB-SalNet achieves state-of-the-art performance. The same method can be adapted to process 3D images, demonstrating correctness and generalization ability of our method.
2. Related Work
2.1. Pre-Trained Methods in Biomedical Images
2.2. Unsupervised Biomedical Image Segmentation
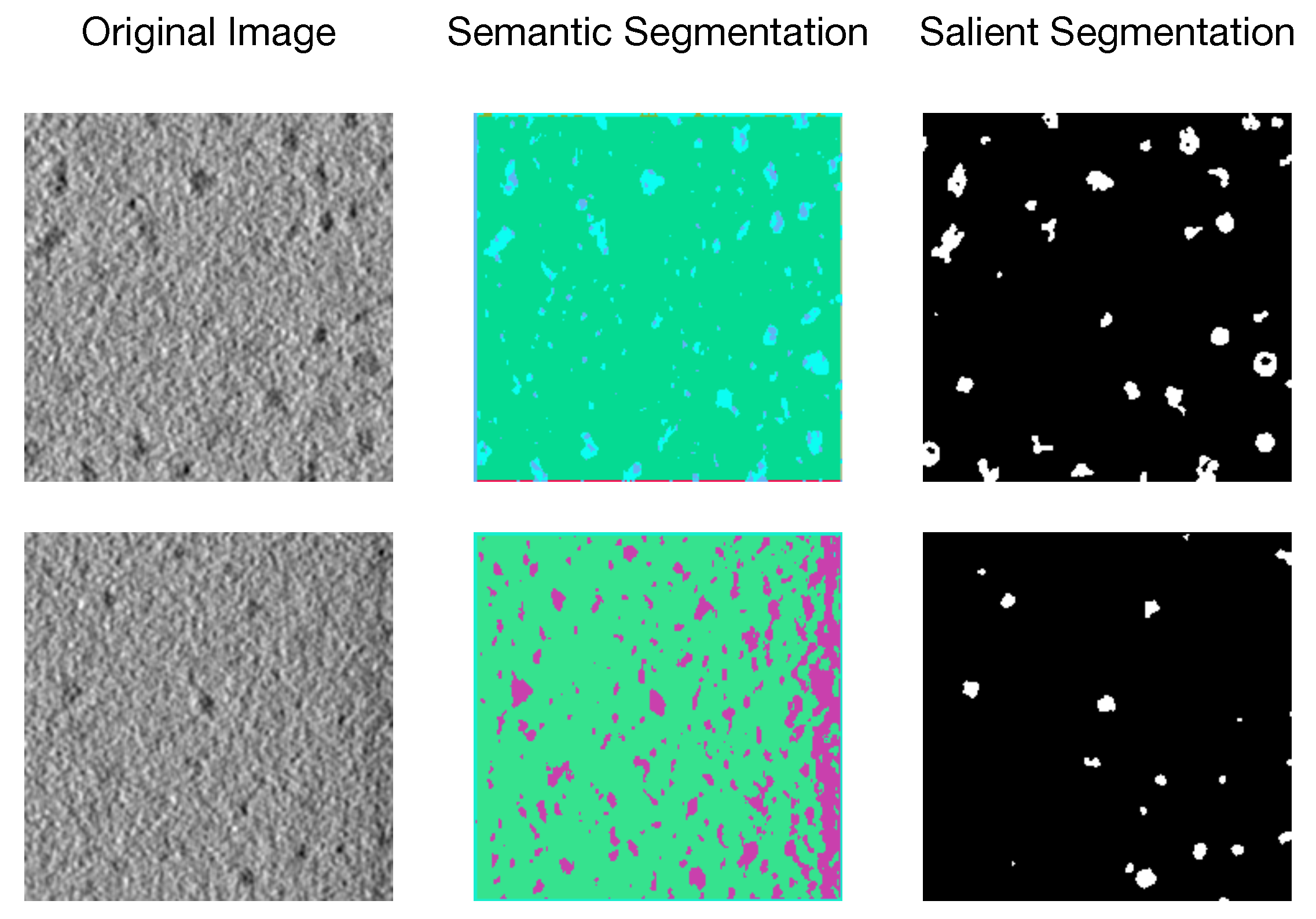
2.3. Salient Segmentation
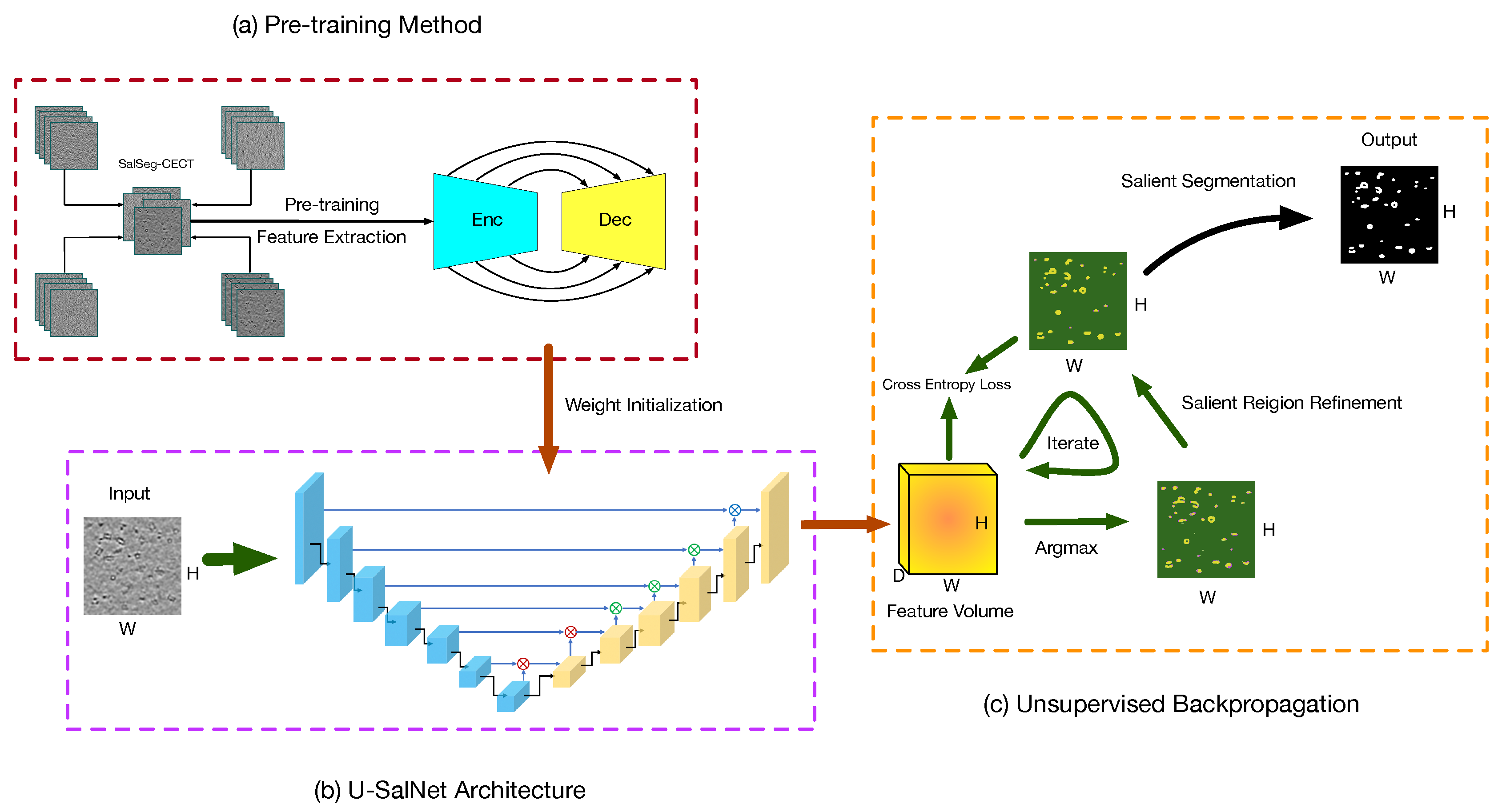
3. PUB-SalNet
3.1. Pre-Training Method
3.2. U-SalNet Architecture
3.3. Unsupervised Backpropagation
| Algorithm 1 Unsupervised Backpropagation Algorithm |
| Require: Original biomedical image |
| Ensure: Salient segmentation results |
| 1: // Initialize backbone parameters |
| 2: // Initialize classifier parameters |
| 3: |
| 4: for do |
| 5: if then |
| 6: |
| 7: |
| 8: |
| 9: |
| 10: |
| 11: //predict salient labels |
| 12: for do |
| 13: |
| 14: for |
| 15: end for |
| 16: |
| 17: |
| 18: end if |
| 19: end for |
4. Experiments
4.1. Datasets Setting
4.2. Implementation Details
4.3. Evaluation Metrics
4.4. Quantitative Evaluation
4.4.1. Comparison with State-of-the-Art
4.4.2. Ablation Study
4.4.3. Parameter Sensitivity Analysis
4.5. Qualitative Evaluation
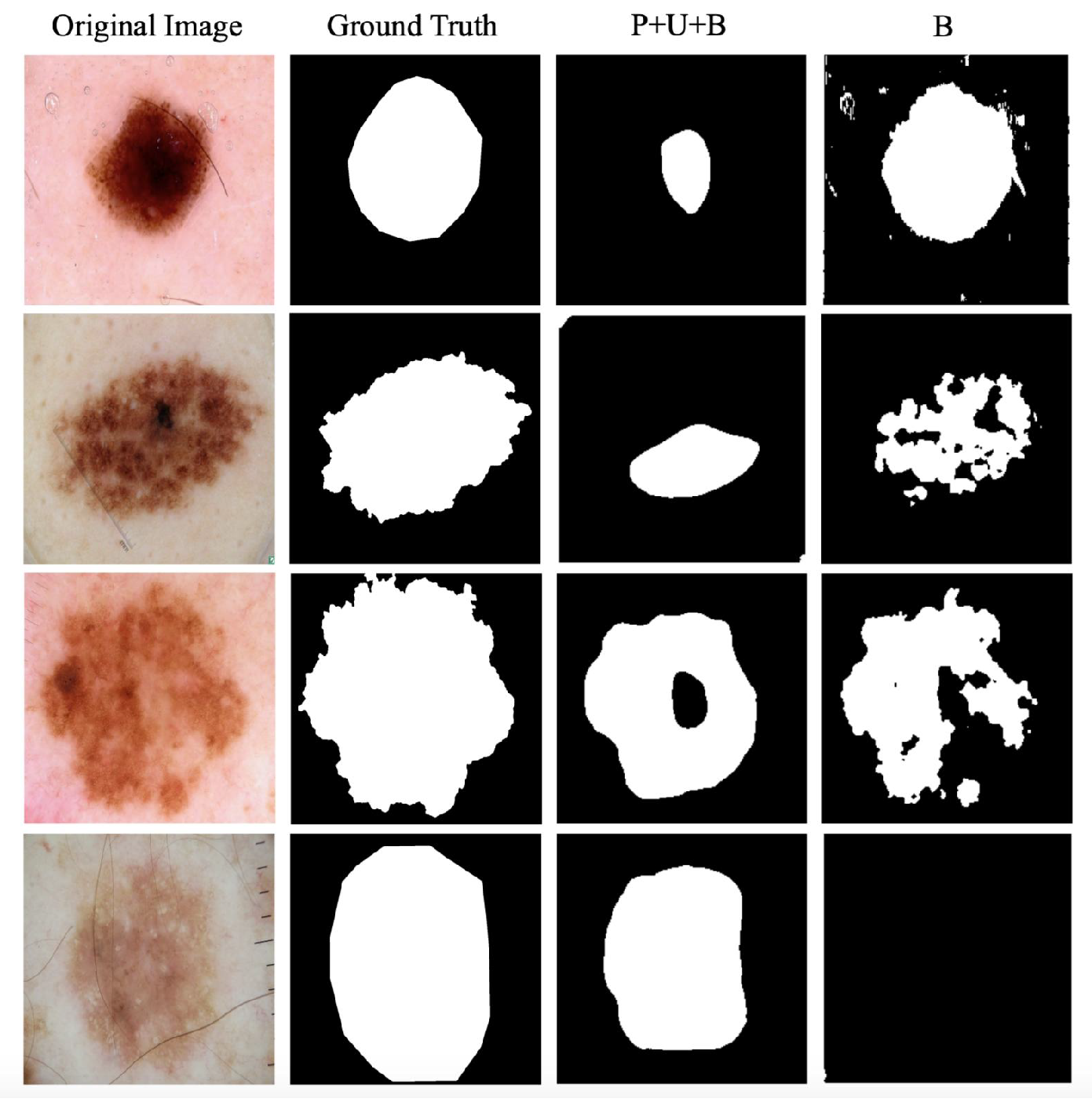
4.6. Case Study on the ISBI Challenge
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lee, J.G.; Jun, S.; Cho, Y.W.; Lee, H.; Kim, G.B.; Seo, J.B.; Kim, N. Deep learning in medical imaging: General overview. Korean J. Radiol. 2017, 18, 570–584. [Google Scholar] [CrossRef] [PubMed]
- Kanezaki, A. Unsupervised image segmentation by backpropagation. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 1543–1547. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]
- Geiger, A.; Lenz, P.; Urtasun, R. Are we ready for autonomous driving? the kitti vision benchmark suite. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 July 2012; pp. 3354–3361. [Google Scholar]
- Yuan, J.; Gleason, S.S.; Cheriyadat, A.M. Systematic benchmarking of aerial image segmentation. IEEE Geosci. Remote Sens. Lett. 2013, 10, 1527–1531. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; pp. 91–99. [Google Scholar]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.K.; Vikas, S.; Sarma, R.R.; Shenoy, G.; Kumar, R. Segmentation and Classification of CT Renal Images Using Deep Networks. In Soft Computing and Signal Processing; Springer: Singapore, 2019; pp. 497–506. [Google Scholar]
- Raza, K.; Singh, N.K. A tour of unsupervised deep learning for medical image analysis. arXiv 2018, arXiv:1812.07715. [Google Scholar]
- Moriya, T.; Roth, H.R.; Nakamura, S.; Oda, H.; Nagara, K.; Oda, M.; Mori, K. Unsupervised segmentation of 3D medical images based on clustering and deep representation learning. In Medical Imaging 2018: Biomedical Applications in Molecular, Structural, and Functional Imaging; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; Volume 10578, p. 1057820. [Google Scholar]
- Croitoru, I.; Bogolin, S.V.; Leordeanu, M. Unsupervised Learning of Foreground Object Segmentation. Int. J. Comput. Vis. 2019, 127, 1279–1302. [Google Scholar] [CrossRef]
- Ilyas, T.; Khan, A.; Umraiz, M.; Kim, H. SEEK: A Framework of Superpixel Learning with CNN Features for Unsupervised Segmentation. Electronics 2020, 9, 383. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef]
- Chen, S.; Ma, K.; Zheng, Y. Med3D: Transfer Learning for 3D Medical Image Analysis. arXiv 2019, arXiv:1904.00625. [Google Scholar]
- Honkanen, M.K.; Matikka, H.; Honkanen, J.T.; Bhattarai, A.; Grinstaff, M.W.; Joukainen, A.; Kröger, H.; Jurvelin, J.S.; Töyräs, J. Imaging of proteoglycan and water contents in human articular cartilage with full-body CT using dual contrast technique. J. Orthop. Res. 2019, 37, 1059–1070. [Google Scholar] [CrossRef]
- Xia, X.; Kulis, B. W-net: A deep model for fully unsupervised image segmentation. arXiv 2017, arXiv:1711.08506. [Google Scholar]
- Chen, J.; Frey, E.C. Medical Image Segmentation via Unsupervised Convolutional Neural Network. Available online: https://openreview.net/pdf?id=XrbnSCv4LU (accessed on 10 May 2020).
- Pan, F.; Shin, I.; Rameau, F.; Lee, S.; Kweon, I.S. Unsupervised Intra-domain Adaptation for Semantic Segmentation through Self-Supervision. arXiv 2020, arXiv:2004.07703. [Google Scholar]
- Hu, K.; Liu, S.; Zhang, Y.; Cao, C.; Xiao, F.; Huang, W.; Gao, X. Automatic segmentation of dermoscopy images using saliency combined with adaptive thresholding based on wavelet transform. Multimed. Tools Appl. 2019, 1–18. [Google Scholar] [CrossRef]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Visin, F.; Kastner, K.; Cho, K.; Matteucci, M.; Courville, A.; Bengio, Y. Renet: A recurrent neural network based alternative to convolutional networks. arXiv 2015, arXiv:1505.00393. [Google Scholar]
- Liu, N.; Han, J.; Yang, M.H. PiCANet: Learning pixel-wise contextual attention for saliency detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3089–3098. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. Slic Superpixe, Technical Report. Available online: https://infoscience.epfl.ch/record/149300 (accessed on 10 May 2020).
- Pei, L.; Xu, M.; Frazier, Z.; Alber, F. Simulating cryo electron tomograms of crowded cell cytoplasm for assessment of automated particle picking. BMC Bioinform. 2016, 17, 405. [Google Scholar] [CrossRef]
- Gutman, D.; Codella, N.C.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Fan, D.P.; Gong, C.; Cao, Y.; Ren, B.; Cheng, M.M.; Borji, A. Enhanced-alignment measure for binary foreground map evaluation. arXiv 2018, arXiv:1805.10421. [Google Scholar]
- Fan, D.P.; Cheng, M.M.; Liu, Y.; Li, T.; Borji, A. Structure-measure: A new way to evaluate foreground maps. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 4548–4557. [Google Scholar]
- Itti, L.; Koch, C.; Niebur, E. A model of saliency-based visual attention for rapid scene analysis. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 1254–1259. [Google Scholar] [CrossRef]
- Zhai, Y.; Shah, M. Visual attention detection in video sequences using spatiotemporal cues. In Proceedings of the 14th ACM international conference on Multimedia, Santa Barbara, CA, USA, 23–27 October 2006; pp. 815–824. [Google Scholar]
- Hou, X.; Zhang, L. Saliency detection: A spectral residual approach. In Proceedings of the 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007; pp. 1–8. [Google Scholar]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; Volume 70, pp. 3319–3328. [Google Scholar]
- Smilkov, D.; Thorat, N.; Kim, B.; Viégas, F.; Wattenberg, M. Smoothgrad: Removing noise by adding noise. arXiv 2017, arXiv:1706.03825. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar]
- Bojarski, M.; Choromanska, A.; Choromanski, K.; Firner, B.; Jackel, L.; Muller, U.; Zieba, K. VisualBackProp: Efficient visualization of CNNs. arXiv 2016, arXiv:1611.05418. [Google Scholar]
- Pan, J.; Ferrer, C.C.; McGuinness, K.; O’Connor, N.E.; Torres, J.; Sayrol, E.; Giro-i Nieto, X. Salgan: Visual saliency prediction with generative adversarial networks. arXiv 2017, arXiv:1701.01081. [Google Scholar]

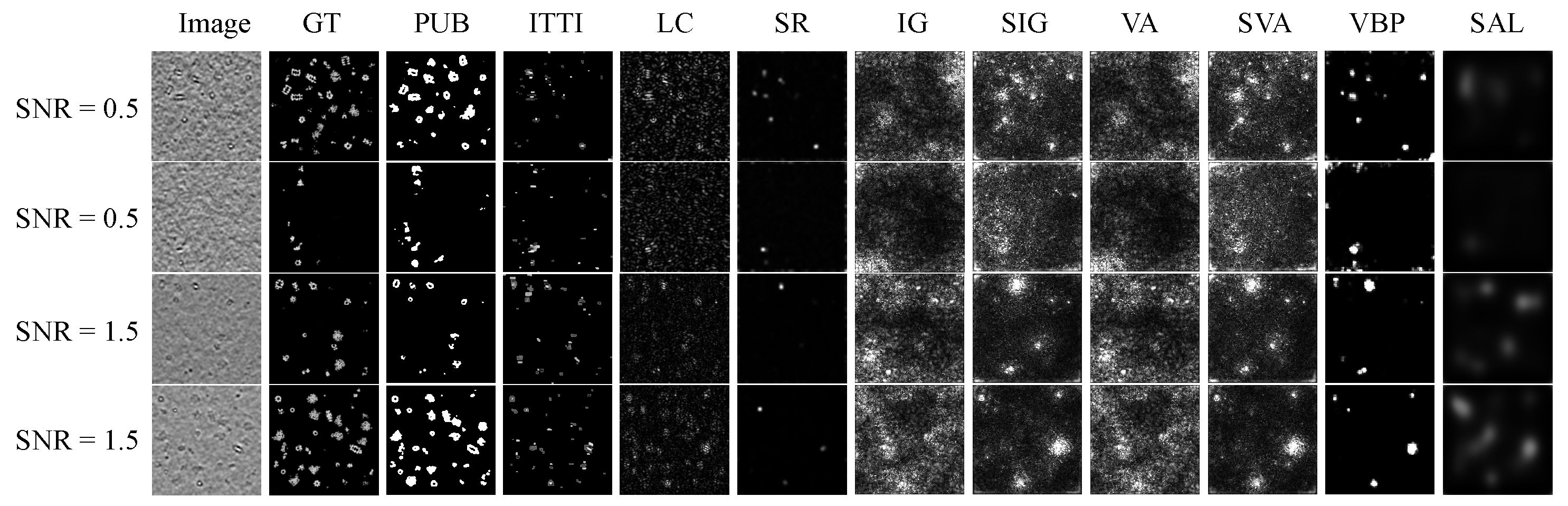

| Data Set | SNR = 0.5 | SNR = 1.5 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Metric | |||||||||
| Method | |||||||||
| Itti [30] | 0.1277 | 0.4759 | 0.3811 | 0.4445 | 0.1206 | 0.6396 | 0.4639 | 0.4781 | |
| LC [31] | 0.1626 | 0.3277 | 0.4466 | 0.4846 | 0.1463 | 0.4615 | 0.4369 | 0.5022 | |
| SR [32] | 0.1340 | 0.2535 | 0.3020 | 0.4406 | 0.1316 | 0.3439 | 0.2911 | 0.4423 | |
| IG [33] | 0.2843 | 0.1713 | 0.4775 | 0.4262 | 0.2978 | 0.1848 | 0.4739 | 0.4322 | |
| SIG [34] | 0.2623 | 0.2647 | 0.4959 | 0.4781 | 0.2310 | 0.3387 | 0.5134 | 0.5177 | |
| VA [35] | 0.2843 | 0.1713 | 0.4775 | 0.4262 | 0.2978 | 0.1848 | 0.4739 | 0.4322 | |
| SVA [34] | 0.2625 | 0.2647 | 0.4957 | 0.4779 | 0.2305 | 0.3414 | 0.5129 | 0.5186 | |
| VBP [36] | 0.1295 | 0.3049 | 0.4033 | 0.4527 | 0.1224 | 0.4588 | 0.4053 | 0.4717 | |
| SalGAN [37] | 0.1427 | 0.1984 | 0.3126 | 0.4411 | 0.1585 | 0.2367 | 0.4090 | 0.4629 | |
| PUB-SalNet | 0.0914 | 0.6573 | 0.7036 | 0.6494 | 0.0762 | 0.7426 | 0.7522 | 0.7209 | |
| Improvement | ↓ 28.43% | ↑ 38.12% | ↑ 41.88% | ↑ 34.01% | ↓ 36.82% | ↑ 16.10% | ↑ 46.51% | ↑ 39.00% | |
| Data Set | SNR = 0.5 | SNR = 1.5 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Metric | |||||||||
| Method | |||||||||
| B | 0.1461 | 0.1628 | 0.3692 | 0.4230 | 0.1433 | 0.1628 | 0.3960 | 0.4223 | |
| U+B | 0.2870 | 0.1628 | 0.4834 | 0.3585 | 0.2677 | 0.1628 | 0.5130 | 0.3693 | |
| P+B | 0.1063 | 0.5631 | 0.5906 | 0.5661 | 0.0949 | 0.6551 | 0.5947 | 0.5979 | |
| P+U | 0.1104 | 0.6214 | 0.6306 | 0.6506 | 0.0973 | 0.7544 | 0.7465 | 0.7617 | |
| P+U+B | 0.0914 | 0.6573 | 0.7036 | 0.6494 | 0.0762 | 0.7426 | 0.7522 | 0.7209 | |
| Data Set | SNR = 0.5 | SNR = 1.5 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Metric | |||||||||
| Method | |||||||||
| PUB-SalNet-B20 | 0.0984 | 0.6347 | 0.6964 | 0.6428 | 0.0793 | 0.7239 | 0.7447 | 0.7124 | |
| PUB-SalNet-B40 | 0.0961 | 0.6396 | 0.6945 | 0.6443 | 0.0766 | 0.7318 | 0.7320 | 0.7107 | |
| PUB-SalNet-B60 | 0.0945 | 0.6437 | 0.6710 | 0.6358 | 0.0774 | 0.7218 | 0.7294 | 0.7032 | |
| PUB-SalNet-B80 | 0.0943 | 0.6543 | 0.7221 | 0.6598 | 0.0783 | 0.7327 | 0.7340 | 0.7065 | |
| PUB-SalNet-B100 | 0.0914 | 0.6573 | 0.7036 | 0.6494 | 0.0762 | 0.7426 | 0.7522 | 0.7209 | |
| Data Set | ISBI 2017 Skin | |||
|---|---|---|---|---|
| Metric | ||||
| Method | ||||
| B | 0.3136 | 0.3378 | 0.4140 | |
| P+U+B | 0.3498 | 0.3378 | 0.4674 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, F.; Jiang, Y.; Zeng, X.; Zhang, J.; Gao, X.; Xu, M. PUB-SalNet: A Pre-Trained Unsupervised Self-Aware Backpropagation Network for Biomedical Salient Segmentation. Algorithms 2020, 13, 126. https://doi.org/10.3390/a13050126
Chen F, Jiang Y, Zeng X, Zhang J, Gao X, Xu M. PUB-SalNet: A Pre-Trained Unsupervised Self-Aware Backpropagation Network for Biomedical Salient Segmentation. Algorithms. 2020; 13(5):126. https://doi.org/10.3390/a13050126
Chicago/Turabian StyleChen, Feiyang, Ying Jiang, Xiangrui Zeng, Jing Zhang, Xin Gao, and Min Xu. 2020. "PUB-SalNet: A Pre-Trained Unsupervised Self-Aware Backpropagation Network for Biomedical Salient Segmentation" Algorithms 13, no. 5: 126. https://doi.org/10.3390/a13050126
APA StyleChen, F., Jiang, Y., Zeng, X., Zhang, J., Gao, X., & Xu, M. (2020). PUB-SalNet: A Pre-Trained Unsupervised Self-Aware Backpropagation Network for Biomedical Salient Segmentation. Algorithms, 13(5), 126. https://doi.org/10.3390/a13050126





