Abstract
Salient segmentation is a critical step in biomedical image analysis, aiming to cut out regions that are most interesting to humans. Recently, supervised methods have achieved promising results in biomedical areas, but they depend on annotated training data sets, which requires labor and proficiency in related background knowledge. In contrast, unsupervised learning makes data-driven decisions by obtaining insights directly from the data themselves. In this paper, we propose a completely unsupervised self-aware network based on pre-training and attentional backpropagation for biomedical salient segmentation, named as PUB-SalNet. Firstly, we aggregate a new biomedical data set from several simulated Cellular Electron Cryo-Tomography (CECT) data sets featuring rich salient objects, different SNR settings, and various resolutions, which is called SalSeg-CECT. Based on the SalSeg-CECT data set, we then pre-train a model specially designed for biomedical tasks as a backbone module to initialize network parameters. Next, we present a U-SalNet network to learn to selectively attend to salient objects. It includes two types of attention modules to facilitate learning saliency through global contrast and local similarity. Lastly, we jointly refine the salient regions together with feature representations from U-SalNet, with the parameters updated by self-aware attentional backpropagation. We apply PUB-SalNet for analysis of 2D simulated and real images and achieve state-of-the-art performance on simulated biomedical data sets. Furthermore, our proposed PUB-SalNet can be easily extended to 3D images. The experimental results on the 2d and 3d data sets also demonstrate the generalization ability and robustness of our method.
1. Introduction
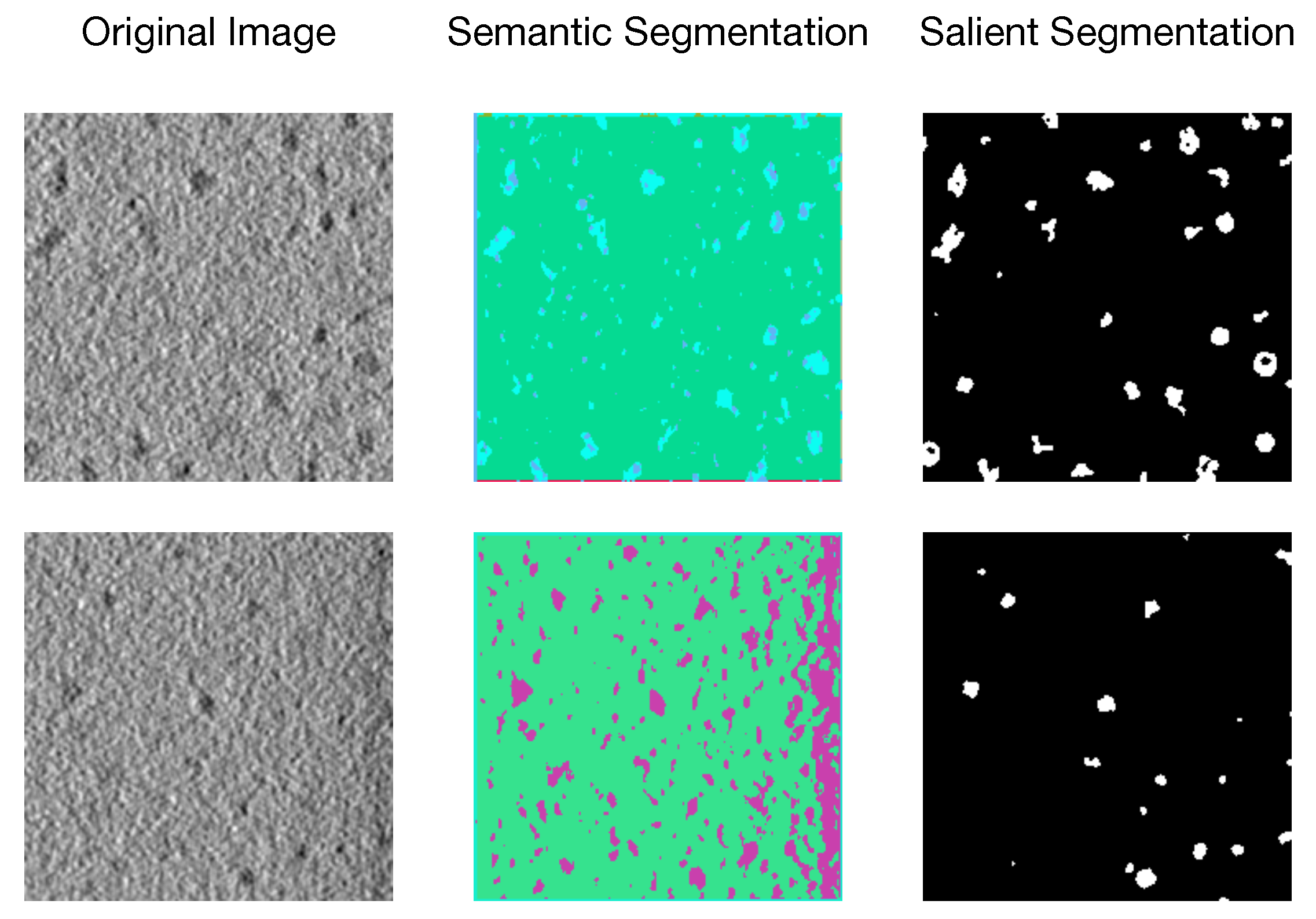
Biomedical image segmentation has drawn attention due to its widespread applications in computer-aided diagnosis and intelligent medical programs [1], among which salient segmentation refers to pixel-level annotation for regions of interest (e.g., organelle, substructures, and lesions) on biomedical images (e.g., Cellular Electron Cryo-Tomography (CECT) 3D images, Computed Tomography (CT), and Magnetic Resonance Imaging (MRI)). An example of semantic segmentation [2] and salient segmentation with unsupervised methods on CECT images is shown in Figure 1. Although image segmentation is widely used in many fields (such as Natural Scenes [3], Autonomous Driving [4], and Aerial Imaging [5]), we discover that traditional segmentation methods [6,7,8,9] cannot handle CECT images properly due to heterogeneous salient objects, various SNR, and low resolution of the data set, while salient segmentation can efficiently and effectively capture objects of interest to people and mask off irrelevant regions. However, accurate salient segmentation is challenging due to different shapes and sizes of the region of interest and diversity of images produced by various biomedical imaging devices.
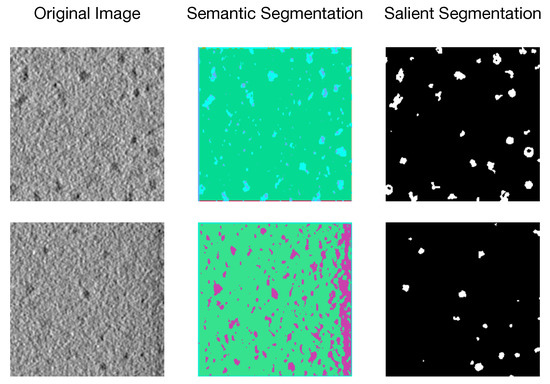
Figure 1.
An unsupervised example of semantic segmentation and salient segmentation on CECT images.
Recently, current salient segmentation methods on biomedical images [10] mostly use improved convolutional neural networks (CNNs) and its variants, which rely on supervised learning that require labor-intensive annotation of large data sets by experts. Furthermore, such methods are strictly limited by the quality of data sets. They are vulnerable to problems of model generalization and extensibility when facing adversarial training. In contrast, unsupervised learning not only derives insights directly from the data but also uses them to make data-driven decisions. It is more practical and robust for some complex tasks in biomedical areas, such as saliency detection and image segmentation. Therefore, it is crucial to work out an effective unsupervised salient segmentation method for biomedical images.
However, in recent years, few works have looked into unsupervised salient segmentation on biomedical images due to the complexity of biological structures. The authors in [11] systematically reviewed current unsupervised models for biomedical image segmentation. More recently, a unified unsupervised approach based on clustering and deep representation learning was designed by [12]. The authors in [13] proposed a teacher–student unsupervised learning system. The teacher performs unsupervised object discovery and, at the same time, multiple students with various network architectures are trained to ensure a better diversity. The authors [14] utilized the the squeeze and excitation network to capture interdependencies between channels and iteratively enable the CNN to learn the target generated by K-Means clustering. These methods are either based on super-pixel clustering with posterior selection or dependent on carefully optimized network hyper-parameters, which indicates that they are in need of human interference and are not completely unsupervised.
In this paper, we propose the PUB-SalNet, which is a self-aware network based on pre-training and attentional backpropagation. In order to build a high-performance automatic biomedical salient segmentation model to improve computer-aided diagnosis and other biomedical image analyzing tasks, we design a processing pipeline with three major modules: (1) A pre-training method specially designed for biomedical images; (2) The U-SalNet model, which selectively attends to salient objects via fusing two attention mechanisms into U-Net; (3) An unsupervised self-aware backpropagation method based on superpixels, which iteratively updates the parameters of U-SalNet. Quantitative and qualitative experiments comparing our performance with state-of-the-art salient segmentation methods demonstrate that the proposed PUB-SalNet outperforms all existing unsupervised methods and achieves state-of-the-art performance on the simulated biomedical data sets. Furthermore, PUB-SalNet can also be easily extended to 3D images. The experimental results on 2D and 3D biomedical data sets show generalization ability and robustness of the proposed method. Our main contributions are summarized as follows:
- We propose the novel PUB-SalNet model for biomedical salient segmentation, which is a completely unsupervised method utilizing weights and knowledge from pre-training and attention-guided refinement during back propagation.
- We aggregate a new biomedical data set called SalSeg-CECT, featuring rich salient objects, different SNR settings, and various resolutions, which also serves for pre-training and fine-tuning for other complex biomedical tasks.
- Extensive experiments show that the proposed PUB-SalNet achieves state-of-the-art performance. The same method can be adapted to process 3D images, demonstrating correctness and generalization ability of our method.
The rest of the paper is organized as follows. We review related works in the next section. In Section 3, we describe the PUB-SalNet and the completed processing pipeline for salient segmentation. Quantitative and qualitative experiments are discussed in detail in Section 4. Lastly, in Section 5, we conclude our method and future works.
2. Related Work
2.1. Pre-Trained Methods in Biomedical Images
Many works have shown that the pre-training method along with adequate fine-tuning is superior to training from scratch, with a lower risk of over-fitting and being less dependent on the size of the training set [15]. However, in the biomedical area, it is extremely challenging to build an effective pre-trained model due to the difficulty of data acquisition and annotation by experts. Only a few pre-trained models are related to biomedical images, among which the most famous is Ref. [16]’s MedicalNet. They collect data from some medical challenges and build the 3DSeg-8 data set with diverse modalities to train MedicalNet, and then transfer it to other segmentation and classification tasks and achieve state-of-the-art performance. However, their work is demanding of high-quality medical images with various scan regions, target organs, and pathologies, which is not applicable to cellular image analysis.
With recent breakthroughs in CECT 3D imaging technology [17], it is now possible for researchers to deeply look into and comprehend the macromolecular structure of the cell, which is more meaningful to biomedical fundamental studies. In order to solve the challenges above, we present a pre-trained model intended for biomedical images featuring a low SNR, low resolution, and rich salient objects.
2.2. Unsupervised Biomedical Image Segmentation
Unsupervised segmentation for biomedical images is very promising yet challenging. The authors in [18] concatenate two fully convolutional networks together into an autoencoder. The encoder produces a k-way pixel-wise prediction while the decoder reconstructs the image. Both the normalized cutting loss of the segmentation map and the reconstruction error are jointly minimized during training. However, we find the training very difficult to converge due to the inappropriate combination of the loss functions. The authors in [19] use the classic ACWE (Active Contours Without Edges) method as an supervision for the CNN-based segmentation model. However, for biomedical images, finding contours is a difficult task due to great variance in image quality. The authors in [20] used domain adaptation to minimize the inter-domain and intra-domain gap in three steps, which was also naturally an unsupervised and pre-trained approach to produce segmentation labels, but also requires high quality images, and resemblance in the two domains [21] proposes an unsupervised skin lesion segmentation method to combine color and brightness saliency maps into enhanced fusion saliency. Although it shows good results on dermoscopy images, it relies too much on coloring and contrast information and cannot effectively perform salient segmentation on grey-scale images (such as CECT). The authors in [2] optimize the pixel labels using a common CNN network while their parameters are iteratively updated by a gradient descent to unify labels within a superpixel. However, their model is trained every time on a natural image and displays randomness in predictions. It cannot utilize knowledge from a large training set. In addition, it does not work well when applied to noisy biomedical images. In order to solve these problems and adapt to salient segmentation on biomedical images, we load weights pre-trained on an assembled biomedical data set and present the U-SalNet model to extract significant features.
2.3. Salient Segmentation
Current salient segmentation methods on biomedical images treat the problem as a binary (namely salient and non-salient) segmentation task, identifying the label (foreground or background) of pixels. Recently, the authors in [22] propose an attention gate (AG) model to focus on significant targets for medical image analysis. Through AG, the model can ignore the background in images while highlighting the salient objects meaningful to the medical segmentation task. Specifically, AG extracts local information from a denser layer in the decoder and then uses it as a gating signal for the current layer to combine low-level features from the encoder network with the decoded features. However, it does not explicitly make use of global interference when producing attention maps, resulting in their saliency maps ignoring global contrast mechanisms. Different from their work, our proposed U-SalNet model applies both global and local self-attention to better integrate saliency information.
3. PUB-SalNet
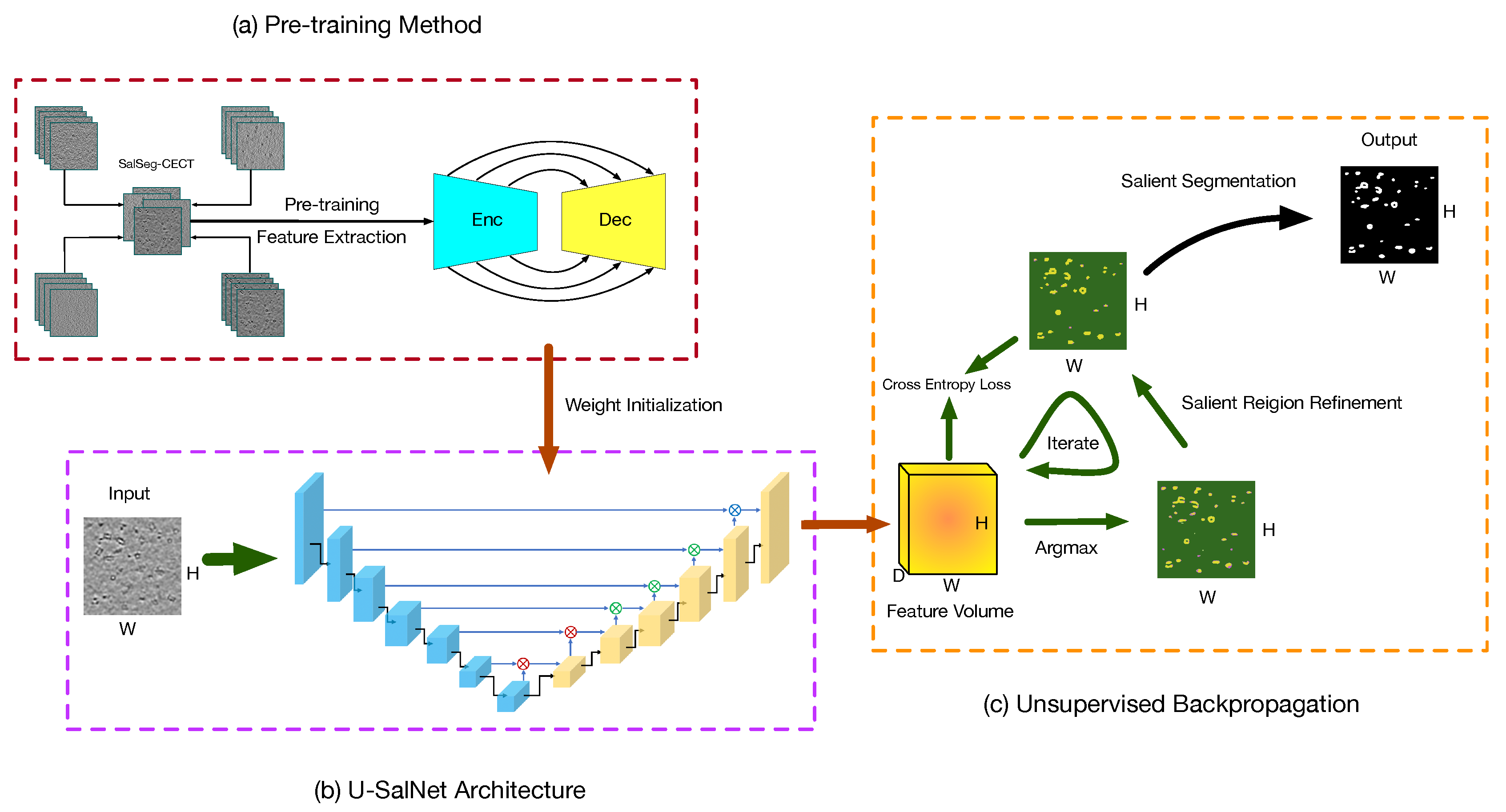
In this section, we describe our proposed PUB-SalNet. The goal of our work is to build a high-performance automatic salient segmentation model appropriate for biomedical tasks without ground-truth labels to improve computer-aided diagnosis. To reach this target, we design a processing pipeline with three major steps, as shown in Figure 2. In the first step, we aggregate a new biomedical data set called SalSeg-CECT. We then pre-train a deep feature extraction model specially designed for biomedical images, which can be used as a backbone module to initialize model parameters and to boost other tasks without data annotations. In the second step, we present the U-SalNet model on the basis of the U-Net architecture to learn to selectively attend to salient segmentation objects. The network includes global attention and local attention to facilitate learning saliency through global contrast and local similarity. In the last step, we jointly refine the salient regions together with feature representations from U-SalNet, with the parameters updated by unsupervised attentional backpropagation. Details of each step are explained in the following sections.
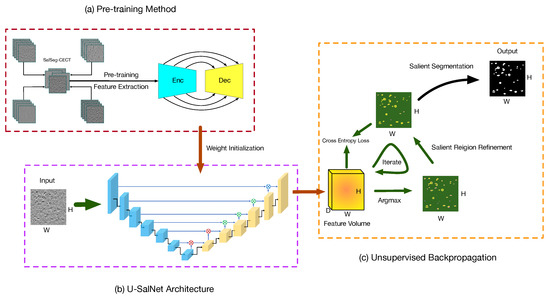
Figure 2.
The overview framework of our proposed method. The processing pipeline consists of three main steps: (a) pre-training on the SalSeg-CECT data set; (b) prediction using the U-SalNet model; (c) unsupervised attentional backpropagation iterating on single images. The Enc and Dec stand for the encoder and decoder. The ⨂, ⨂, and ⨂ denote the global attention mechanism, local attention mechanism and convolutional decoding, respectively.
3.1. Pre-Training Method
Inspired by Ref. [16]’s work, we aggregate a large data set from several simulated CECT data sets with rich salient objects, different SNR settings, and various resolutions, which is called SalSeg-CECT and will be described in Section 4. Based on the SalSeg-CECT data set, we then pre-train a model specially designed for biomedical images, as is shown in the red dashed-line box in Figure 2a. Our goal is to learn robust feature representations which can benefit training on biomedical data by utilizing a pre-trained network on the SalSeg-CECT data set. In this work, for deep feature extraction on biomedical data sets, we adopt the common encoder–decoder architecture to train our backbones of the network. Particularly, we choose the U-Net model as the basic structure on 2D images, and the V-Net for 3D volumes. The significant differences of SalSeg-CECT images from natural images come from the low SNR, limited tilt projection range (the missing wedge effect), and crowded nature of intracellular structures. Therefore, our pre-trained method is different from the current common pre-trained models. To the best of our knowledge, we are the first to pre-train a model on biomedical CECT data.
3.2. U-SalNet Architecture
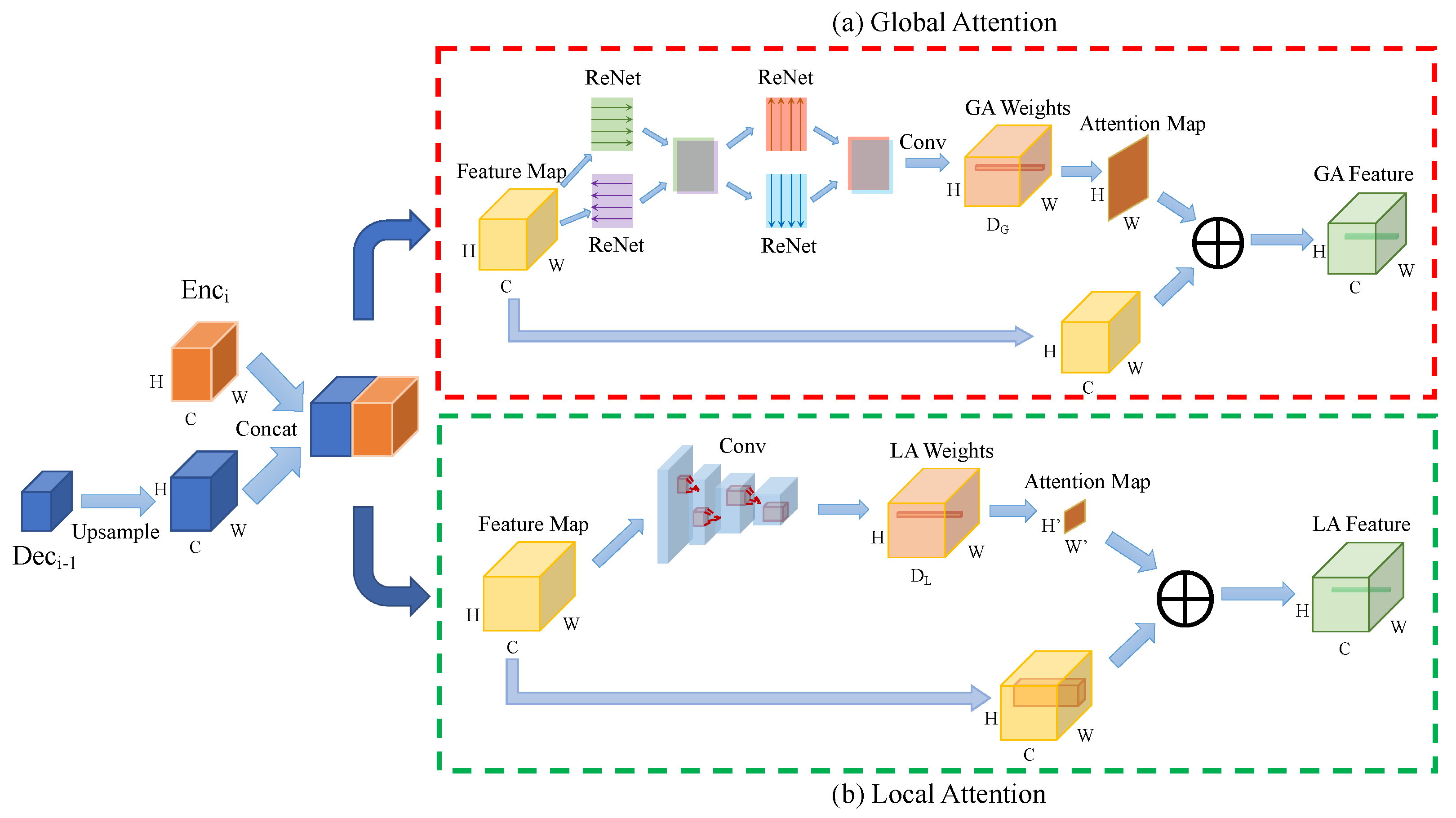
Our U-SalNet model includes two attentional mechanisms: global attention and local attention, which are integrated into the U-Net architecture, as is shown in the purple dashed-line box in Figure 2b. U-SalNet aims to selectively segment salient objects from the background by generating an attention map at each pixel. We apply the two modes of attention to refining salient regions in biomedical images. The detailed U-SalNet architecture is shown in Figure 3. We first upsample each level of feature maps in the decoder branch and concatenate them with their corresponding levels of features in the encoder branch. After concatenating the outputs from encoder and decoder, we get a feature map as the input to the attention module, where C, W, H denote the channels, width, and height, respectively.
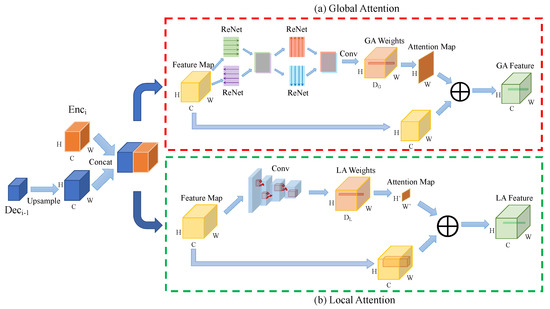
Figure 3.
The architecture of our U-SalNet model. (a) Global Attention and (b) Local Attention corresponds to ⨂ and ⨂ from Figure 2b, respectively. GA and LA stand for Global Attention and Local Attention. Conv means the convolution operation. ⨁ stands for weighted summation over the feature map.
For global attention, as is shown in Figure 3a, to generate attention over the whole feature map for each pixel, we first apply four ReNet models [23] to sweep across a feature volume both horizontally and vertically along two directions to concentrate global information. Next, a convolution operation is performed to transform this feature map to channels, where . At the same time, the feature vector at each pixel is normalized via softmax to obtain global attention weights , as is shown in Equation (1). Here, .
In order to generate the global attention feature , as is shown in Equations (2) and (3), the features at all locations in the whole feature map are summed with weights according to . refers to the salient correlation between the object pixel and the pixel at the location . is the Conv feature at ( in . has the same size with .
For the local attention, as is shown in Figure 3b, we consider a local feature cube centered at with width and height . is derived from convolution layers with a reception field of in the original feature map for each location. Similar to global attention, the features in are weightedly summed by to construct , as is shown in Equation (4):
It is worth noticing that, although our attention mechanism is similar to [24]’s work, they only consider the serial combination of global and local attention. Our U-SalNet focuses more on fusing two attention modules through convolutional decoding. In addition, their model adopts deep supervision, as is shown in Equation (5), where denotes the true saliency map and is the predicted saliency map. In addition, their experiments are all based on natural images and do not apply to biomedical images. On the contrary, our proposed U-SalNet architecture not only applies to complex biomedical images featuring a low SNR, low resolution, and rich salient objects but also achieves complete unsupervision. We will further describe the unsupervised attentional backpropagation algorithm in the next section.
3.3. Unsupervised Backpropagation
The proposed unsupervised backpropagation algorithm is shown in Algorithm 1. For the salient segmentation task, we consider two aspects: (1) predicting salient objects through current network parameters; and (2) training network parameters through current salient predictions. Accordingly, we can obtain salient regions by a forward pass through the neural network and use the gradient descent to optimize the backward pass of the network at the same time. In order to update the parameters, we adopt stochastic gradient descent (SGD) with momentum to backpropagate results of the cross-entropy loss calculated between outputs of the model and the refined salient object labels . In the backpropagation step, we use the SLIC algorithm [25] implemented in scipy for the method. is unified within every superpixel in an image, which is obtained through voting of labels inside a single superpixel and taking the majority of labels as the result. Different from Ref. [2]’s work, their prediction results are random and often fail on biomedical images. For solving these problems, our network parameters are initialized with the pre-trained method mentioned above. Furthermore, two attentional modules of the U-SalNet are combined to further refine the results, which achieve self-awareness. Finally, the forward–backward process is iterated I times to generate the final prediction of salient objects . The probabilities were trained in a self-supervised manner using . The orange dashed-line box in Figure 2c illustrates the proposed algorithm to train our U-SalNet model.
| Algorithm 1 Unsupervised Backpropagation Algorithm |
| Require: Original biomedical image |
| Ensure: Salient segmentation results |
| 1: // Initialize backbone parameters |
| 2: // Initialize classifier parameters |
| 3: |
| 4: for do |
| 5: if then |
| 6: |
| 7: |
| 8: |
| 9: |
| 10: |
| 11: //predict salient labels |
| 12: for do |
| 13: |
| 14: for |
| 15: end for |
| 16: |
| 17: |
| 18: end if |
| 19: end for |
4. Experiments
4.1. Datasets Setting
Our SalSeg-CECT data set includes 36,000 2D and 72,000 3D CECT images, which are generated with four levels of SNR (0.1, 0.5, 1.0 and 1.5), various resolutions, and missing wedge effects. The procedure of simulation uses the same simulator as in [26], which simulates tomographic images by imitating the actual tomography reconstruction process using macromolecular complexes of known densities. For 2D images, tomograms are sliced into grayscale images into three dimensions with a resulting width and height of 200 pixels. Our augmentation includes random flipping and cropping. For 3D images, tomograms are segmented into volumes of and their values normalized before being fed into models. The test set is generated independently with macromolecular complexes different from the training set with SNR = 0.5 and SNR = 1.5. To verify the ability of generalization of PUB-SalNet, we also apply it to the ISBI data set [27] and visualize our results in Figure 6.
4.2. Implementation Details
All our networks are implemented with PyTorch. Three NVIDIA GTX 1080 Ti GPU with 11 GB GPU memory each are used for pre-training and testing. In the pre-training step, we choose batch size = 4 for both 2D and 3D settings. For 2D, we use the SGD optimizer with learning rate = 0.01, momentum = 0.9 and weight decay = 0.0005, while, for 3D, the learning rate is and momentum is 0.99. In case of memory explosion, we apply global attention twice followed by local attention three times and a convolutional decoding layer in the decoder of U-SalNet. The default number and compactness of superpixels are 10,000 and 100. The maximum number of backpropagation iterations is set to 1000. The label of the majority of pixels is regarded as “non-salient”. The initial number of classes to be decreased is set to 100 as default. If the algorithm does not converge to two classes of labels after 1000 iterations, all the labels except for the non-salient type will be counted as “salient”.
4.3. Evaluation Metrics
To compare the quantitative results generated by different methods, here we use four popular metrics to evaluate our model against other unsupervised methods.
Region Similarity F. To measure the similarity of matching regions from two salient segmentation maps, F is defined as:
where to balance between and .
Pixel-wise Accuracy .F does not consider true negative saliency predictions. We define the normalized () mean absolute error (MAE) between predicted salient segmentation maps and ground truth masks as:
where W and H are the width and height of images, respectively.
Enhanced Alignment Measure E. Proposed by [28], using the enhanced alignment matrix to measure the two properties (pixel-level matching and image-level statistics) of a binary map, E is defined as:
where h and w are the height and width of the map, respectively.
Structural Similarity S.S proposed by [29] evaluates the structural similarity by considering both regions and objects. Since saliency of potential spacial structures is crucial to biomedical images, we additionally use S to comprehensively evaluate the structural similarity of biomedical images.
4.4. Quantitative Evaluation
Table 1, Table 2 and Table 3 show quantitative evaluation results on the simulated biomedical test set, which we will detailedly discuss in the following three subsections.

Table 1.
Comparison of performance of ten unsupervised methods with four metrics on the simulated biomedical test sets. stands for Mean Absolute Error (MAE), F for region similarity, E for the enhanced alignment measure, and S for structural similarity. Lower is better for , and higher is better for the other three metrics. The results are calculated according to Equations (6)–(8). The best performance of each metric is in bold and the second best is underlined. The improvements of our PUB-SalNet over the best of other methods in relative percentage is shown in the last row.

Table 2.
Quantitative comparisons between different combination of modules from our PUB-SalNet model. B stands for a single unsupervised backpropagation module; U+B stands for U-SalNet architecture with B; P+B means B based on the pre-training method; P+U means U-SalNet based on the pre-training method, note that this is actually not an unsupervised method; P+U+B is our proposed PUB-SalNet.

Table 3.
The quantitative comparison of parameter sensitivity analysis under four metrics. , F, E and S are the same as Table 1. For PUB-SalNet-BX, X stands for the initial number of classes to be decreased, as is in Function Init() parameter in Algorithm 1.
4.4.1. Comparison with State-of-the-Art
As is shown in Table 1, we compare our PUB-SalNet model to nine other state-of-the-art unsupervised methods. We demonstrate through experimental results that our proposed PUB-SalNet outperforms all existing unsupervised methods with a great margin (such as for E-Measure on and on ) and achieves new state-of-the-art performance.
4.4.2. Ablation Study
To demonstrate the effectiveness of the proposed PUB-SalNet model, we compare quantitative results of different combinations of modules from our method, as is shown in Table 2. B stands for backpropagation from [2]’s work, which serves as our baseline because it is a classic unsupervised image segmentation method using deep learning. The experimental results in Table 2 shows that three parts of our PUB-SalNet functions together and are all indispensable. It is even competitive compared to the supervised method.
4.4.3. Parameter Sensitivity Analysis
In order to demonstrate the robustness of our proposed model, we also construct an experiment of parameter sensitivity analysis. The quantitative comparison under four metrics is shown in Table 3. Our experiments show a deviation within 1–3% on the evaluation of the four metrics, which indicates that the parameters of our model have little influence to salient segmentation results.
4.5. Qualitative Evaluation
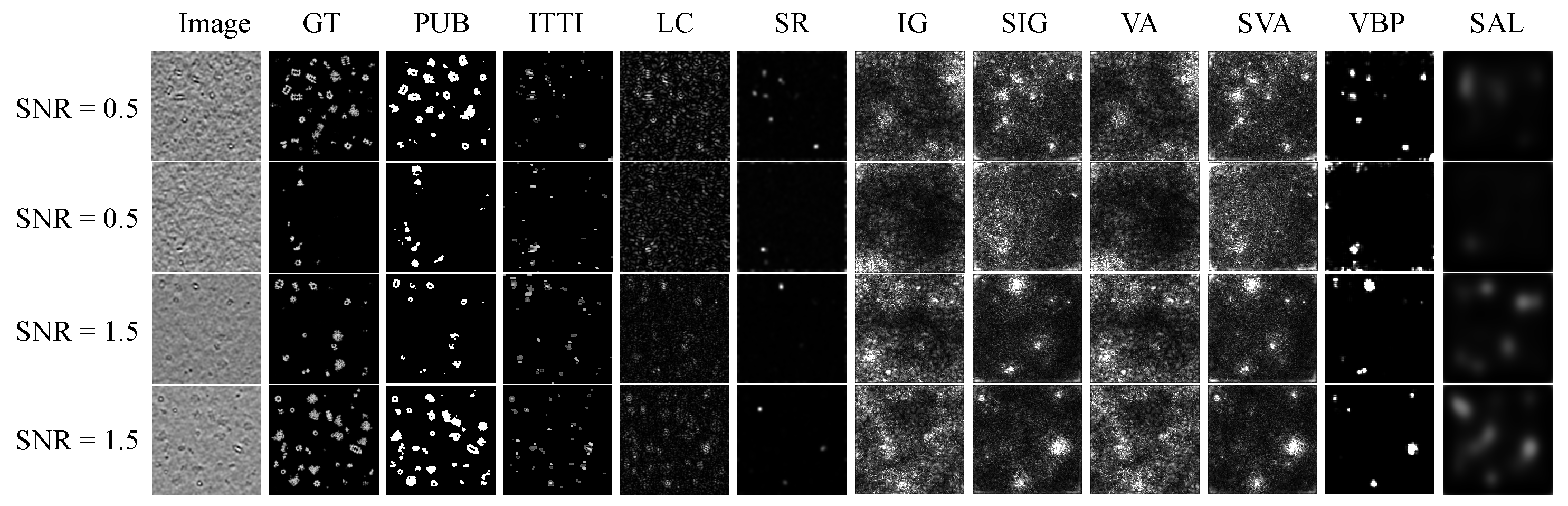
Figure 4 demonstrates the saliency maps predicted by nine unsupervised saliency detection methods on our testing data set with and . Traditional algorithms sometimes are able to detect multiple salient objects due to their superior capabilities of capturing low-level contrasts of features. When facing grayscale biomedical data sets with low SNR, deep learning methods are not as promising as we expected, which highlights blurry regions with richer contextual information. The performance of PUB-SalNet outperforms all other unsupervised methods on salient segmentation. We also present our results on the 3D CECT test set in Figure 5, which proves the generalization ability of our model. Our model is capable of detecting salient objects under various settings and can be effectively extended to 3D image processing tasks.
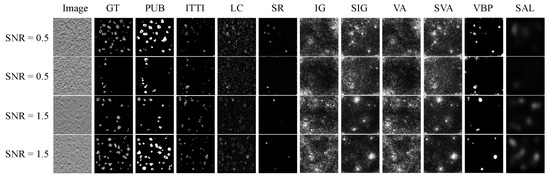
Figure 4.
Qualitative visual results of ten unsupervised methods on the simulated biomedical data set with SNR = 0.5 and 1.5. GT stands for ground truth images, PUB is PUB-SalNet, and the other nine methods are referenced in Table 1.

Figure 5.
Visualization of 3D salient segmentation by PUB-SalNet on a 3D subvolume of size from the CECT test set. The pictures are obtained using UCSF Chimera, which displays the isosurface of the four corresponding 3D images; (a) is the original image with a threshold of 0 (b) is the ground truth of macro-molecular structures (c) is our prediction (d) demonstrates that the predicted salient region greatly overlaps with the ground truth macro-molecular structure.
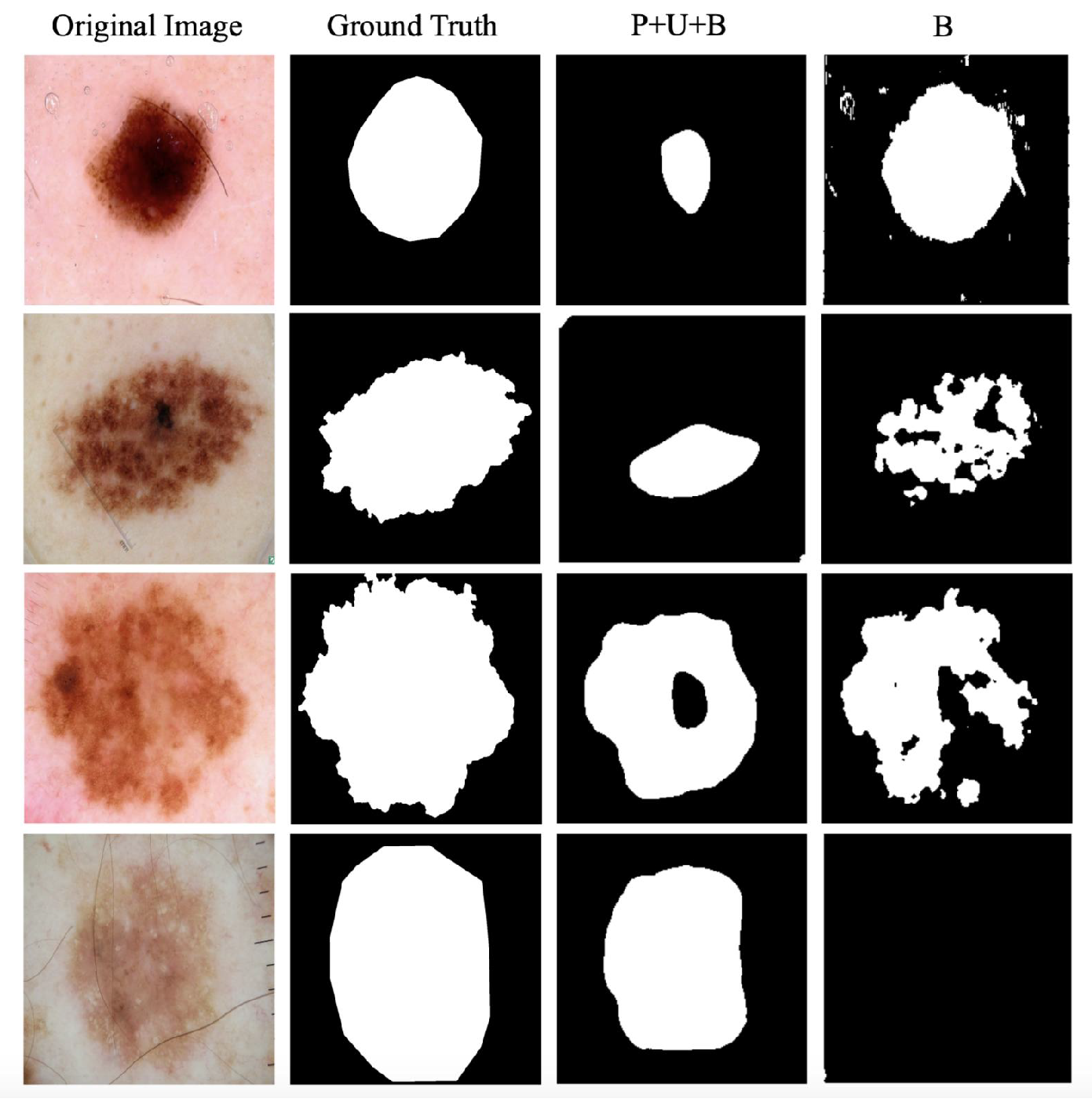
4.6. Case Study on the ISBI Challenge
A case study of 2D salient segmentation by PUB-SalNet and the B module on ISBI 2017 Challenge on Skin Lesion Analysis [27] is shown in Figure 6. To the best of our knowledge, we are the first to conduct unsupervised salient segmentation on the ISBI challenge. Comparison of the strong baseline model (B, for backpropagation only) and our proposed PUB-SalNet under three metrics is shown in Table 4. B module outperforms P+U+B by predicting shapes and edges with more accuracy. However, with the lack of global features, it falsely captures small differences in color within a piece of illness. It can also be easily perturbed by impurities or foreign matters, as is shown in the first two examples. P+U+B can produce smoother results, although sometimes fails to match the target in shape. P+U+B benefits from abundant semantic information and produces better results in the last two rows, while B focuses on wrong patches of color and cannot detect saliency on a global scale.
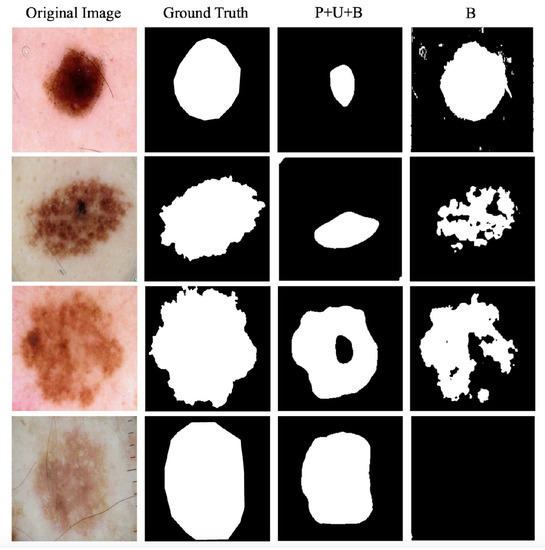
Figure 6.
Case study of 2D salient segmentation by PUB-SalNet and the B module on the ISBI Challenge [27].

Table 4.
The performance comparison of the strong baseline model (B, for backpropagation only) and our proposed PUB-SalNet under three metrics on the ISBI Challenge [27]. , F and E are the same as in Table 1. The best performance of each metric is in bold.
5. Conclusions
In this paper, we propose a completely unsupervised self-aware network based on pre-training and attentional backpropagation for biomedical salient segmentation, namely PUB-SalNet. We compare our PUB-SalNet model to nine other state-of-the-art unsupervised methods. We demonstrate through experimental results that our proposed PUB-SalNet outperforms all existing unsupervised methods with a great margin (such as for E-Measure on and on ) and achieves new state-of-the-art performance. The experimental results on the 2D and 3D data also display the generalization ability and robustness of our method. In the future, we will integrate our salient segmentation method into other complex biomedical tasks, such as biomedical image registration and quantification of uncertainty in segmentation.
Author Contributions
Conceptualization, M.X., F.C. and Y.J.; Methodology, F.C. and Y.J.; Software, F.C. and Y.J.; Validation, F.C. and Y.J.; Formal analysis, F.C. and Y.J.; Investigation, F.C. and Y.J.; Resources, X.Z.; Data processing, F.C., Y.J. and X.Z.; Writing—original draft preparation, F.C. and Y.J., and M.X.; Writing—review and editing, X.G. and J.Z.; Visualization, F.C. and Y.J.; Supervision, M.X.; Project administration, M.X.; Funding acquisition, M.X., X.G. and J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by U.S. National Institutes of Health (NIH) grant P41 GM103712. This work was supported by U.S. National Science Foundation (NSF) grants DBI-1949629 and IIS-2007595. X.Z. was supported by a fellowship from Carnegie Mellon University’s Center for Machine Learning and Health. This work was supported by the King Abdullah University of Science and Technology (KAUST) Office of Sponsored Research (OSR) under Award No. URF/1/2602-01 and URF/1/3007-01.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lee, J.G.; Jun, S.; Cho, Y.W.; Lee, H.; Kim, G.B.; Seo, J.B.; Kim, N. Deep learning in medical imaging: General overview. Korean J. Radiol. 2017, 18, 570–584. [Google Scholar] [CrossRef] [PubMed]
- Kanezaki, A. Unsupervised image segmentation by backpropagation. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 1543–1547. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]
- Geiger, A.; Lenz, P.; Urtasun, R. Are we ready for autonomous driving? the kitti vision benchmark suite. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 July 2012; pp. 3354–3361. [Google Scholar]
- Yuan, J.; Gleason, S.S.; Cheriyadat, A.M. Systematic benchmarking of aerial image segmentation. IEEE Geosci. Remote Sens. Lett. 2013, 10, 1527–1531. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; pp. 91–99. [Google Scholar]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.K.; Vikas, S.; Sarma, R.R.; Shenoy, G.; Kumar, R. Segmentation and Classification of CT Renal Images Using Deep Networks. In Soft Computing and Signal Processing; Springer: Singapore, 2019; pp. 497–506. [Google Scholar]
- Raza, K.; Singh, N.K. A tour of unsupervised deep learning for medical image analysis. arXiv 2018, arXiv:1812.07715. [Google Scholar]
- Moriya, T.; Roth, H.R.; Nakamura, S.; Oda, H.; Nagara, K.; Oda, M.; Mori, K. Unsupervised segmentation of 3D medical images based on clustering and deep representation learning. In Medical Imaging 2018: Biomedical Applications in Molecular, Structural, and Functional Imaging; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; Volume 10578, p. 1057820. [Google Scholar]
- Croitoru, I.; Bogolin, S.V.; Leordeanu, M. Unsupervised Learning of Foreground Object Segmentation. Int. J. Comput. Vis. 2019, 127, 1279–1302. [Google Scholar] [CrossRef]
- Ilyas, T.; Khan, A.; Umraiz, M.; Kim, H. SEEK: A Framework of Superpixel Learning with CNN Features for Unsupervised Segmentation. Electronics 2020, 9, 383. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef]
- Chen, S.; Ma, K.; Zheng, Y. Med3D: Transfer Learning for 3D Medical Image Analysis. arXiv 2019, arXiv:1904.00625. [Google Scholar]
- Honkanen, M.K.; Matikka, H.; Honkanen, J.T.; Bhattarai, A.; Grinstaff, M.W.; Joukainen, A.; Kröger, H.; Jurvelin, J.S.; Töyräs, J. Imaging of proteoglycan and water contents in human articular cartilage with full-body CT using dual contrast technique. J. Orthop. Res. 2019, 37, 1059–1070. [Google Scholar] [CrossRef]
- Xia, X.; Kulis, B. W-net: A deep model for fully unsupervised image segmentation. arXiv 2017, arXiv:1711.08506. [Google Scholar]
- Chen, J.; Frey, E.C. Medical Image Segmentation via Unsupervised Convolutional Neural Network. Available online: https://openreview.net/pdf?id=XrbnSCv4LU (accessed on 10 May 2020).
- Pan, F.; Shin, I.; Rameau, F.; Lee, S.; Kweon, I.S. Unsupervised Intra-domain Adaptation for Semantic Segmentation through Self-Supervision. arXiv 2020, arXiv:2004.07703. [Google Scholar]
- Hu, K.; Liu, S.; Zhang, Y.; Cao, C.; Xiao, F.; Huang, W.; Gao, X. Automatic segmentation of dermoscopy images using saliency combined with adaptive thresholding based on wavelet transform. Multimed. Tools Appl. 2019, 1–18. [Google Scholar] [CrossRef]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Visin, F.; Kastner, K.; Cho, K.; Matteucci, M.; Courville, A.; Bengio, Y. Renet: A recurrent neural network based alternative to convolutional networks. arXiv 2015, arXiv:1505.00393. [Google Scholar]
- Liu, N.; Han, J.; Yang, M.H. PiCANet: Learning pixel-wise contextual attention for saliency detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3089–3098. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. Slic Superpixe, Technical Report. Available online: https://infoscience.epfl.ch/record/149300 (accessed on 10 May 2020).
- Pei, L.; Xu, M.; Frazier, Z.; Alber, F. Simulating cryo electron tomograms of crowded cell cytoplasm for assessment of automated particle picking. BMC Bioinform. 2016, 17, 405. [Google Scholar] [CrossRef]
- Gutman, D.; Codella, N.C.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Fan, D.P.; Gong, C.; Cao, Y.; Ren, B.; Cheng, M.M.; Borji, A. Enhanced-alignment measure for binary foreground map evaluation. arXiv 2018, arXiv:1805.10421. [Google Scholar]
- Fan, D.P.; Cheng, M.M.; Liu, Y.; Li, T.; Borji, A. Structure-measure: A new way to evaluate foreground maps. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 4548–4557. [Google Scholar]
- Itti, L.; Koch, C.; Niebur, E. A model of saliency-based visual attention for rapid scene analysis. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 1254–1259. [Google Scholar] [CrossRef]
- Zhai, Y.; Shah, M. Visual attention detection in video sequences using spatiotemporal cues. In Proceedings of the 14th ACM international conference on Multimedia, Santa Barbara, CA, USA, 23–27 October 2006; pp. 815–824. [Google Scholar]
- Hou, X.; Zhang, L. Saliency detection: A spectral residual approach. In Proceedings of the 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007; pp. 1–8. [Google Scholar]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; Volume 70, pp. 3319–3328. [Google Scholar]
- Smilkov, D.; Thorat, N.; Kim, B.; Viégas, F.; Wattenberg, M. Smoothgrad: Removing noise by adding noise. arXiv 2017, arXiv:1706.03825. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar]
- Bojarski, M.; Choromanska, A.; Choromanski, K.; Firner, B.; Jackel, L.; Muller, U.; Zieba, K. VisualBackProp: Efficient visualization of CNNs. arXiv 2016, arXiv:1611.05418. [Google Scholar]
- Pan, J.; Ferrer, C.C.; McGuinness, K.; O’Connor, N.E.; Torres, J.; Sayrol, E.; Giro-i Nieto, X. Salgan: Visual saliency prediction with generative adversarial networks. arXiv 2017, arXiv:1701.01081. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).