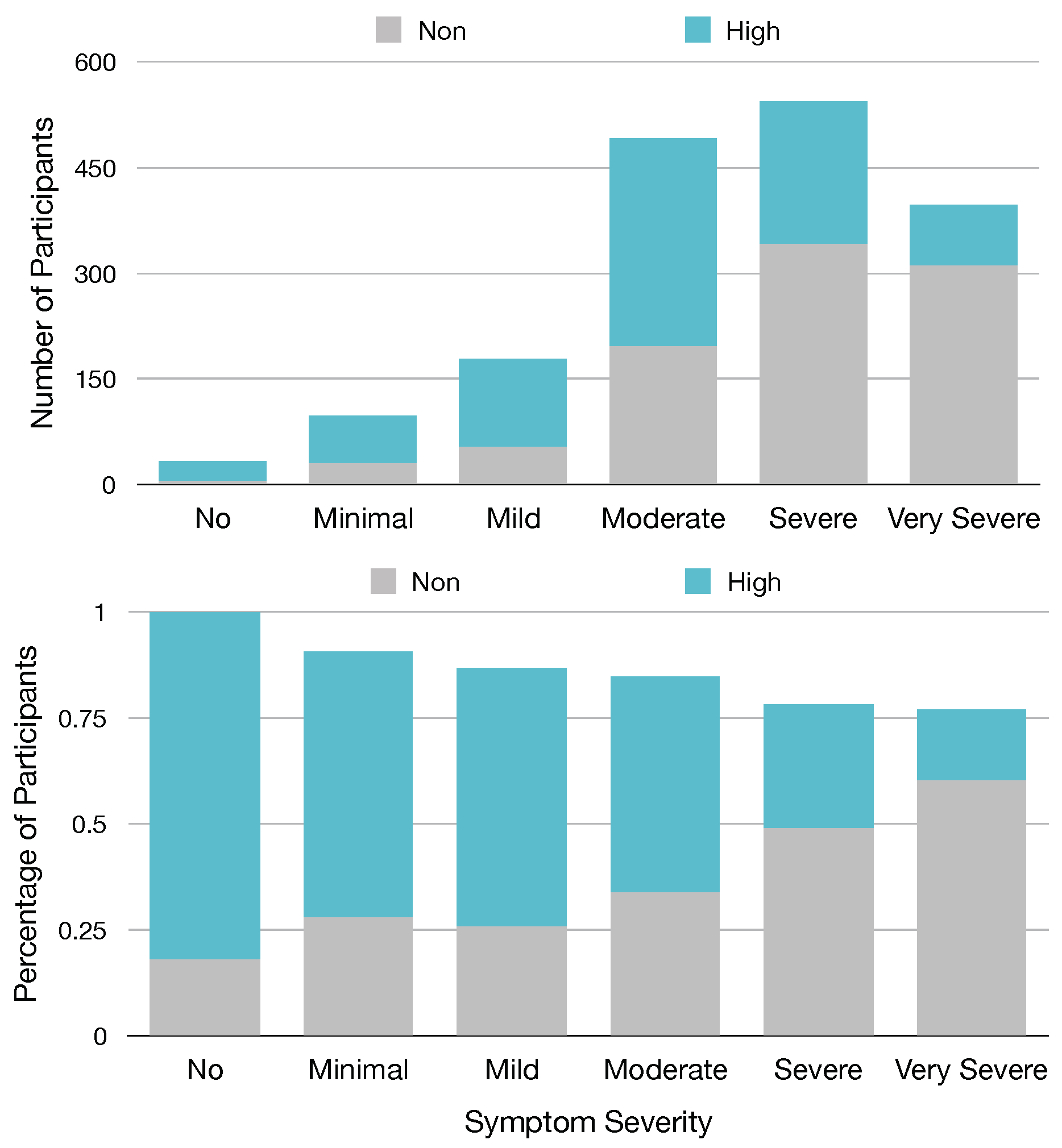1. Introduction
Lyme disease is the most common vector-borne disease in the United States. The CDC estimates that 300,000 people in the U.S. (approximately
of the population) are diagnosed with Lyme Disease each year [
1], a rate 1.5 times higher than breast cancer [
2], and six times higher than HIV/AIDS [
3].
In its early, or acute, form, the disease may cause a hallmark erythema migrans (EM) rash and/or flu-like symptoms such as fever, malaise, fatigue, and generalized achiness [
4]. A significant proportion of patients with Lyme disease develop chronic debilitating symptoms that persist in the absence of initial treatment or following short-course antibiotic therapy [
5]. This condition is commonly referred to as post-treatment Lyme disease or as chronic or persistent Lyme disease. In this paper, we refer to these patients as having persistent Lyme disease.
It is estimated that as many as
of those diagnosed and treated early remain ill after treatment [
5]. However, despite the high incidence and severity of Lyme disease, little research has been done, both clinically and analytically [
6,
7,
8,
9]. The result has been a stagnant and controversial research environment with little innovation and a costly lack of understanding or consensus. Physicians still do not know the best way to diagnose or treat Lyme, how it progresses, or why some patients respond to treatment and others do not.
Motivating questions. We are motivated by questions that interest both physicians and patients to better inform treatment approaches and to identify factors that might predict treatment response.
MyLymeData. Founded over 30 years ago, LymeDisease.org (LDo) is a national 501(c)(3) non-profit dedicated to advocacy, research, and education. LDo has conducted surveys with the Lyme disease patient community since 2004, and published the results in peer reviewed journals. In November 2015, LDo launched MyLymeData, a patient registry. MyLymeData has enrolled over 13,000 patients and continues to grow. Participants are asked hundreds of questions regarding their health history, diagnosis, symptoms, and treatment.
The first study using data from the registry was published in 2018. That study focused on treatment response variation among patients and identified a subgroup of high treatment responders using the Global Rating of Change Scale (GROC), a widely used and highly validated treatment response measurement [
10]. The GROC survey questions assess the degree to which participants reported that their condition improved, worsened, or remained unchanged following antibiotic treatment. We assign participants to class labels based on their GROC responder status. We label each participant as a high responder if they experienced substantial improvement, a low responder if they experienced slight improvement, and a non-responder if they worsened or remained unchanged. Medically, a major goal is to understand what patient attributes, protocols, or circumstances lead to patient improvement.
Machine learning techniques. One challenge facing medical experts looking to derive insights from the data are that the high-dimensional structure of the data obscures the relationship between patient features and their GROC responder status. In this work, we apply various machine learning models in order to measure the efficacy of individual features (survey question responses) in classifying the patients’ GROC responder status, and to identify both meaningful and redundant information within the survey responses. We apply simple
wrapper approaches to feature selection [
11].
We aggregate the results of several approaches to select a final subset of features we find most relevant to patients’ GROC responder status, thereby highlighting what patient attributes and protocols are most likely associated with improved patient well being. These findings point to areas where additional analysis might prove useful. Related work. There are many approaches for feature selection, where one seeks to identify the most salient features for a predictive model. These are roughly divided into wrapper and filter methods; wrapper methods evaluate subsets of features in a predictive model, while filter methods select features without information from the predictive model [
11]. The feature selection approaches we apply are simple wrapper approaches. We also compare to results of popular supervised feature extraction approaches, which aim to build a small number of features that capture the most salient data information for the predictive model [
12]. We compare to the results produced by semi-supervised nonnegative matrix factorization [
13] and supervised principal component analysis [
14]. Application of feature selection and feature extraction techniques to biological and medical data to yield interpretable predictive models has a long history; see, e.g., [
15,
16,
17,
18,
19].
Organization. In
Section 2, we describe the MyLymeData dataset and preprocessing steps, and introduce the techniques and models that we use throughout the paper. In
Section 3.1, we run all the models on the full MyLymeData dataset to evaluate the potential of each model to predict GROC labels using all of the data features. In
Section 3.2, we apply the models to individual features to identify the features that are most important in predicting GROC labels and we form a subset of top features by aggregating the results of all the models. In
Section 3.3, we evaluate the predictive ability of this subset of top features in comparison to the full dataset. Finally, in
Section 4, we discuss our results and their implication to Lyme Disease treatment protocols.
2. Data and Methods
Here, we describe our experimental setup, the MyLymeData set, and the models and methods we use. We note that the methods described are not new or novel and that our focus is not to propose new methods, but instead to describe a new and useful application to this survey data set.
2.1. Experimental Setup
All experiments are run on a MacBook Pro 2015 with a 2.5 GHz Intel Core i7 and a MacBook Pro 2018 with a 2.9 GHz Intel Core i9. We use Matlab version R2019b and Python version 3.7.3 to run experiments. We create neural network models using Tensorflow; our network architecture is detailed in
Section 2.6.3. We run uni-variate linear regression and calculate entropy using the Python SciPy library; we use the scipy.stats.linregress() function for uni-variate linear regression and the stats.entropy() function to calculate entropy. We run multivariate linear regression, support vector machine (SVM), Decision Tree, and
k-nearest neighbors (KNN) models using the Python scikit-learn library; we run multivariate linear regression using the sklearn.linear_model.LinearRegression() function, we run the SVM using the sklearn.svm.SVC() function, we run the Decision Tree with the sklearn.tree.DecisionTreeClassifier() function and criterion = “entropy” parameter, and we run the KNN model with the sklearn.neighbors.KNeighborsClassifier() function. We experimentally choose optimal values of the margin regularizer for support vector machine, max depth for decision trees, and
k value for KNN (see
Section 2.5 for details). The source code for experiments is publicly available at
github.com/jvendrow/Feature-Selection-on-MyLymeData.
2.2. The Dataset
We use data from Phase 1 of the MyLymeData patient registry, a private Lyme disease patient survey data set. Participants include respondents who report being US residents diagnosed with Lyme disease. We look only at participants who satisfy all of the following criteria:
Participant has persistent Lyme disease, which consists of patients who have experienced persistent symptoms for at least six months after antibiotic treatment.
Participant responded that they were unwell.
Participant answered the GROC (Global Rating of Change) survey questions.
We assign each participant a label based on their response to GROC, as previously described in [
10]. As asked, the GROC question produces a 17-point Likert scale. It is a two-part question asking first if the patient is “better”, “worse”, or “unchanged”. Patients who responded that they were better or worse are asked to specify the degree of improvement ranging from “almost the same” to “a very great deal better/worse”. “Almost the same” responses for better or worse were combined with the unchanged response. As modified, the resultant 15 point Likert scale ranges between −7 and 7, with 0 as the midpoint for unchanged. We separate participants into three categories:
Non-responders, who answered between −7 and 0, indicating there was no improvement.
Low responders, who answered between 1 and 3, indicating there was slight improvement.
High responders, who answered between 4 and 7, indicating there was substantial improvement.
The dataset consists of 2162 participants who satisfy the necessary criteria and 215 features (question responses) drawn from the MyLymeData survey. Each participant is assigned a label indicating non-responder, low responder, or high responder. The dataset has a total of 947 non-responders, 396 low responders, and 819 high responders. In our experiments, we investigate only the non-responders and high-responders. We choose not to include the low responders in our study because of the small number of low responders, and because this middle group could make it difficult to identify features that separate the non-responders from high responders. We also subsample from the dataset in order to produce an evenly sized groups of non-responders and high responders, and use this subsample for all experiments. As we describe in
Section 2.3 and
Section 2.5, we randomize this split repeatedly over many trials in order to capture all the information from the dataset. The 215 features cover diagnostic factors (such as delays to diagnosis, stage of diagnosis, or presence of coinfections), treatment approach, individual antibiotic use and duration of use, alternative treatments, symptoms (severity, presence at time of diagnosis, and three worst), type of clinician, and degree of functional impairment. We refer to this dataset as MLD (MyLymeData), and we refer to all participants with a specific label as a class of participants.
In order to improve the survey-taking experience for participants, the format of the survey used a branching structure to direct only relevant questions to participants. Thus, for many of the features in our analysis, only a subset of participants provided a response. For every such feature, we group all participants who did not respond to the feature together with a unique response.
For our figures and tables, we provide abbreviations of the relevant features. A name with a number following it indicates a series of questions within the same subtopic, and here we note such features with an “i”. “Bio_Sex” indicates biological sex. “Sx_Dx_i” indicates symptoms present at diagnosis. “Tick” indicates the presence of a tick bite. “Sx_Sev_i” indicates the severity of specific symptoms. “Sx_Top_i” indicates a specific symptom as being in the top 3 symptoms. “Abx” indicates whether the participant is currently taking antibiotics and/or alternative treatments. “Abx_Not_i” indicates the reasons that a participant is currently not taking antibiotics. “Abx_Dur” indicates the duration of the current antibiotic treatment protocol. “Abx_Eff” indicates the effectiveness of the current antibiotic treatment protocal. “Abx_Oral,” “Abx_IM,” and “Abx_IV” indicate whether the current antibiotic protocal includes oral, IM, and/or IV antibiotics, respectively. “Abx_i” indicates whether the participant is currently taking a specific oral antibiotic. “Abx_IM_i” indicates whether the participant is currently taking a specific IM antibiotic. “Abx_IV_i” indicates whether the participant is currently taking a specific IM antibiotic. “Alt_Tx_Eff_i” and “Alt_Tx_Sfx_i” indicate the effectiveness of and side effects of current alternative treatment approaches, respectively. “Med_i” indicates whether a participant is taking a specific non-antibiotic medication. “Provider_i” indicates whether a participant’s Lyme disease is being treated by a specific type of healthcare provider. “Wrk_Day” indicates the number of times a participant went to work but was unable to fully concentrate because of not feeling well. “Provider_Vsts” indicates the number of times that a participant visited a healthcare provider for their Lyme disease. “Rely_Others” indicates whether a participant currently relies on the help of others without pay. “Word” indicates a participant’s main form of work. “Disab_Pay” indicates the status of disability payments due to Lyme disease. “Education” includes a participant’s highest level of education. “Mis_Dx” indicates whether a participant was misdiagnosed with another condition. These features are more fully described in
Table A1 of the
Appendix A.
2.3. Subsampling
Most of the machine learning techniques that we use require balanced class sizes to produce accurate results. For this reason, for all of our experiments, we run our models on a subsample of the data, produced by selecting participants from MLD so that there are an even number of non-responders and high responders. Thus, this dataset has 819 participants with each label. We run our models over many different random subsamples of the dataset. More details for subsamples are included in the description of our training and testing procedure in
Section 2.5.
2.4. Individual Feature Importance
One way we assess the contribution of each feature to the predictive ability of our classification models is by evaluating the accuracy of the model given only a single feature. We run our classification models on each individual feature using the same experimental setup that we use for the full data set, including re-performing a hyperparameter sweep for each feature to account for the variations in the structure of the data set when only one feature is used.
2.5. Training/Testing Setup
Here, we describe our setup for training and testing for our classification models. First, we randomly subsample the dataset for an equal number of non-responders and high responders, and create a random 80/20 training and testing split. We select hyperparameters by performing cross-validation with five splits across the training set for each hyperparameter. Once the hyperparameters are chosen, we retrain our model on the full training set and perform a final evaluation on the test set. For the neural network model, one needs a maximum number of epochs at which to stop training to avoid overfitting. Thus, after our cross-validation process, we re-split the training data 80/20 into training and validation. We train our neural network on the training set, and choose the model at the epoch that maximizes the validation accuracy before performing final evaluation on the test set. We repeat this full process 10 times, randomizing both the train/test split and the subsampling step, and take the average of the test accuracies.
2.6. Models
Our machine learning methods are all supervised learning methods, meaning that the model aims to learn a mapping from an input, in this case our participants and their responses, to an output, in this case, the three classes we identified. We use two distinct supervised learning methods: regression and classification. Regression models produce a continuous value that numerically approximates the output, while classification models produce a discrete output, in this case one of the three classes. In our classification models, we calculate accuracy A as
where
denotes the total amount of participants and
denotes the total amount of participants that were predicted correctly.
2.6.1. Linear Regression
Linear regression is a regression model that attempts to produce the best affine hyperplane to fit a dataset, (see, e.g., [
12], Section 3.1) and references therein. The model computes the optimal affine hyperplane that minimizes the sum of the squares of the distances of the points from their projections onto the hyperplane along the dependent variable coordinate subspaces.
2.6.2. SVM
A support vector machine (SVM) is a popular and widely used classification model in the area of machine learning, (see, e.g., [
12], Section 7) and references therein. The model attempts to separate datapoints of different classes with an affine hyperplane. The SVM aims to find the optimal hyperplane by reducing both the amount of points classified incorrectly and the distance of these incorrectly classified points from the hyperplane. During our training procedure, we perform hyperparameter selection on the margin regularizer parameter, which varies the penalty to misclassifying examples versus the size of the margin.
2.6.3. Neural Network
We train a neural network model with two dense layers, (see, e.g., [
12], Section 5.1) and references therein. In
Figure 1, we display the architecture of our neural network. Each hidden layer has 30 nodes. We use a softmax output layer for multiclass classification, and, between layers, we include batch normalization and dropout layers. We compile our model with an Adam optimizer [
20]. During our training procedure, we perform a hyperparameter search over the learning rate (see
Section 2.5 for details).
2.6.4. Entropy and Decision Tree
In order to create the decision tree, we use the entropy metric to measure the importance of each feature ([
12], Section 1.6). The goal of entropy is to calculate the randomness in the data, so we measure feature importance by calculating the decrease in entropy after the data are split by the feature. Let
X be a discrete random variable that takes on a value of one of the two labels with probability
where
denotes the total participants and
denotes the participants with label i. Then, we measure entropy as
Using this criterion, we measure the importance of a feature by comparing the entropy of the dataset to the conditional entropy of the dataset after the dataset is split based on the participants’ responses to this question. The conditional entropy for a given feature is the sum of the entropy calculated on the subset of the data restricted by each possible feature response (value) multiplied by the fraction of participants with that feature response (value) ([
12], Section 1.6). We refer to the decrease in entropy (the difference between entropy on the full dataset and the conditional entropy) as information gain.
We create a decision tree that assesses feature importance using the entropy criterion, (see, e.g., [
12], Section 14.4) and references therein. To create this tree, the scikit-learn model places the most important features highest in the tree to improve its ability to split the data based on class labels, so at every node the function uses the feature that most effectively decreases entropy. In order to prevent overfitting with the decision tree, during our training process, we run hyperparameter selection on the depth of the tree.
2.6.5. k-Nearest Neighbors
The KNN algorithm classifies an example by looking at the
k points nearest to it and selecting the most common label amongst these points, (see, e.g., [
12], Section 2.5) and references therein. We measure the distance between points using Euclidean distance. The most important decision when training this model is the choice of
K, so we perform hyperparameter selection during our training procedure to choose
K experimentally.
3. Results
First, we run each of our models on the complete dataset in order to evaluate the potential of each model to predict GROC labels using all of the data features. We then run our models on individual features in order to identify features that are important in predicting GROC labels, and we aggregate these results into a subset of “key" features. Finally, we run our models on the two subsets of only the key features and only the remaining features to measure the effectiveness of our identified key feature set at predicting GROC labels.
3.1. General Performance
We first run each of our models on the MLD dataset to evaluate the potential of each model to predict GROC labels using all of the data features. This gives us a measure to compare against in determining the importance of each feature in the next section.
In
Table 1, we list the results of running each model on the MLD dataset. We list classification accuracies for our classification models, and the
value for the linear regression model. For classification accuracies, we also list the accuracy of the model on each of the two classes: non-responders and high responders.
We see that all of our models achieve a substantial classification accuracy, with the highest accuracy of 0.747 produced by SVM. Additionally, each model shows some variation in the accuracies for non-responders and high responders, but overall the classification accuracies were similar for both classes, though slightly higher for non-responders. We also see that the linear regression produced a substantial coefficient of 0.440.
These regression and classification results suggest to us that there is a substantial relationship between a participant’s GROC class and certain survey responses. Our goal in the next section will be to identify the specific features with the most significant relationship to GROC class.
3.2. Identifying Key Features
Here, we run our models on individual features in order to identify those that are most important in predicting GROC labels. We list notable results here and will highlight those features that are ranked highly by several models.
We use the following metrics for feature importance: (1)
value from linear regression, (2) information gain (entropy), (3) SVM test accuracy, (4) KNN test accuracy, and (5) neural network test accuracy. We calculate
values by running a separate single-variable regression for each feature. In
Section 2.6.4, we describe the process for calculating information gain and, in
Section 2.4, we provide the details for our experimental setup for calculating individual feature importance using our classification models.
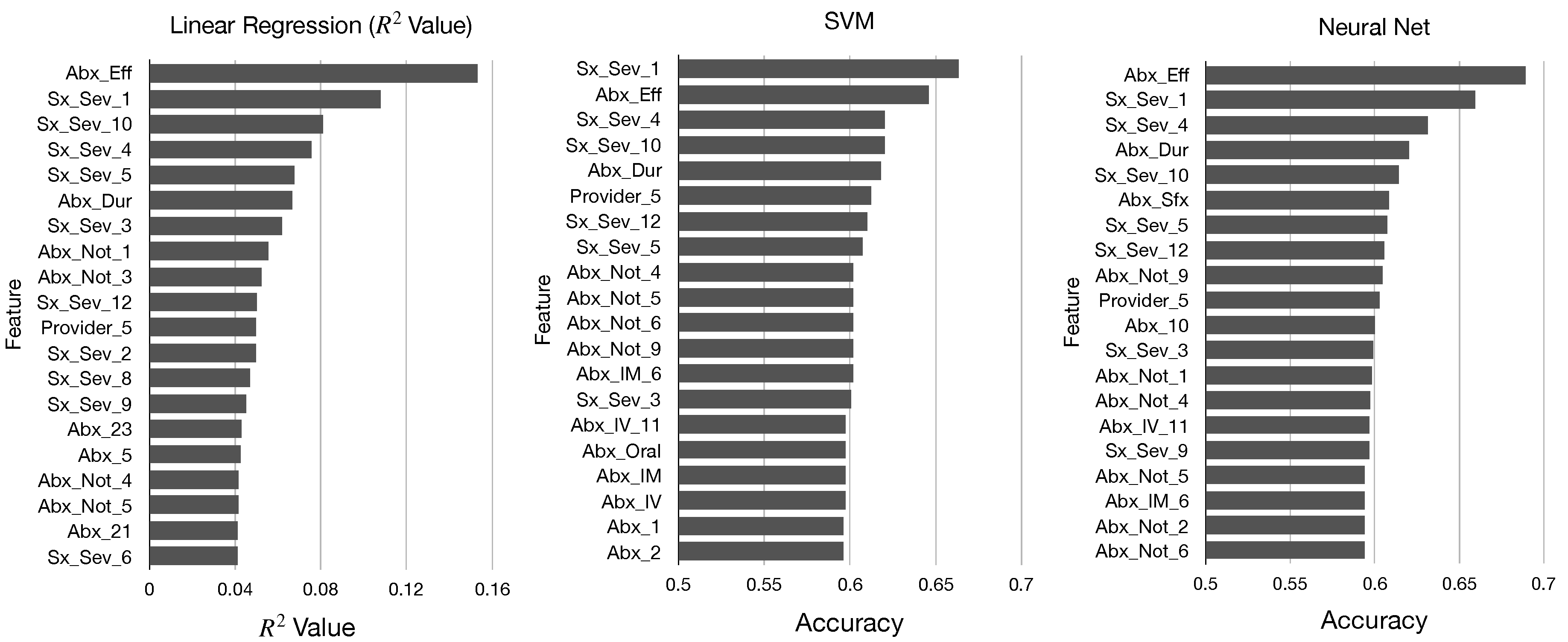
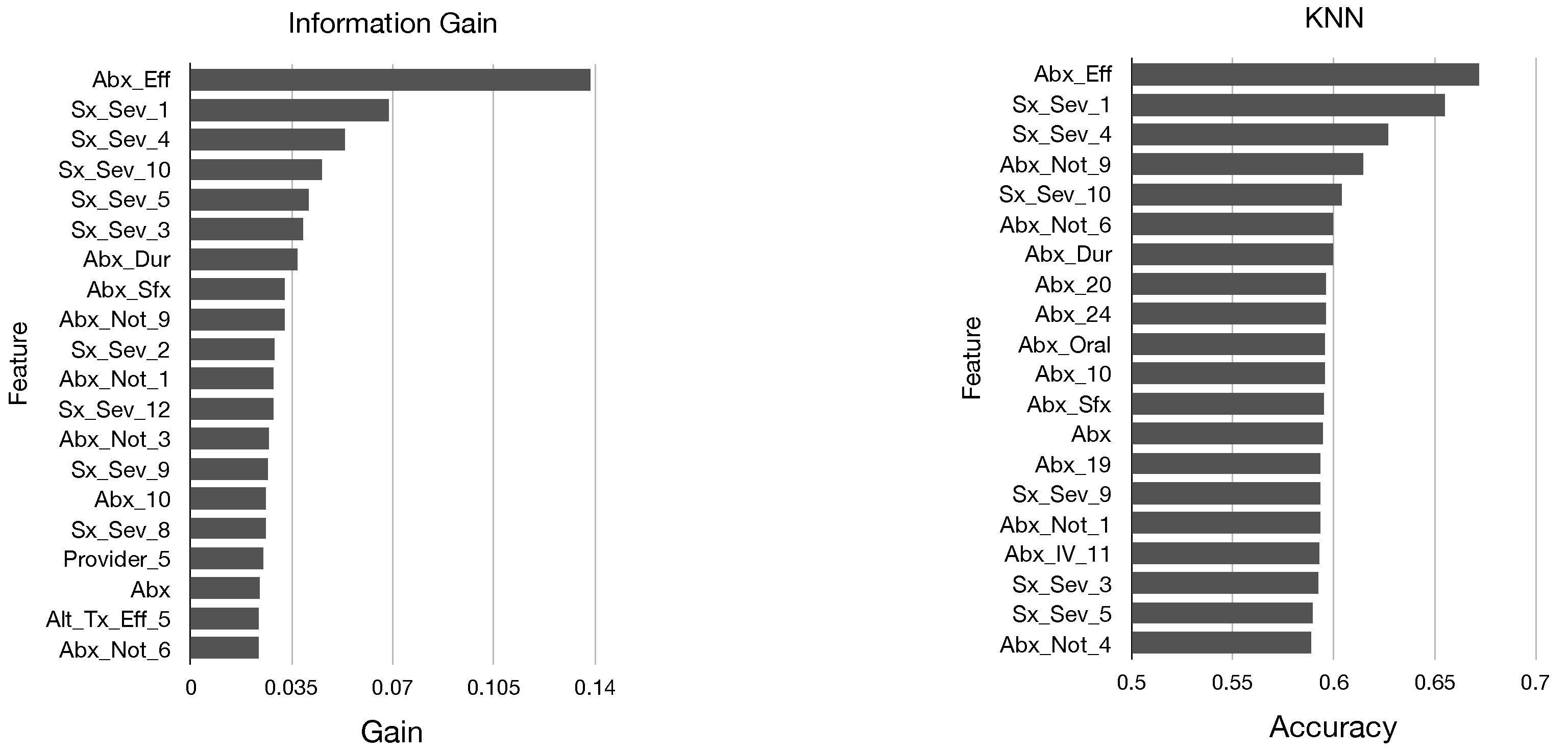
In
Figure 2 and
Figure 3, we display the top 20 features in sorted order identified by each of these five metrics. We determine that there are many similarities in the top features identified by each metric; all of our models identified mainly features pertaining to antibiotics and symptom severity, and the features Abx_Eff and Sx_Sex_1 were near the top for each model.
Top Features
From the results of our models, we now create a ranking of the most important features in our dataset for predicting GROC labels. To do this, we first define
to be the ranking of feature
i by model
m. Since we have 215 features, note that
for all
. For each model, the metric we use to produce the rankings among the features is the metric used to order the features in
Figure 2 and
Figure 3. In order to aggregate these rankings, we take the simple approach of averaging the ranking of each feature by all of our models. Let
be the average rank of feature
i. Then,
This aggregates our rankings into a single score where smaller values indicate more important features. In
Table 2, we show the top 30 features sorted by value.
3.3. Restricting to Key Features
In order to demonstrate the significance of the top 30 features (displayed in
Table 2) that we have identified through our previous experiments as being important for predicting GROC labels, we run experiments using a dataset with only these 30 features and a dataset with all but these 30 features. We refer to the dataset of the 30 most important features as Top MLD, and we refer to the dataset of 185 remaining features as Bottom MLD.
In
Table 3, we list the results for running each model on Top MLD, Bottom MLD, and MLD, where the results for MLD are repeated from
Table 1 for ease of comparison. We list classification accuracies for our classification models, and the
value for linear regression. See
Section 2.1 for experimental design details.
Based on our model, we see that the 30 features we identified (represented above by Top MLD) compare closely to or outperform MLD, suggesting that they hold a significant portion of the information which yields each model their predictive ability for the GROC label. These results also suggest possible redundancy in the dataset given the large portion of features that contain a small amount of the information used by the models for their predictive ability. The results could also offer intuition for LymeDisease.org, the creators of this survey, about what features to focus on in designing future surveys, as well as intuition for other future survey designers.
3.4. Comparison to Supervised Feature Extraction
In the previous sections, we have performed supervised feature
selection approaches to identify 30 key features for predicting GROC response labels. A related, but distinct, set of approaches are supervised feature
extraction techniques, which seek to identify salient collections of features (topics) which are correlated and contribute significantly to predicting response variables in a predictive model. In particular, we present the results of semi-supervised nonnegative matrix factorization (SSNMF) [
21] and supervised principal component analysis (SPCA) [
14] on our data set of 215 features. In these experiments, we apply these models with three topics to our data and present below the ten features which are most significantly represented in each topic.
In
Table 4 and
Table 5, we display the results of running SSNMF with three topics and SPCA with three topics, respectively, on MLD. For each topic formed by one of the models, we display the features with the highest presence in that topic. For SPCA, a feature can contribute either positively or negatively to a topic, so we order features based on the magnitude of presence and mark with [+] or [−] whether this feature contributes positively or negatively to the topic. We note that the identified topics (collections of features) bear significant overlap with our identified set of 30 key features. This is unsurprising as both SSNMF and SPCA seek to extract features that not only span the data, but which provide data representations that can be classified into GROC response classes accurately. In addition to this overlap, we also see clear grouping amongst features, with separate grouping of features relating to symptoms and features relating to antibiotics.
4. Discussion
Here, we explore and analyze the results from the previous sections.
4.1. Predictive Significance of the MyLymeData Dataset
Our models were able to achieve a highest test accuracy of 0.747 on MLD using an SVM for predicting the two selected classes of GROC response. We demonstrate that a significant portion of the predictive information from the dataset comes from only 30 of the 215 features. In fact, for two classification models, our top dataset of 30 features performed better than the full dataset.
4.2. Antibiotics
Of the 30 top features that we identified from the models, 21 of these features related directly to antibiotics, which suggests that many factors relating to antibiotics, including the effectiveness of antibiotic treatment (Abx_Eff), the length of the current treatment protocol (Abx_Dur), and the reasons why a participant is not taking antibiotic (Abx_Not), are important predictors of a participant’s GROC class.
By most of our models, Abx_Eff was the most important feature by a large margin. This is expected because the effectiveness of the current antibiotic treatment may reflect response to antibiotic therapy generally, which GROC measures. This suggests a very close relationship between antibiotic treatment and GROC label. This also yields intuitive evidence that the models are successfully selecting the most important features, since the information offered by Abx_Eff should make it a top feature.
The fact that most of the features identified in the top 30 were related to the use of antibiotic treatment is important because there is currently an on-going debate about whether antibiotics are useful for treating persistent Lyme disease. Our analysis suggests that antibiotic related questions may be the most important in predicting a patient’s treatment response for those with persistent Lyme disease. This topic is explored in greater detail in a companion study analyzing the role of specific features identified in the top 30 in connection with treatment response [
22].
4.3. Symptoms
Of the top 30 features, we identified from our models that eight of these features are from the 13 named Sx_Sev_i that ask about the current severity of specific symptoms, suggesting that these symptoms or the severity of these symptoms are important predictors of GROC class. Based on our ranking metric, the second most important predictor of GROC label is feature Sx_Sev_1, which asks the current severity of fatigue symptoms. To visualize this relationship, in
Figure 4, we provide a chart relating participants’ responses to Sx_Sev_1 with a GROC label.
Figure 4 demonstrates a clear relationship between Fatigue severity and GROC treatment response label, as it shows that participants who report no minimal or mild symptom severity for fatigue are most often high responders, while those with severe or very severe fatigue are most often non-responders. This suggests that the severity of fatigue symptoms could be a useful metric in determining GROC for Lyme disease patients, which matches this result in our experiment.
4.4. Feature Selection vs. Extraction Methods
In
Section 3.3, we applied two feature extraction methods, SSNMF and SPCA, and compare the results to our feature selection methods. One advantage of these techniques is that one can see how the identified features relate to one another and how they relate to the classes. Specifically, of the six topics formed between the two methods, five of these topics either contained features solely concerning antibiotics or solely concerning symptoms. In SPCA, a feature can contribute to a topic either positively or negatively, which yields additional information; specifically, we see in Topic 1 of SPCA in
Table 5 that questions about antibiotic use contribute positively, while questions about reasons for not taking antibiotics contribute negatively. It makes sense that these groups correlate together as the first set of questions is given only to participants taking antibiotics and the second set is given only to participants not taking antibiotics.
One disadvantage of feature extraction methods, however, is that it can be more challenging to identify which features from these computed topics individually contribute most to the GROC response class. Furthermore, the computed topics can include large groups of features. For example, in both experiments, the topics identified are not sparse and contain small nonzero values for all features. This is an advantage of the simple feature selection techniques applied previously, which identify single important features.
4.5. Branching
Although feature Abx appears only on spot 20 on our list of top features, a more comprehensive analysis reveals the true importance of this feature. Feature Abx identifies whether the participant is taking antibiotics, and this feature is used for branching purposes, so features concerning specifics of antibiotic treatment are only asked to patients who indicated in this feature that they are currently taking antibiotics. Thus, any feature that relies on the branching effect of Abx will inherently contain all the information contained in Abx, since for any such feature all patients who are not taking antibiotics would be grouped together. Given the number of features amongst the top features that relate to antibiotics, this suggests that the Abx feature is really one of the most important, and the importance of many other features takes advantage of the information offered by Abx.
The branching structure also affects the importance and interpretability of the top feature set by yielding some features with substantial predictive information unrelated to the purpose of the question being asked. Within the top feature set, those that appear to be most affected by this are the Abx_i features and Abx_IM_6. Upon further inspection, these features have little significance aside from maintaining the branching information from Abx that identifies which participants are currently taking antibiotics, yet the little information they add after this split make them more important than Abx based on information content alone. On one hand, this indicates that the subject matter of these features may not be as important as the analysis initially suggests. However, this does not take away from the purpose of these features within the top features list, which is to hold as much significant information as possible in a small subset of the features, since these features do hold important information from the branching in Abx.
4.6. Machine Learning for Survey Data
Here, we outline two important factors one should consider when applying machine learning methods to a survey dataset.
One consideration comes in choosing the models and metrics of evaluation. Within the goal of finding important features in a data set, one can either evaluate the predictive information from a single feature, or remove a single feature from the data set and measure the effects on performance, as in an ablation study. The former metric measures the predictive information from a single feature, while the later metric measures the amount of information unique to the specific features in comparison to the other features. We measured only the first, but the second approach could be equally interesting.
When applying machine learning models to survey data for feature selection, it is also important to contextualize the data and results within the branching structure, which can affect the meaning and interpretation of the results. We describe the effects of the branching structure on our analysis in the previous section.
4.7. Comparison with Companion Study
In a companion study performed in parallel to this work [
22], the authors use more traditional statistical methods and investigative analysis on the MyLymeData dataset. The results of our classification and regression models agree with many of its findings. Specifically, this study found that the features most indicative of level of treatment response were a participant’s use of antibiotics and symptom severity (specifically, fatigue), aligning closely with our analysis of the results from
Section 3. This study also found that longer treatment durations are associated with high treatment response, aligning with our ranking of feature Abx_Dur corresponding to duration of antibiotic treatment protocol as one of the top 5 features. Additionally, the study suggests that high treatment response is associated with close medical care provided by clinicians whose practices focus on tick-borne disease, aligning with our ranking of feature Provider_5 concerning the type of healthcare provider treating the patient’s Lyme disease within the top 10 features.